Abstract
The arylomycins are a class of natural-product antibiotics that act via the inhibition of type I signal peptidase (SPase), and we have found in diverse bacteria that their activity is limited by the presence of a resistance-conferring Pro residue in SPase that reduces inhibitor binding. We have also demonstrated that Staphylococcus epidermidis, which lacks this Pro residue, is extremely susceptible to the arylomycins. Here, to further explore the potential utility of the arylomycins, we report an analysis of the activity of a synthetic arylomycin derivative, arylomycin C16, against clinical isolates of S. epidermidis and other coagulase-negative staphylococci (CoNS) from distinct geographical locations. Against many important species of CoNS, including S. epidermidis, S. haemolyticus, S. lugdunensis, and S. hominis, we find that arylomycin C16 exhibits activity equal to or greater than that of vancomycin, the antibiotic most commonly used to treat CoNS infections. While the susceptibility was generally correlated with the absence of the previously identified Pro residue, several cases were identified where additional factors also appear to contribute.
The coagulase-negative staphylococci (CoNS) are a heterogeneous group of at least 15 different species of Gram-positive bacteria that have emerged in recent decades as important nosocomial pathogens (10, 31). A particularly problematic species is Staphylococcus epidermidis, which is responsible for a growing number of infections among hospital patients with compromised immune systems and is especially notorious for forming biofilms that adhere to surgical equipment and other hospital surfaces and indwelling devices (3, 18). Methicillin was traditionally the first-line antibiotic against CoNS, but its widespread use has resulted in resistance in 50% to 80% of CoNS infections and 75 to 90% of nosocomial S. epidermidis infections (18). As a result, vancomycin is now the first line agent for treating CoNS infections; however, isolates with reduced susceptibility to vancomycin have also been observed (11, 26), and the emergence of enterococci harboring mobile elements that confer vancomycin resistance has raised concerns that resistance might be transferred to S. epidermidis and/or other CoNS (17, 28). These concerns continue to motivate the search for new antibiotics that are active against CoNS, especially S. epidermidis.
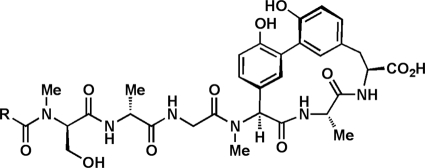
The arylomycins (Fig. 1) are a novel class of natural-product antibiotics that act by inhibiting bacterial type I signal peptidase (SPase) (19, 25). SPase is a Ser-Lys dyad protease that removes N-terminal signal sequences from preproteins following their translocation across the cytoplasmic membrane (5, 20). SPase is an attractive target for antibiotic therapy because it is conserved, essential, and located in the relatively accessible outer leaflet of the cytoplasmic membrane. Furthermore, because bacterial SPase acts via a catalytic mechanism that is distinct from that of its eukaryotic homologues, the arylomycins are unlikely to exhibit mechanistic toxicity in humans (5, 20).
FIG. 1.
Structures of arylomycin A2 (R = iso-C12) and arylomycin C16 (R = iso-C16) [iso-C12 = CH2(CH2)7CH(CH3)2 and iso-C16 = CH2(CH2)11CH(CH3)2].
