Abstract
Populations of the cyanobacterium Planktothrix comprise multiple coexisting oligopeptide chemotypes that can behave differently in nature. We tested whether this population subdivision can, in principle, be driven by parasitic chytrid fungi, which are almost neglected agents of Planktothrix mortality. Two chytrid strains, Chy-Lys2009 and Chy-Kol2008, were isolated from Planktothrix-dominated lakes in Norway. The two strains shared 98.2% and 86.2% of their 28S and internal transcribe spacer rRNA gene sequences, respectively. A phylogenetic analysis placed them in the order Rhizophydiales family Angulomycetaceae. Chy-Lys2009 and Chy-Kol2008 could completely lyse Planktothrix cultures within days, while they failed to infect other filamentous cyanobacteria. The effect on Planktothrix was chemotype dependent, and both chytrid strains showed distinct chemotype preferences. These findings identify chytrid fungi infecting Planktothrix as highly potent and specialized parasites which may exert strong selective pressure on their hosts. According to established hypotheses on host-parasite coevolution, parasitism with the above properties may result in subdivision of Planktothrix populations into coexisting chemotypes and periodic shifts in the relative Planktothrix chemotype composition. These predictions are in agreement with field observations. Moreover, a genetic analysis verified the co-occurrence of Chy-Lys2009 and Chy-Kol2008 or related chytrid strains along with distinct Planktothrix chemotypes in at least one water body. Our findings are consistent with a scenario where chytrid parasitism is one driving force of Planktothrix population subdivision, which in turn leads to polymorphism in parasitic chytrid fungi. Future studies should test the validity of this scenario under field conditions.
The coexistence of conspecific lineages within a given population is a common trait in many microorganisms. In cyanobacteria, such population subdivision is not well understood. Specifically, the mechanisms that cause cyanobacteria to differentiate or to maintain homogeneity are far from established. Population subdivision is best studied in the picocyanobacteria Synechococcus and Prochlorococcus (4, 21, 32), which show niche partitioning among ecotypes, thought to occur at a comparatively low level of 16S rRNA gene sequence similarity (95 to 97%). The coexistence of genetically distinct lineages was also found in populations of more complex cyanobacteria (3, 14, 25). Most authors proposed abiotic factors as the driving force of cyanobacterial subpopulation evolution and dynamics.
Recently, we reported population subdivision in the filamentous freshwater cyanobacterium Planktothrix at a level of >99% 16S rRNA gene sequence similarity (35, 36). Due to differences in rates of occurrence and sequences of genes encoding nonribosomal and ribosomal oligopeptide synthesis, strains of Planktothrix may posses distinct cellular patterns of oligopeptides (38, 49). This allows delimitation of oligopeptide chemotypes, henceforth referred to as chemotypes. A field study in Norwegian Lake Steinsfjorden identified four coexisting Planktothrix chemotypes that differed considerably in seasonal dynamics, depth distribution, and participation in loss processes (35). Shifts in the relative chemotype composition occurred periodically and started with a significant loss of biomass of one chemotype, followed by the growth of others. There was no correlation between temperature or the availability of light, the amount of phosphorus or nitrogen, and the relative chemotype composition of the local Planktothrix population. A second study identified the same chemotypes in six Norwegian and two Finnish lakes of vastly dissimilar nutrient loadings and morphologies (36), implying once again that the chemotype composition of Planktothrix populations is not controlled by abiotic conditions. On the basis of the above findings, we proposed a top-down approach to explain chemotype evolution and dynamics; i.e., we suggested that chemotypes are functional groups that form in response to and that are controlled by biotic factors causing loss of Planktothrix biomass rather than abiotic factors controlling its growth (36).
A follow-up on this hypothesis appears to be worthwhile in several respects. Oligopeptides and chemotypes occur throughout the phylum of cyanobacteria (49). Knowledge on Planktothrix chemotype evolution and dynamics may therefore be generalizable and may provide insights into the yet unknown function of cyanobacterial oligopeptide production. Studies on cyanobacterial population subdivision often focus on abiotic conditions, although ecological considerations call for a broader view (42). The well-defined Planktothrix chemotypes provide a promising model for investigating the role of biotic loss processes in cyanobacterial population subdivision. The formation of long filaments largely protects Planktothrix against generalist grazers (29). The implications of biotic factors for cyanobacterial population subdivision can therefore be studied in a simple setting where only a limited number of specialized grazers and parasites play a role.
One group of specialized parasites, the parasitic chytrid fungi, is commonly believed to inflict significant mortality on numerous types of organisms, including Planktothrix (8, 13, 40), but is nevertheless virtually overlooked by present-day research on cyanobacterial ecology. The parasitic potency of chytrid fungi is proven most impressively by the disease chytridiomycosis, which threatens amphibian diversity at a global scale (15). Early studies on chytrid-cyanobacteria interrelations suggested that chytrid fungi have narrow host ranges, usually encompassing only one or a few related species, and that there is a chytrid to most if not all cyanobacteria (41). In nature, chytrid infection of cyanobacteria is considered omnipresent (33).
Chytrid fungi form zoospores that employ a flagellum and probably chemotaxis to find their hosts (17). In the case of chytrid fungi infecting Planktothrix, the zoospore encysts at the surface of a suitable host and penetrates it and the walls between neighboring cells of the same filament by forming a rhizoid of up to 150 μg in length (8). Host cells are digested by release of extracellular enzymes from the rhizoid (17), most likely including serine proteases of the trypsin and chymotrypsin types (44). Later, a sporangium is formed, in which new zoospores are produced.