Despite the apparent accessibility, essentiality, and conservation of SPase, initial reports suggested that the arylomycins were active against only a few Gram-positive bacteria, including Streptococcus pneumoniae, Rhodococcus opacus, and Brevibacillus brevis (15, 25), and not against other important Gram-positive pathogens or against any Gram-negative bacteria. However, after reporting the first synthesis of an arylomycin, arylomycin A2, as well as the synthetic derivative arylomycin C16 (Fig. 1), we found that each potently inhibits the growth of S. epidermidis (24) and that S. epidermidis evolves resistance to the arylomycins by mutating residue 29 of one of its two SPases, SpsIB, from Ser (Ser29) to Pro (Pro29) (29). Moreover, a Pro residue is naturally present at the analogous position in the homologous SPases of the pathogens Staphylococcus aureus, Escherichia coli, and Pseudomonas aeruginosa, and we showed that it imparts resistance by reducing the affinity with which the arylomycins bind. Furthermore, we found that a remarkably diverse range of both Gram-positive and Gram-negative bacteria whose SPases lack a Pro at the analogous position are susceptible to the arylomycins, including Staphylococcus epidermidis, Streptococcus pyogenes, Helicobacter pylori, Chlamydia trachomatis, and some strains of Francisella tularensis (29). In total, the results suggest that the identified SPase polymorphism is a major contributor to naturally occurring arylomycin resistance. However, we also showed that Yersinia pestis and some strains of S. aureus are susceptible to the arylomycins despite the presence of an analogous Pro, while others, such as many of the Lactobacillales, Clostridia, and Bacteriodetes, are resistant despite its absence, implying that in some cases, susceptibility must depend on additional factors, such as variable levels of toxicity associated with the inhibition of protein secretion.
The potent activity of the arylomycins against a strain of S. epidermidis (RP62A) suggests that they might be useful in the treatment of this and perhaps other CoNS. Here, to examine the spectrum of activity of the arylomycins against clinical isolates of S. epidermidis and other CoNS, we report the activity of arylomycin C16 against two panels of isolates from hospitals in geographically diverse locations and compare the activity to that of vancomycin. The results reveal that the arylomycins have potent antibacterial activity against a range of important CoNS species whose SpsIB orthologs lack the previously identified resistance-conferring Pro, while less activity is observed against species where Pro is present. While we generally observed similar susceptibilities for different isolates within a species, significant differences were observed in several cases, with one atypical instance of susceptibility resulting from the presence of a Ser in place of the resistance-conferring Pro. Significant differences in susceptibility between isolates of the same species are usually observed with clinically deployed antibiotics where selection for resistance has occurred during therapy (2, 9, 14, 21), and therefore, these results may be relevant to understanding the natural evolution of arylomycin resistance in nature.
MATERIALS AND METHODS
A total of 282 nonduplicate, clinical isolates of CoNS, identified to species level, were obtained from the London Health Services Centre, London, Ontario, Canada (13). Of these, 143 isolates were S. epidermidis, while the remaining 139 consisted of 12 groups comprised of various numbers of isolates of Staphylococcus capitis, Staphylococcus caprae, Staphylococcus cohnii, Staphylococcus haemolyticus, Staphylococcus lugdunensis, Staphylococcus saprophyticus, Staphylococcus simulans, Staphylococcus warneri, and Staphylococcus hominis. This panel of CoNS is a subset of a larger panel whose susceptibility to quinupristin-dalfopristin, linezolid, telithromycin, and vancomycin has been reported (13). A second set of nonduplicate clinical isolates of CoNS not identified to species level was obtained from the Department of Microbiology, University of California San Diego Medical Center Hillcrest, San Diego, CA. These strains were collected from patients with infections of various coagulase-negative bacteria between April and June of 2008.
Arylomycin C16 was synthesized as described previously (24). The MICs of arylomycin C16 were determined for isolates from both sets of CoNS using a modified CLSI agar dilution method. Bacterial inocula were prepared from log-phase cultures or from suspensions of colonies grown on solid medium, diluted appropriately, and spotted onto tryptic soy agar at a concentration of 1 × 104 CFU/spot. Spots were then incubated for 24 h at 37°C, and the MIC was defined as the concentration at which there was no visible growth. MICs of vancomycin were also determined, in an identical manner, to provide a reference activity. Several of the isolates demonstrated heterogeneous growth on solid medium, and in these cases, individual colonies of each morphotype were analyzed separately and identified to species level based on sequencing and phylogenetic analysis of a portion of the dnaJ gene, which has previously been demonstrated to serve as a sensitive measure to identify staphylococci to species level (see Fig. S1 in the supplemental material) (27). Species-level identification was also confirmed by dnaJ sequence analysis for isolates whose arylomycin MICs deviated from the MIC50 by more than 4-fold (see Fig. S1 in the supplemental material). Additionally, the spsIB genes from these isolates were sequenced to determine whether any polymorphisms were present that could account for the observed variation in susceptibility. Primers for the PCR amplification of spsIB genes were designed based on regions of highly conserved sequence within the upstream open reading frame SERP0551 (S. epidermidis RP62A numbering) and the downstream rexB gene (see Fig. S2 in the supplemental material). Degeneracies were included at positions that varied within the sequenced strains of the CoNS in order to maximize the likelihood of annealing (see Table S1 in the supplemental material).