The way in which chytrid fungi live suggests a link between Planktothrix chemotype performance in nature and chytrid parasitism. Many of the more than 600 known oligopeptides produced by cyanobacteria and used here to define Planktothrix chemotypes are potent inhibitors of serine proteases, such as chymotrypsin and trypsin (37), and may therefore interfere with proteases released by chytrid fungi during rhizoid formation and growth. Other oligopeptides are potentially toxic to eukaryotes in general (9). Such compounds may suppress chytrid infection at almost any stage. Due to distinct cellular oligopeptide patterns of Planktothrix chemotypes, the outcome of chytrid parasitism may well be chemotype dependent, which would provide a possible explanation for the distinct behavior of Planktothrix chemotypes in nature. Furthermore, the subdivision of Planktothrix populations into chemotypes may be a consequence of coevolution between Planktothrix and parasitic chytrid fungi.
Here we provide a laboratory test of whether chytrid parasitism can, in principle, be a driving force of Planktothrix population subdivision into chemotypes and chemotype dynamics. We test whether (i) chytrid parasitism is chemotype dependent, (ii) the parasitic potency of chytrid fungi is strong enough to exert significant selective pressure at the chemotype level, (iii) chytrid fungi can differ in their chemotype preferences, (iv) chytrid fungi have a host range that is narrow enough to result in coevolution with Planktothrix only, and (v) chytrid fungi with different chemotype preferences coexist in nature.
MATERIALS AND METHODS
Planktothrix chemotypes, their origin, and culture conditions.
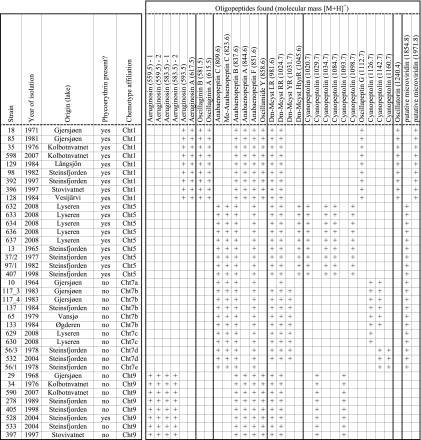
A detailed overview of the Planktothrix strains that were considered in this study and their properties is given in Fig. 1. The strains represented four chemotypes that coexisted in lakes throughout southeastern Norway and probably other areas in Scandinavia and Finland (36) and that showed distinct behavior in the field (35). To reflect the wide distribution of the four chemotypes, Planktothrix strains from nine lakes in southeastern Norway and Finland were considered. To retain consistency, the chemotype designation of the present report follows that of Rohrlack and coworkers (36); i.e., the chemotypes are henceforth referred to as Cht1, Cht5, Cht7, and Cht9. Cht7 is further subdivided into Cht7a to Cht7e, to reflect minor variations in cellular oligopeptide patterns (Fig. 1).
FIG. 1.
Properties of Planktothrix sp. strains that were used in this study. Shown for each strain are its chemotype affiliation, its identification number in the NIVA culture collection, when and from which water body it was isolated, if it contained the pigment phycoerythrin, and its cellular oligopeptide composition. For each oligopeptide, the name and its molecular mass (in daltons, in parentheses) are given. The oligopeptides are sorted by structural class. Mcyst, microcystin. Strains with phycoerythrin correspond to P. rubescens, and those without the pigment correspond to P. agardhii. Lakes Långsjön and Vesijärvi are situated in Finland. All other lakes are situated in southeastern Norway. All data are taken from Rohrlack and coworkers (36).
The Planktothrix sp. strains were grown in nonaxenic batch cultures using Z8 medium (24). The cultures were maintained in 50-ml glass flasks at 17°C. Light at a photon flux density of 3 to 5 μmol m−2 s−1 was supplied by warm-white fluorescent tubes at a light-dark cycle of 12 h/12 h. Subcultures used to isolate and maintain chytrid fungi or to run infection experiments were taken at the late logarithmic growth phase. At that stage, the optical densities of cultures measured at 800 nm exceeded 0.1.
According to current taxonomy (43), the Planktothrix strains of the present study belonged to two species, Planktothrix rubescens and Planktothrix agardhii, which are distinguishable by the presence of the accessory pigment phycoerythrin in P. rubescens and the resulting red color of its filaments. However, a 16S rRNA sequence similarity of >99% and considerable overlap in fatty acid composition, morphology (43), multiple genetic markers (20), oligopeptide chemotypes, and ecology (35) contradict the taxonomic discrimination of P. rubescens and P. agardhii. Here, we therefore treat the two groups to be conspecific and refer to them as Planktothrix sp.
Isolation and maintenance of chytrid fungi.
In July 2008, a field sample with a high density of Planktothrix sp. strains showing some chytrid infection was taken from Lake Kolbotnvatnet (southeastern Norway). It was kept at room temperature until infection had reached its peak. Then a small amount of this sample was transferred to a dense Planktothrix sp. NIVA-CYA98 culture, which after 2 weeks was used to infect a fresh host culture and so on. After several cycles, the chytrid strain Chy-Kol2008 was established by inoculating a fresh host culture with a single host filament bearing only one undehisced chytrid sporangium. The same approach was used to isolate the second chytrid strain, Chy-Lys2009, from Lake Lyseren (southeastern Norway) with Planktothrix sp. NIVA-CYA634 as the host. This was done using an environmental sample that was taken in June 2009.