To determine with high confidence the history of speciation within the CoNS examined in this study, a phylogenetic analysis was performed using regions of four essential genes that have previously been validated as speciation markers (16S rRNA genes, rpoB, groEL, and dnaJ) (see Table S2 in the supplemental material) (1, 6, 7, 27). For each protein-coding gene, published DNA sequences from strains identified to species level were translated into protein sequences and aligned using MUSCLE (8). The DNA sequences were then mapped onto the amino acid alignment using the program TranAlign (23). Aligned 16S rRNA gene sequences were obtained from the Ribosomal Database Project (4). For each organism, the four aligned sequences were concatenated, and phylogenetic analysis was conducted by the maximum-likelihood method with PhyML 3.0 (12), using the HKY85 nucleotide substitution model, 20 rate categories, and SPR (subtree pruning and regrafting) branch improvement. The phylogeny of SPase genes was determined similarly, using sequences deposited in the NCBI database as well as sequences obtained in the present study. Trees were displayed using MEGA4 (30).
RESULTS
A total of 143 isolates of S. epidermidis from the London Health Services Center were analyzed. The MIC50 and MIC90 of arylomycin C16 against these 143 S. epidermidis isolates were found to be 0.5 μg/ml and 1 μg/ml, respectively (Table 1). These susceptibilities are similar to that reported previously for the S. epidermidis laboratory strain RP62A (MIC = 0.5 μg/ml) (24) and compare favorably to the MIC50 and MIC90 of vancomycin, both of which we found to be 2 μg/ml, in agreement with values reported previously (13). Additionally, the range of MICs observed among the 143 isolates was narrow, with the most resistant isolate having an arylomycin C16 MIC of 4 μg/ml.
TABLE 1.
MICs of arylomycin C16 and vancomycin for coagulase-negative staphylococcus strains, identified to species level, obtained from the London Health Services Centre
| Species | No. of isolates | MIC (μg/ml) of: |
|||||
|---|---|---|---|---|---|---|---|
| Arylomycin C16 |
Vancomycin |
||||||
| MIC50 | MIC90 | Range | MIC50 | MIC90 | Range | ||
| S. epidermidis | 143 | 0.5 | 1 | 0.12-4 | 2 | 2 | 1-2 |
| S. lugdunensis | 10 | 0.25 | 0.25 | 0.03-32 | 2 | 2 | 2 |
| S. hominis | 17 | 0.25 | 0.5 | 0.06-0.5 | 2 | 4 | 1-4 |
| S. haemolyticus | 10 | 2 | 2 | 1-2 | 2 | 2 | 2-4 |
| S. simulans | 12 | 2 | >64 | 2->64 | 1 | 2 | 1-2 |
| S. caprae | 10 | 8 | 16 | 4-16 | 2 | 2 | 1-2 |
| S. cohnii | 27 | 8 | 16 | 4-16 | 2 | 2 | 1-2 |
| S. capitis | 24 | 16 | 32 | 4-64 | 2 | 2 | 1-2 |
| S. warneri | 19 | 64 | >64 | 2->64 | 4 | 4 | 1-4 |
| S. saprophyticus | 10 | >64 | >64 | >64 | 1 | 2 | 1-2 |
| S. xylosusa | 4 | ||||||
| S. aureusa | >64b | ||||||
Reference strains S. xylosus ATCC 29971 and S. aureus NCTC 8325 are included for comparison.
From reference 24.
The remaining 9 species of CoNS examined displayed arylomycin C16 susceptibilities ranging from highly susceptible to resistant (Table 1). Specifically, we found that S. haemolyticus, S. lugdunensis, and S. hominis are highly susceptible to arylomycin C16, with MIC50 and MIC90 values of 0.25 to 2 μg/ml. In contrast, we found that S. capitis, S. caprae, and S. cohnii are only moderately susceptible, with MIC50 values between 8 and 16 μg/ml and MIC90 values between 16 and 32 μg/ml, and that S. warneri and S. saprophyticus are resistant, with MIC50 values equal to or greater than 64 μg/ml and MIC90 values of more than 64 μg/ml. Finally, while many S. simulans isolates were sensitive to the arylomycins (MIC50 of 2 μg/ml), a significant number of isolates were highly resistant, resulting in a MIC90 of >64 μg/ml.
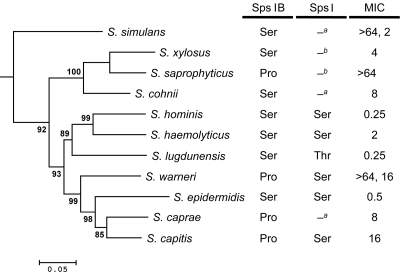
Under laboratory conditions, S. epidermidis RP62A evolves arylomycin resistance by mutation of Ser29 to Pro29 in SpsIB, one of two active SPases in this organism; in the related organism S. aureus, as well as in the more distantly related E. coli and P. aeruginosa, an analogous Pro29 is naturally present and contributes significantly to arylomycin resistance (29) (here and throughout the remainder of the manuscript, unless otherwise indicated, Ser29 and Pro29 refer to the residue in SpsIB orthologs at the position corresponding to 29 in S. epidermidis). In contrast, we did not observe mutations in the second S. epidermidis SPase gene, spsI, in any of the resistant isolates. To determine whether similar mutations might be responsible for the differences in arylomycin susceptibility observed among the different CoNS, we examined the sequences of their spsI and spsIB genes (Fig. 2; also see Table S3 in the supplemental material). To identify SPase sequences, we used the NCBI BLAST interface to search the sequenced genomes of S. capitis, S. haemolyticus, S. hominis, S. lugdunensis, S. saprophyticus, and S. warneri, and found that, except for S. saprophyticus, each possesses both SpsI and SpsIB homologs. We also included the recently reported sequence of the spsIB gene from Staphyloccus xylosus (22), which does not have an SpsI SPase (S. Leroy, personal communication). Because no correlation between susceptibility and the sequence of spsI was observed and because S. epidermidis evolves resistance by mutation of spsIB, we PCR amplified and sequenced the spsIB orthologs from S. caprae, S. cohnii, and S. simulans, whose genomes have not been fully sequenced (see the supplemental material). Overall, the data support the model that Pro29 contributes significantly to arylomycin resistance within CoNS. S. lugdunensis, S. haemolyticus, S. hominis, and a significant percentage of S. simulans isolates are extremely susceptible, and these species lack Pro29. In contrast, the CoNS species most resistant to the arylomycins, S. warneri and S. saprophyticus, each harbor an SPase with Pro29. The correlation between Pro29 and arylomycin susceptibility is less clear for species that display intermediate susceptibilities; for example, while S. caprae and S. capitis have Pro29 and S. cohnii has Ser29, all three display MIC50 values between 4 and 16 μg/ml.
FIG. 2.
Phylogenetic relationship, SPase sequence, and arylomycin susceptibility of CoNS species. The tree was constructed from the concatenated DNA sequence of several conserved genes using the method of maximum likelihood (see Materials and Methods). Macrococcus caseolyticus was used as an outgroup to root the tree, and alternative likelihood ratio test values are indicated at the branches to reflect the confidence of the branching pattern. The scale bar length corresponds to 0.05 substitutions per nucleotide position. For each species, the amino acids in SpsIB and in SpsI at the positions corresponding to residue 29 in S. epidermidis SpsIB are indicated. MIC values reflect the MIC50 except for S. simulans and S. warneri, where the values listed correspond to peaks in a bimodal distribution of MICs. a and b indicate that the presence and/or sequence of an SpsI homologue is unknown (a) or that the species does not have an SpsI homologue (b).