Chy-Kol2008 and Chy-Lys2009 were maintained using cultures of Planktothrix sp. strains NIVA-CYA98 and NIVA-CYA634 as hosts, respectively, in 50-ml culture vessels at 20°C. Continuous light at a photon flux density of 3 to 5 μmol m−2 s−1 was supplied by warm-white fluorescent tubes. Fresh host cultures were inoculated when the infection had peaked.
Suspensions of zoospores to be used for DNA extraction and in infection experiments were prepared by filtering severely infected host cultures through 10-μm-pore-size gauze. The filtrate was inspected microscopically to ensure the viability of the zoospores. Their density was determined by a light microscope using a blood cell counting chamber and a sample fixed with Lugol's solution. All suspensions used in experiments contained >2 × 105 zoospores per milliliter.
Characterization of chytrid strains Chy-Kol2008 and Chy-Lys2009.
The different stages of chytrid infection were studied by light microscopy during routine chytrid culture maintenance with Planktothrix sp. strains NIVA-CYA98 and NIVA-CYA634 as hosts. In addition, sequence information for parts of the rRNA operon, the internal transcribed spacers (ITSs), and a part of the 28S gene was used to phylogenetically compare Chy-Kol2008 and Chy-Lys2009 with each other and with other related members of the class Chytridiomycetes for which sequences were available from public databases. DNA was extracted from infected host biomass and zoospore suspensions using a MoleStrips DNA tissue kit (Mole Genetics, Oslo, Norway). PCR was performed using the primers ITS5 (50) and ITS4Chytrid (28) for the ITS and primers 5.8S-R and LR7 (47) for the 5′ end (1,386 bp) of the 28S gene. PCRs were performed in a 25-μl reaction volume using BD Advantage polymerase with 10× BD Advantage SA buffer, 0.2 mM deoxynucleoside triphosphates, and 200 nM each primer. The thermocycling conditions were 94°C for 1 min, followed by 35 cycles of denaturing (94°C for 30 s), annealing (55°C for 30 s), and extension (68°C for 1 min [ITS] or 2 min [28S gene]) and a final extension step (68°C for 2 min). The PCR products were sequenced both ways on a 3730XL genetic analyzer (ABI, Carlsbad, CA), using a BigDye Terminator (version 3.1) cycle sequencing kit.
The sequences of Chy-Kol2008 and Chy-Lys2009 were aligned with similar sequences identified by a BLAST search. Separate alignments were made for each of the gene regions using the automatic function in the MAFFT program (23) and were manually inspected in BioEdit (version 7.0.9) software (18). The phylogeny was estimated using the neighbor-joining method in the software MEGA (version 4) (45) and applying 2,000 bootstrap resamplings.
Infection experiments with Planktothrix sp. strains
The effect of chytrid strains Chy-Kol2008 and Chy-Lys2009 on 35 Planktothrix sp. strains representing chemotypes Cht1, Cht5, Cht7a to Cht7e, and Cht9 was studied. The experiments were run in 48-well microtiter plates by mixing in each case 1 ml Planktothrix sp. suspension with 0.1 ml zoospore suspension. The tests were run in triplicate for all Planktothrix sp. and chytrid strains. The microtiter plates were kept at 20°C and under continuous light at a photon flux density of 3 to 5 μmol m−2 s−1 for 7 days. Then the severity of chytrid infection was quantified by light microscopic inspection at ×120 magnification, according to the following scale: 0, no sporangia, no active zoospores; 1, up to 10% of host filaments with sporangia, active zoospores present; 2, 10 to 50% of host filaments with sporangia, active zoospores present; 3, 50 to 90% of host filaments with sporangia, active zoospores present; 4, 90 to 100% of host filaments with sporangia, active zoospores present; 5, final stage of infection, with most host filaments disintegrated, numerous sporangia visible, and active zoospores present or absent. Note that since the average survival time of zoospores was about 1 day, their absence at day 7 of an experiment may indicate resistance of the Planktothrix sp. or an infection that peaked days before. The microscopic analysis did not distinguish between dehisced and undehisced sporangia.
Infection experiments with cyanobacteria other than Planktothrix sp.
To test whether the host range of chytrid strains Chy-Kol2008 and Chy-Lys2009 extended beyond Planktothrix sp., their effect on 28 strains representing 15 Anabaena, Aphanizomenon, and Tychonema species was investigated. Only species that resemble Planktothrix sp. in morphology and that can coexist with Planktothrix sp. in the plankton of European lakes were selected. A detailed overview is given in Table 1. The cyanobacteria were cultured and the infection experiments were run as described above for Planktothrix sp. Planktothrix sp. strains NIVA-CYA98 and NIVA-CYA634 served as positive controls.
TABLE 1.
Cyanobacteria other than the Planktothrix sp. used here to determine the host range of parasitic chytrid fungi
| Strain | Species | Yr of isolation | Origina |
|---|---|---|---|
| NIVA-CYA82 | Anabaena circinalis | 1980 | Lake Steinsfjorden |
| NIVA-CYA418 | Anabaena crasa | 2000 | Lake Storvatnet |
| NIVA-CYA74 | A. crassa | 1980 | Lake Østensjøvatnet |
| NIVA-CYA147 | Anabaena danica | 1984 | Lake Langen |
| NIVA-CYA269/2 | Anabaena flos-aquae | 1990 | Lake Frøylandsvatnet |
| NIVA-CYA267/4 | A. flos-aquae | 1990 | Lake Fammestadtjønni |
| NIVA-CYA419/2 | Anabaena inaequalis | 2000 | Lake Storvatnet |
| NIVA-CYA83/1 | Anabaena lemmermannii | 1981 | Lake Edlandsvatnet |
| NIVA-CYA281/1 | A. lemmermannii | 1990 | Lake Storavatnet |
| NIVA-CYA266/1 | A. lemmermannii | 1990 | Lake Bergesvatnet |
| NIVA-CYA270/1 | A. lemmermannii | 1990 | Lake Arefjordvatnet |
| NIVA-CYA335 | A. lemmermannii | 1993 | Lake Hallevatnet |
| NIVA-CYA426 | A. lemmermannii | 2000 | Lake Storvatnet |
| NIVA-462/2 | A. lemmermannii | 2003 | Lake Søndre Heggelivatnet |
| NIVA-CYA66 | Anabaena planctonica | 1979 | Lake Langsævatnet |
| NIVA-CYA416 | Anabaena solitaria | 1999 | Lake Bergemsvatnet |
| NIVA-CYA226/1 | Anabaena spiroides | 1986 | Lake Holstadvatnet |
| NIVA-CYA323 | Anabaena subcylindrica | 1993 | Lake Fuggdal |
| NIVA-CYA626 | Aphanizomenon flos-aquae | 2004 | Lake Melangsee, Germany |
| NIVA-CYA641 | A. flos-aquae | 2008 | Lake Årungen |
| NIVA-CYA103 | Aphanizomenon gracile | 1982 | Pond in Vingrom |
| NIVA-CYA365 | Aphanizomenon klebahnii | 1996 | Lake Balaton, Hungary |
| NIVA-CYA364 | A. klebahnii | 1996 | Lake Balaton, Hungary |
| NIVA-CYA69 | Tychonema bornetii | 1976 | Lake Mjøsa |
| NIVA-CYA95 | T. bornetii | 1982 | River Glåma |
| NIVA-CYA265/1 | T. bornetii | 1990 | Lake Vikevatnet |
| NIVA-CYA33/1 | Tychonema bourrellyi | 1976 | Lake Mjøsa |
| NIVA-CYA102/1 | Tychonema sp. | 1981 | Lake Edlandsvatnet |
Strains were isolated from lakes in Norway, unless stated otherwise.
Test for co-occurrence of chytrid fungi with distinct chemotype preferences in nature.
A co-occurrence of parasitic chytrid fungi with dissimilar Planktothrix sp. chemotype preferences would be of importance in several respects. It would indicate that chytrid parasitism can cause recurring shifts in the relative chemotype composition of Planktothrix sp. populations. It would also be an indication that the subdivision of Planktothrix sp. populations into chemotypes exerts diversifying selective pressure on parasitic chytrid fungi. We used freeze-dried historic samples from Norwegian Lake Steinsfjorden to test whether strains Chy-Kol2008 and Chy-Lys2009 or closely related strains co-occurred in nature. The samples were collected by use of a plankton net between 1997 and 1999, a period with massive blooms of Planktothrix sp. The presence of Cht1, Cht5, and Cht9 during this period was proven by isolation and analysis of Planktothrix sp. strains (Fig. 1). DNA was extracted from about 2 mg of each sample using a MoleStrips DNA tissue kit (Mole Genetics). The presence of the Chy-Kol2008 and Chy-Lys2009 ITS genotypes was verified by quantitative PCR (qPCR) with an ABI 7500 real-time PCR system, and amplification was detected using a Mesa Blue qPCR kit for SYBR assay (Eurogentec, Liege, Belgium). Specific PCR primers were designed on the basis of the highly variable ITS region, where sufficient variation was found to clearly discriminate between Chy-Kol2008 and Chy-Lys2009 and other sequences in the NCBI database. The primers and sequences were as follows: for Chy-Kol2008, primers Chy_KolF (AACTTTAGAGTAACAGCGATA) and Chy_KolR (GGTGTAACAGCCTGAAAT), and for Chy-Lys2009, primers Chy_LysF (AGTGTGAAAGGGAGTTGATATAG) and Chy_LysR (GGGACTGATGTTTGGTTGA).
Statistics.
On the basis of infection data for 35 Planktothrix sp. strains and both chytrid strains, a Euclidian distance matrix was created. A second distance matrix was calculated on the basis of oligopeptide occurrence in Planktothrix sp. Mantel's test was used to check whether the two matrixes were significantly correlated, i.e., whether chytrid infection was chemotype dependent. The Mantel test was repeated for individual oligopeptide groups (aeruginosins, anabaenopeptins, cyanopeptolins, microcystins, microviridins, oscillaginins, and oscillatorin). To compare the host preferences of Chy-Kol2008 and Chy-Lys2009, their capacity to infect 35 Planktothrix sp. strains was used to calculate individual Euclidian distance matrixes. These were compared by Mantel's test. A significant correlation would indicate identical or similar host preferences. Distance matrixes were calculated and Mantel tests were performed using the program GenAlEx (version 6.4) (31). The outcome of infection experiments with cyanobacteria other than Planktothrix sp. made it unnecessary to run a statistical analysis.
Nucleotide sequence accession numbers.
All organisms of the present study were deposited at the Norwegian Institute for Water Research (NIVA) Culture Collection of Cyanobacteria and Algae. The DNA sequences of chytrid strains Chy-Kol2008 and Chy-Lys2009 were deposited in the EMBL database under accession numbers FR670787 and FR670788, respectively.