Most of the species examined have the narrow range of susceptibilities to arylomycin C16 that is expected for an antibiotic that has not been used clinically (2, 9, 14, 21) (Table 1). However, for several species, we found a greater range in susceptibilities or we found outliers with susceptibilities that differed significantly from those of the other isolates (Table 1; also see Fig. S3 in the supplemental material). For example, one isolate of S. lugdunensis is significantly more susceptible than the species average (its MIC is 8-fold below the MIC50), while a second isolate is unusually resistant (its MIC is 128-fold above the MIC50). Sequencing of the spsIB genes revealed identical sequences at the amino acid level (including at residue 29) (see Fig. S4 in the supplemental material). In addition, a bimodal distribution of MICs was observed for isolates of S. warneri and S. simulans, with peaks at 16 μg/ml and >64 μg/ml and at 2 μg/ml and >64 μg/ml, respectively (see Fig. S3 in the supplemental material). Sequencing the spsIB genes from representative isolates of each group revealed nearly identical SPase sequences and no variation at residue 29 (see Fig. S4 in the supplemental material). Finally, we identified a particularly susceptible isolate of S. warneri, with a MIC that is 32-fold below the species MIC50. Interestingly, the particularly susceptible strain of S. warneri had Ser29 as opposed to Pro29, which is typically present in S. warneri isolates, further supporting the genotype/phenotype correlation.
The observed differences in MICs that cannot be accounted for by differences in SpsIB sequence, both within and between species, suggest that additional factors contribute to arylomycin susceptibility. To determine whether these additional factors are shared among related organisms, we examined MICs as a function of phylogeny (Fig. 2). The phylogenetic analysis, based on four highly conserved genes (see Materials and Methods), suggests that S. simulans was the earliest species to diverge from the common ancestor of the examined CoNS, with the other species forming two groups, one containing S. xylosus, S. saprophyticus, and S. cohnii and the second containing the remaining species. Notably, SpsI SPases have only been shown to be present in the second group, although their presence cannot be ruled out in the unsequenced species.
S. saprophyticus, which has Pro29 and is extremely resistant to the arylomycins (MIC50 > 64 μg/ml), is most closely related to S. cohnii, which lacks Pro29, and yet is only moderately susceptible to the arylomycins (MIC50 = 8 μg/ml). To test whether other species that are closely related to S. saprophyticus and S. cohnii share this intrinsically lower susceptibility, we determined the MIC of a typed strain of S. xylosus, which is closely related to S. saprophyticus but, like S. cohnii, lacks Pro29. As expected, the strain of S. xylosus is moderately susceptible to the arylomycin (MIC = 4 μg/ml). Thus, this group of related bacteria appears to have a lower basal level of arylomycin susceptibility. In contrast, S. epidermidis, S. hominis, S. lugdunensis, and S. haemolyticus, which lack Pro29 and display extreme arylomycin susceptibility (MIC50 values of 0.25 to 2 μg/ml), as well as S. capitis and S. caprae, which have Pro29 but remain moderately susceptible (MIC50 values of 8 to 16 μg/ml), are more related to one another than to the other CoNS species examined. Despite the fact that some S. warneri isolates are extremely resistant, the remaining isolates have MICs of ∼16 μg/ml, which is consistent with the level of resistance observed for the other CoNS with Pro29 within the more susceptible phylogenetic group.
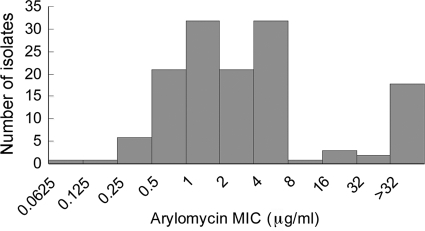
Finally, we evaluated the activity of arylomycin C16 against strains from a CoNS panel that has not been identified to the species level (obtained from the University of California San Diego Medical Center) and observed MIC50 and MIC90 values of 2 μg/ml and >32 μg/ml, respectively (Fig. 3). In addition to revealing a portion of resistant isolates, the distribution of MICs shows significant variations in the susceptibility of the remaining strains. Although interpretation of these results is complicated by the lack of species identification, the MIC distribution is consistent with a model in which both the presence of Pro29 and a second, yet-to-be-determined factor combine to yield tiers of arylomycin susceptibility.