RESULTS
Chytrid morphology and course of infection revealed by light microscopy during routine chytrid culture maintenance.
Strains Chy-Kol2008 and Chy-Lys2009 had identical morphologies and infection patterns that were in agreement with those described by Canter and Lund (8) for Rhizophidium megarrhizum (see Fig. S1 in the supplemental material). Zoospores encysted on the apex or at points of fracture of Planktothrix sp. trichomes. Sporangia were formed at the same locations. The simultaneous encystment of multiple zoospores at one site and the occurrence of multiple sporangia per host filament suggested infection of host trichomes by several parasite individuals. The rhizoids could be best seen in partially decomposed Planktothrix sp. filaments and pervaded several host cells. The effect of chytrid parasitism on Planktothrix sp. strains NIVA-CYA98 and NIVA-CYA634, used for routine maintenance of Chy-Kol2008 and Chy-Lys2009, respectively, was disastrous. During the first days after inoculation of a fresh host culture, encysted fungal zoospores and sporangia were observed in increasing numbers. Infected filaments fragmented rapidly, providing new sites for zoospore encystment. After a few days, the entire host culture was overrun by fungal zoospores. Host cultures were completely lysed within days (for NIVA-CYA634 when it was infected with Chy-Lys2009) or a few weeks (for NIVA-CYA98 when it was infected with Chy-Kol2008).
Phylogenetic analysis of chytrid strains Chy-Lys2009 and Chy-Kol2008.
Analysis of Chy-Lys2009 DNA from two replicates each of zoospores and infected host biomass yielded identical sequences for both the ITS and the 28S regions, suggesting that Chy-Lys2009 represented a single rRNA genotype. The same was found for Chy-Kol2008. The sequence identities between Chy-Kol2008 and Chy-Lys2009 were 98.2% in the 28S region and 86.2% in the ITS region.
To classify our chytrid strains relative to the current taxonomy of the chytrid order Rhizophydiales, the 28S sequences from Chy-Lys2009 and Chy-Kol2008 were aligned with 103 chytrid sequences of Rhizophydiales identified by a BLAST search. The alignment had 865 characters and 323 variable sites (75 singletons). The aligned ITS data included 17 sequences and had 742 characters with 272 variable characters (23 singletons). For both rRNA regions, the neighbor-joining method clustered Chy-Kol2008 and Chy-Lys2009 together in one clade with 100% bootstrap support (see Fig. S2 in the supplemental material). This clade was placed within the recently described Rhizophydiales family Angulomycetaceae (26) with 80% (28S) and 62% (ITS) bootstrap support. The morphology of Chy-Kol2008 and Chy-Lys2009 was consistent with that of members of the family Angulomycetaceae.
Infection experiments with Planktothrix sp.
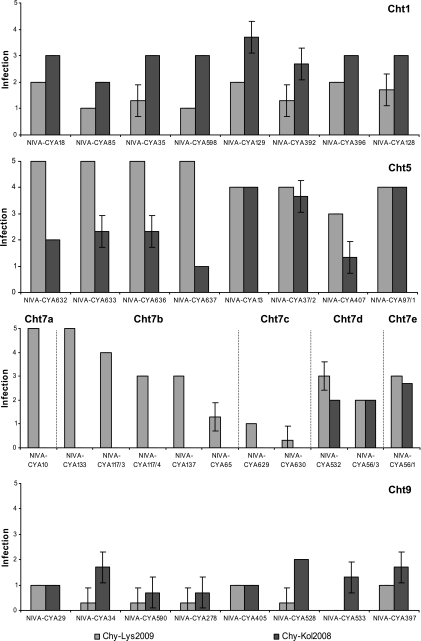
Seven days after addition of chytrid zoospores, the condition of the Planktothrix sp. ranged from healthy without signs of infection to completely lysed with numerous fungal sporangia present. The severity of infection was correlated to the cellular composition of oligopeptides; i.e., the effect of Chy-Lys2009 and Chy-Kol2008 on Planktothrix sp. was chemotype dependent (r = 0.47, P < 0.001) (Fig. 2). When each structural class of oligopeptides is considered individually, the correlation between the severity of infection and the occurrence of oligopeptides was best for the cyanopeptolin class (r = 0.56, P < 0.001), followed by anabaenopeptins (r = 0.40, P < 0.001), microcystins (r = 0.36, P < 0.001), microviridins (r = 0.27, P = 0.002), and aeruginosins (r = 0.16, P = 0.014). No such correlation was found for oscillaginins and oscillatorin. The effects of Chy-Lys2009 and Chy-Kol2008 on Planktothrix sp. strains were uncorrelated (r = 0, P = 0.541); i.e., the two chytrid strains had different chemotype preferences. The infectious capability of chytrid strain Chy-Lys2009 was highest for Planktothrix sp. chemotypes Cht5, Cht7a, Cht7b, Cht7d, and Cht7e. Cht1 was infected to some extent, while Cht7c and Cht9 were nearly or actually resistant to Chy-Lys2009. Chytrid strain Chy-Kol2008 efficiently infected Planktothrix sp. chemotypes Cht1, Cht5, Cht7d, and Cht7e. Cht9 was infected to some extent, and Cht7a to Cht7c were resistant to Chy-Kol2008.
FIG. 2.
Infection intensity in Planktothrix sp. strains representing chemotypes Cht1, Cht5, Cht7a to Cht7e, and Cht9 exposed to chytrid strains Chy-Lys2009 and Chy-Kol2008 for 7 days. Intensity was measured according to the following scale: 0, no sporangia, no active zoospores, 1, up to 10% of host filaments with sporangia, active zoospores present; 2, 10 to 50% of host filaments with sporangia, active zoospores present; 3, 50 to 90% of host filaments with sporangia, active zoospores present; 4, 90 to 100% of host filaments with sporangia, active zoospores present; 5, final stage of infection, most host filaments disintegrated, numerous sporangia visible, and active zoospores present or absent. Each column represents the mean value of tree replicates with the respective standard deviation.
Infection experiments with cyanobacteria other than Planktothrix sp.
Chytrid strains Chy-Lys2009 and Chy-Kol2008 failed to infect any of the 15 Anabaena, Aphanizomenon, and Tychonema species considered in this study. The positive controls showed, as expected, severe chytrid infection.
Test for co-occurrence of chytrid fungi with distinct chemotype preferences in nature.
The Chy-Lys2009 and Chy-Kol2008 ITS genotypes co-occurred in a sample that was collected from Lake Steinsfjorden on 12 July 1999. The Chy-Lys2009 ITS genotype was also detected on all other sampling dates (31 July 1997, 17 September 1997, 17 March 1998, 17 August 1998, 28 July 1998). The melting temperature curves for the Chy-Lys2009 PCR product were identical for all samples and differed from those for Chy-Kol2008.
DISCUSSION
Results of the present study identify chytrid fungi infecting Planktothrix sp. to be highly potent and specialized parasites with chemotype-dependent infectiousness. They possess all properties required to inflict significant mortality on host populations, which is in good agreement with previous field observations (8, 11). Chytrid fungi may exert strong selective pressure on Planktothrix sp. that may lead to reciprocal adaptation. According to the red queen hypothesis, exposure to parasites with narrow host ranges may select for an increase in host diversity (5, 12, 19). The population subdivision into coexisting Planktothrix sp. chemotypes in the presence of fungal parasites with chemotype-dependent infectiousness is compatible with this idea.
The kill-the-winner concept of Thingstad and Lignell (46) provides another hypothesis regarding the subdivision of Planktothrix sp. populations. Originally developed to describe interrelations of viruses and bacteria, the concept suggests that parasites maintain or increase host diversity by infecting mostly the host with the highest abundance, i.e., the winner of an ongoing competition for resources. This forces competing hosts into coexistence. Adapted to the chemotype-dependent chytrid parasitism on Planktothrix sp. and given the co-occurrence of chytrid fungi with distinct chemotype preferences, the kill-the-winner concept predicts the coexistence of multiple Planktothrix sp. chemotypes competing for the same growth resources. It also foresees periodic shifts in the relative chemotype composition. These predictions are in agreement with field observations in Scandinavia (35, 36) and Central Europe (48).
Host diversification may have different effects on parasites. They may be forced into a generalist strategy where exploitation of the host is suboptimal (52), or they may become diversified themselves. Our results suggest that the latter may have occurred in the chytrid-Planktothrix sp. relationship. The chytrid strains Chy-Lys2009 and Chy-Kol2008 were found to be specialists with distinct chemotype preferences. An analysis of field material suggests that Chy-Lys2009 and Chy-Kol2008 or related strains co-occurred in Norwegian Lake Steinsfjorden along with at least three Planktothrix sp. chemotypes. This situation is consistent with a scenario where Planktothrix population subdivision leads to polymorphism in parasitic chytrid fungi.
In an earlier study, we identified gene flow between Planktothrix sp. populations in southeastern Norway (36). Here we found indications for the same process in their chytrid parasites. Chy-Lys2009 and Chy-Kol2008 were originally isolated from two lakes in southeastern Norway (Lake Lyseren and Lake Kolbotnvatnet). Genetic data suggest that the two studied chytrids or closely related strains also occur in Lake Steinsfjorden. The easiest way to explain this redetection in different water bodies is to assume a gene flow between chytrid populations in southeastern Norway. Moreover, the detection of the Chy-Lys2009 rRNA ITS genotype in all Planktothrix sp. samples taken from Lake Steinsfjorden during a 3-year period points to an omnipresence of parasitic chytrid fungi in nature. Both similar patterns of dispersal of a Planktothrix sp. and its chytrid parasites and the presumed omnipresence of parasitic chytrid fungi during times of Planktothrix sp. dominance suggest a close ecological and evolutionary relationship between host and parasite in nature.
The function of cyanobacterial oligopeptides to their producers is, despite many suggestions, still ambiguous. It has often been proposed that these substances may be allelochemicals (2, 9). Some oligopeptides are acutely toxic to grazers of cyanobacteria (6, 27). Since oligopeptides, once synthesized, remain largely intracellular (51), this toxic effect on grazers requires ingestion and digestion of the oligopeptide-producing cells. Moreover, there seems to be no mechanism signaling the presence of toxins to the surrounding environment, and so toxin-containing and toxin-lacking cyanobacterial cells are ingested at similar rates (34). It is therefore doubtful that the toxic effect on grazers attacking from outside the cells is of adaptive value to cyanobacteria, while it may well have secondary effects throughout the aquatic community.