FIG. 3.
Distribution of arylomycin susceptibilities of CoNS isolates not identified to species level, obtained from the University of California San Diego Medical Center.
DISCUSSION
Members of the arylomycin class of natural-product antibiotics act via a novel mechanism of action, the inhibition of SPase and, thus, the inhibition of the essential process of protein secretion. We found that, in addition to excellent activity against S. epidermidis (24), arylomycin C16 has potent activity against S. haemolyticus, S. lugdunensis, and S. hominis. Each of these members of the CoNS has an SPase with Ser29, consistent with the central role of this residue in determining arylomycin susceptibility that we observed previously with other bacteria (29).
The remaining species examined range from only moderately susceptible to extremely resistant, and Pro29 clearly makes a significant contribution to resistance in many of these cases. Interestingly, phylogenetic analysis suggests that Ser29 was prevalent during speciation but that Pro29 evolved in at least two independent instances, once in S. saprophyticus and once in the common ancestor of S. caprae and S. capitis (Fig. 2). Pro29 in S. warneri may represent a third instance, although it might instead result from common ancestry with S. caprae and S. capitis; this distinction is beyond the resolution of the current analysis. Interestingly, many synonymous and nonsynonymous mutations differentiate the spsIB genes of the different staphylococcal species, suggesting that ample mutational diversity has been sampled, but only Ser and Pro appear to be tolerated at position 29. Importantly, the multiple independent instances where Pro29 was introduced during staphylococci speciation suggest that there may be a natural selective pressure for Pro29, and the variation within isolates of an extant species suggests that this pressure may have occurred recently and may even still be present. While the nature of the selection pressure is currently unknown, it is tempting to consider that it may be related to the presence of arylomycins in nature (29).
The observed differences in susceptibilities between some of the staphylococcal species reveal that, in addition to SpsIB residue 29, there must be other factors that also contribute to overall susceptibility, and these other factors generally appear to be shared among closely related species, resulting in groups of species having higher or lower basal susceptibilities. One possibility is that the presence of a second SPase gene, orthologs of S. epidermidis spsI, in some species of CoNS could contribute to their different arylomycin susceptibilities. In principle, expressing two arylomycin-sensitive SPase proteins could result in hypersusceptibility if each SPase recognized a different subset of essential preprotein substrates. Alternatively, expressing one sensitive and one resistant SPase could result in either a susceptible or resistant phenotype, depending on the exact mechanism of arylomycin-induced cell death, which is yet to be determined. Interestingly, all of the species with higher basal susceptibilities have two SPases, while S. xylosus (S. Leroy, personal communication) and S. saprophyticus have only one SPase and display lower basal susceptibilities. Consistent with a possible role for a second SPase contributing to arylomycin susceptibility, S. aureus strain 8325 is highly resistant to arylomycin (MIC > 128 μg/ml) (24) despite being closely related to the more susceptible group of CoNS, and this species has only a single SPase, apparently due to a recent deletion of its spsI homologue. Further experiments will be required to elucidate the potential role of multiple SPases in CoNS susceptibilities and/or to identify other factors contributing to susceptibility, such as differences in the composition of secreted proteins or the presence of modifying enzymes. Comparison of resistant and susceptible isolates of the same species may provide a valuable approach to identifying additional factors that contribute to arylomycin susceptibility.