The situation may be different for parasites attacking cyanobacterial cells at least partially from the inside. In this case, oligopeptide production may either stop or slow infection before a host cell is irreversibility harmed, thereby increasing the fitness of this cell. Parasitic chytrid fungi, for instance, use intracellular rhizoids that probably excrete lytic enzymes to extract nutrients from the host and to penetrate its cell walls. In Planktothrix sp., it seems almost inevitable that these processes are disturbed by oligopeptides with protease inhibition capacity, including aeruginosins, anabaenopeptins, cyanopeptolins, microviridins, and oscillaginins (37). Correlations between the occurrence of some of these compounds in Planktothrix sp. and susceptibility to chytrid strains Chy-Kol2008 and Chy-Lys2009 (Fig. 1 and 2) are in agreement with a link between protease inhibitors and the antichytrid defense of Planktothrix sp. The cyanopeptolins in particular seem to be promising candidates for antichytrid agents. It must be underlined here that the present study considered oligopeptide occurrence only. Strains representing the same chemotype can differ to some extent in the cellular oligopeptide concentration and the relative abundance of individual oligopeptides (35). Our findings concerning the correlation between the strength of chytrid infection and oligopeptide composition are therefore somewhat preliminary. A role of protease inhibitors in antiparasite defense of Planktothrix sp. is also supported by the wide distribution of similar defense systems in plants (22) and the presumed participation of protease inhibitors in antichytrid defense of the crawfish frog (Rana areolata) (1).
An antiparasite defense that is based on protease inhibitors usually leads to a coevolutionary arms race (10, 22). It may also be prone to antagonistic pleiotropy; i.e., the production of protease inhibitors may cause resistance to a parasite but at the cost of interference with the host's own metabolism. Here it is important to note that cyanobacterial protease inhibitors can indeed inhibit cyanobacterial proteases (16). Antagonistic pleiotropy may lead to polymorphism in the host population (7). At the same time, antagonistic pleiotropy is under strong stabilizing selection (30), conserving alleles that balance the benefits and costs of an adaptation. The same combination of diversifying and stabilizing selective forces also acts on the Planktothrix sp. gene cluster encoding protease inhibitors of the cyanopeptolin class (39). This combination of selective forces may also explain why Planktothrix sp. populations in southeastern Norway consist of well-delimited chemotypes, which are stable over decades (36). However, despite the above considerations, an involvement of oligopeptide protease inhibitors in defense against parasites remains a hypothesis to be tested in future experiments. Differences in susceptibility of Planktothrix sp. chemotypes to chytrid parasitism may be due to differences in oligopeptide composition, or they may have their cause in a correlation between a yet unknown factor and the oligopeptide fingerprint of Planktothrix sp.
In summary, we propose parasitic chytrid fungi to be one potential driving force of Planktothrix sp. population subdivision and chemotype dynamics. As shown above, this hypothesis is in agreement with established models on host-parasite interactions. It also provides possible explanations for the coexistence of distinct chemotypes in Planktothrix sp. populations, the stability of chemotypes over decades, the occurrence of periodic shifts in the relative chemotype composition, and the lack of a correlation between abiotic environmental conditions and chemotype distribution and dynamics in lakes in Scandinavia and Finland. We also propose that the subdivision of Planktothrix sp. populations into chemotypes may exert diversifying selective pressure on their chytrid parasites. We underline that the above propositions are mainly based on results of laboratory experiments that require validation under field conditions.
Supplementary Material
Acknowledgments
We express our gratitude to Randi Skulberg and Camilla H. C. Hagman, both of the Norwegian Institute for Water Research, for their excellent laboratory assistance.
This work was supported by Norwegian Research Council grant 183360/S30 and by the Norwegian Institute for Water Research.
Footnotes
Published ahead of print on 17 December 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Ali, M. F., et al. 2002. Antimicrobial peptides and protease inhibitors in the skin secretions of the crawfish frog, Rana areolata. Biochim. Biophys. Acta Proteins Proteomics 1601:55-63. [DOI] [PubMed] [Google Scholar]
- 2.Baumann, H. I., and F. Jüttner. 2008. Inter-annual stability of oligopeptide patterns of Planktothrix rubescens blooms and mass mortality of Daphnia in Lake Hallwilersee. Limnologica 38:350-359. [Google Scholar]
- 3.Beard, S. J., P. A. Davis, D. Iglesias-Rodríguez, O. M. Skulberg, and A. E. Walsby. 2000. Gas vesicle genes in Planktothrix spp. from Nordic lakes: strains with weak gas vesicles possess a longer variant of gvpC. Microbiology 146:2009-2018. [DOI] [PubMed] [Google Scholar]
- 4.Becker, S., M. Fahrbach, P. Böger, and A. Ernst. 2002. Quantitative tracing, by Taq nuclease assays, of a Synechococcus ecotype in a highly diversified natural population. Appl. Environ. Microbiol. 68:4486-4494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bell, G. 1982. The masterpiece of nature: the evolution and genetics of sexuality. University of California Press, Berkeley, CA.