Regardless of the origins of arylomycin susceptibility and resistance, it is clear that a broad and clinically important (16) range of CoNS strains are susceptible to the arylomycins. In fact, in these cases, the level of susceptibility compares favorably with the level of susceptibility to vancomycin, the antibiotic currently recommended for treatment of CoNS infections but for which resistance is a major concern. The availability of a novel antibiotic, with a novel mechanism of action, would significantly improve CoNS therapy. Along with previously reported data (29), the data also suggest that if the arylomycin class of natural products could be optimized to overcome the loss in affinity mediated by the presence of the resistance-conferring Pro29, then they would have broad activity against CoNS and, possibly, other bacteria as well.
Supplementary Material
Acknowledgments
We thank S. Leroy and Genoscope (Genomics Institute of CEA, Evry, France) for providing analysis of the unpublished S. xylosus genome sequence.
This work was supported by the Office of Naval Research (awards N000140310126 and N000140810478) and the National Institutes of Health (grant AI081126).
Footnotes
Published ahead of print on 28 December 2010.
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1.Barros, E. M., N. L. Iorio, C. Bastos Mdo, K. R. dos Santos, and M. Giambiagi-deMarval. 2007. Species-level identification of clinical staphylococcal isolates based on polymerase chain reaction-restriction fragment length polymorphism analysis of a partial groEL gene sequence. Diagn. Microbiol. Infect. Dis. 59:251-257. [DOI] [PubMed] [Google Scholar]
- 2.Bowker, K. E., P. Caspers, B. Gaucher, and A. P. MacGowan. 2009. In vitro activities of three new dihydrofolate reductase inhibitors against clinical isolates of gram-positive bacteria. Antimicrob. Agents Chemother. 53:4949-4952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cheung, G. Y., and M. Otto. 2010. Understanding the significance of Staphylococcus epidermidis bacteremia in babies and children. Curr. Opin. Infect. Dis. 23:208-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cole, J. R., et al. 2009. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 37:D141-D145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dalbey, R. E., M. O. Lively, S. Bron, and J. M. van Dijl. 1997. The chemistry and enzymology of the type I signal peptidases. Protein Sci. 6:1129-1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Buyser, M. L., A. Morvan, S. Aubert, F. Dilasser, and N. el Solh. 1992. Evaluation of a ribosomal RNA gene probe for the identification of species and subspecies within the genus Staphylococcus. J. Gen. Microbiol. 138:889-899. [DOI] [PubMed] [Google Scholar]
- 7.Drancourt, M., and D. Raoult. 2002. rpoB gene sequence-based identification of Staphylococcus species. J. Clin. Microbiol. 40:1333-1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Edgar, R. C. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792-1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eliopoulos, G. M., et al. 1986. In vitro and in vivo activity of LY 146032, a new cyclic lipopeptide antibiotic. Antimicrob. Agents Chemother. 30:532-535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Emori, T. G., and R. P. Gaynes. 1993. An overview of nosocomial infections, including the role of the microbiology laboratory. Clin. Microbiol. Rev. 6:428-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garrett, D. O., et al. 1999. The emergence of decreased susceptibility to vancomycin in Staphylococcus epidermidis. Infect. Control Hosp. Epidemiol. 20:167-170. [DOI] [PubMed] [Google Scholar]
- 12.Guindon, S., and O. Gascuel. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52:696-704. [DOI] [PubMed] [Google Scholar]
- 13.John, M. A., C. Pletch, and Z. Hussain. 2002. In vitro activity of quinupristin/dalfopristin, linezolid, telithromycin and comparator antimicrobial agents against 13 species of coagulase-negative staphylococci. J. Antimicrob. Chemother. 50:933-938. [DOI] [PubMed] [Google Scholar]
- 14.Jones, R. N., D. M. Johnson, and M. E. Erwin. 1996. In vitro antimicrobial activities and spectra of U-100592 and U-100766, two novel fluorinated oxazolidinones. Antimicrob. Agents Chemother. 40:720-726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kulanthaivel, P., et al. 2004. Novel lipoglycopeptides as inhibitors of bacterial signal peptidase I. J. Biol. Chem. 279:36250-36258. [DOI] [PubMed] [Google Scholar]
- 16.Marshall, S. A., W. W. Wilke, M. A. Pfaller, and R. N. Jones. 1998. Staphylococcus aureus and coagulase-negative staphylococci from blood stream infections: frequency of occurrence, antimicrobial susceptibility, and molecular (mecA) characterization of oxacillin resistance in the SCOPE program. Diagn. Microbiol. Infect. Dis. 30:205-214. [DOI] [PubMed] [Google Scholar]
- 17.Noble, W. C., Z. Virani, and R. G. Cree. 1992. Co-transfer of vancomycin and other resistance genes from Enterococcus faecalis NCTC 12201 to Staphylococcus aureus. FEMS Microbiol. Lett. 72:195-198. [DOI] [PubMed] [Google Scholar]
- 18.Otto, M. 2009. Staphylococcus epidermidis—the “accidental” pathogen. Nat. Rev. Microbiol. 7:555-567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paetzel, M., J. J. Goodall, M. Kania, R. E. Dalbey, and M. G. Page. 2004. Crystallographic and biophysical analysis of a bacterial signal peptidase in complex with a lipopeptide-based inhibitor. J. Biol. Chem. 279:30781-30790. [DOI] [PubMed] [Google Scholar]
- 20.Paetzel, M., A. Karla, N. C. Strynadka, and R. E. Dalbey. 2002. Signal peptidases. Chem. Rev. 102:4549-4580. [DOI] [PubMed] [Google Scholar]
- 21.Petersen, P. J., N. V. Jacobus, W. J. Weiss, P. E. Sum, and R. T. Testa. 1999. In vitro and in vivo antibacterial activities of a novel glycylcycline, the 9-t-butylglycylamido derivative of minocycline (GAR-936). Antimicrob. Agents Chemother. 43:738-744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Planchon, S., et al. 2007. Proteomic analysis of cell envelope from Staphylococcus xylosus C2a, a coagulase-negative staphylococcus. J. Proteome Res. 6:3566-3580. [DOI] [PubMed] [Google Scholar]
- 23.Rice, P., I. Longden, and A. Bleasby. 2000. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 16:276-277. [DOI] [PubMed] [Google Scholar]
- 24.Roberts, T. C., P. A. Smith, R. T. Cirz, and F. E. Romesberg. 2007. Structural and initial biological analysis of synthetic arylomycin A2. J. Am. Chem. Soc. 129:15830-15838. [DOI] [PubMed] [Google Scholar]
- 25.Schimana, J., et al. 2002. Arylomycins A and B, new biaryl-bridged lipopeptide antibiotics produced by Streptomyces sp. Tü 6075. I. Taxonomy, fermentation, isolation and biological activities. J. Antibiot. (Tokyo) 55:565-570. [DOI] [PubMed] [Google Scholar]
- 26.Schwalbe, R. S., J. T. Stapleton, and P. H. Gilligan. 1987. Emergence of vancomycin resistance in coagulase-negative staphylococci. N. Engl. J. Med. 316:927-931. [DOI] [PubMed] [Google Scholar]
- 27.Shah, M. M., et al. 2007. dnaJ gene sequence-based assay for species identification and phylogenetic grouping in the genus Staphylococcus. Int. J. Syst. Evol. Microbiol. 57:25-30. [DOI] [PubMed] [Google Scholar]
- 28.Sieradzki, K., R. B. Roberts, D. Serur, J. Hargrave, and A. Tomasz. 1999. Heterogeneously vancomycin-resistant Staphylococcus epidermidis strain causing recurrent peritonitis in a dialysis patient during vancomycin therapy. J. Clin. Microbiol. 37:39-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith, P. A., T. C. Roberts, and F. E. Romesberg. 2010. Broad spectrum antibiotic activity of the arylomycin natural products is masked by natural target mutations. Chem. Biol. 17:1223-1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596-1599. [DOI] [PubMed] [Google Scholar]
- 31.Wisplinghoff, H., et al. 2004. Nosocomial bloodstream infections in US hospitals: analysis of 24,179 cases from a prospective nationwide surveillance study. Clin. Infect. Dis. 39:309-317. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.