- 6.Blom, J. F., et al. 2003. Oscillapeptin J, a new grazer toxin of the freshwater cyanobacterium Planktothrix rubescens. J. Nat. Prod. 66:431-434. [DOI] [PubMed] [Google Scholar]
- 7.Bohannan, B. J. M., and R. E. Lenski. 2000. Linking genetic change to community evolution: insights from studies of bacteria and bacteriophage. Ecol. Lett. 3:362-377. [Google Scholar]
- 8.Canter, H. M., and J. W. G. Lund. 1951. Studies on plankton parasites. III. Examples of the interaction between parasitism and other factors determining the growth of diatoms. Ann. Bot. 15:359-371. [Google Scholar]
- 9.Christoffersen, K. 1996. Ecological implications of cyanobacterial toxins in aquatic food webs. Phycologia 35:S42-S50. [Google Scholar]
- 10.Condra, J. H., et al. 1995. In-vivo emergence of HIV-1 variants resistant to multiple protease inhibitors. Nature 374:569-571. [DOI] [PubMed] [Google Scholar]
- 11.Davis, P. A., M. Dent, J. Parker, C. S. Reynolds, and A. E. Walsby. 2003. The annual cycle of growth rate and biomass change in Planktothrix spp. in Blelham Tarn, English Lake District. Freshwat. Biol. 48:852-867. [Google Scholar]
- 12.De Bruin, A., B. W. Ibelings, M. Kagami, W. M. Mooij, and E. Van Donk. 2008. Adaptation of the fungal parasite Zygorhizidium planktonicum during 200 generations of growth on homogeneous and heterogeneous populations of its host, the diatom Asterionella formosa. J. Eukaryot. Microbiol. 55:69-74. [DOI] [PubMed] [Google Scholar]
- 13.Fabbro, L. D., and L. J. Duivenvoorden. 1996. Profile of a bloom of the cyanobacterium Cylindrospermopsis raciborskii (Woloszynska) Seenaya and Subba Raju in the Fitzroy River in tropical Central Queensland. Mar. Freshwat. Res. 47:685-694. [Google Scholar]
- 14.Fewer, D. P., et al. 2009. The non-ribosomal assembly and frequent occurrence of the protease inhibitors spumigins in the bloom-forming cyanobacterium Nodularia spumigena. Mol. Microbiol. 73:924-937. [DOI] [PubMed] [Google Scholar]
- 15.Fisher, M. C., T. W. J. Garner, and S. F. Walker. 2009. Global emergence of Batrachochytrium dendrobatidis and amphibian chytridiomycosis in space, time, and host. Annu. Rev. Microbiol. 63:291-310. [DOI] [PubMed] [Google Scholar]
- 16.Ghosh, S., D. Bagchi, and S. Bagchi. 2008. Proteolytic activity in Microcystis aeruginosa PCC7806 is inhibited by a trypsin-inhibitory cyanobacterial peptide with a partial structure of microviridin. J. Appl. Phycol. 20:1045-1052. [Google Scholar]
- 17.Gleason, F. H., and O. Lilje. 2009. Structure and function of fungal zoospores: ecological implications. Fungal Ecol. 2:53-59. [Google Scholar]
- 18.Hall, T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95-98. [Google Scholar]
- 19.Hamilton, W. D., R. Axelrod, and R. Tanese. 1990. Sexual reproduction as an adaptation to resist parasites (a review). Proc. Natl. Acad. Sci. U. S. A. 87:3566-3573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Humbert, J. F., and B. B. Le. 2001. Genetic diversity in two species of freshwater cyanobacteria, Planktothrix (Oscillatoria) rubescens and P. agardhii. Arch. Hydrobiol. 150:197-206. [Google Scholar]
- 21.Johnson, Z. I., et al. 2006. Niche partitioning among Prochlorococcus ecotypes along ocean-scale environmental gradients. Science 311:1737-1740. [DOI] [PubMed] [Google Scholar]
- 22.Jongsma, M. A., and C. Bolter. 1997. The adaptation of insects to plant protease inhibitors. J. Insect Physiol. 43:885-895. [DOI] [PubMed] [Google Scholar]
- 23.Katoh, K., G. Asimenos, and H. Toh. 2009. Multiple alignment of DNA sequences with MAFFT, p. 39-64. In D. Posada (ed.), Bioinformatics for DNA sequence analysis. Humana Press, New York, NY. [DOI] [PubMed]
- 24.Kotai, J. 1972. Instructions for the preparation of modified nutrient solution Z8 for algae B-11 / 69. Norsk Institutt for Vannforskning, Oslo, Norway.
- 25.Kurmayer, R., G. Christiansen, and I. Chorus. 2003. The abundance of microcystin-producing genotypes correlates positively with colony size in Microcystis sp. and determines its microcystin net production in Lake Wannsee. Appl. Environ. Microbiol. 69:787-795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Letcher, P. M., et al. 2008. Ultrastructural and molecular analyses of Rhizophydiales (Chytridiomycota) isolates from North America and Argentina. Mycol. Res. 112:759-782. [DOI] [PubMed] [Google Scholar]
- 27.Lürling, M., and E. van der Grinten. 2003. Life-history characteristics of Daphnia exposed to dissolved microcystin-LR and to the cyanobacterium Microcystis aeruginosa with and without microcystins. Environ. Toxicol. Chem. 22:1281-1287. [PubMed] [Google Scholar]
- 28.Nikolcheva, L. G., and E. Bärlocher. 2004. Taxon-specific fungal primers reveal unexpectedly high diversity during leaf decomposition in a stream. Mycol. Prog. 3:41-49. [Google Scholar]
- 29.Oberhaus, L., M. Gelinas, B. Pinel-Alloul, A. Ghadouani, and J. F. Humbert. 2007. Grazing of two toxic Planktothrix species by Daphnia pulicaria: potential for bloom control and transfer of microcystins. J. Plankton Res. 29:827-838. [Google Scholar]
- 30.Otto, S. P. 2004. Two steps forward, one step back: the pleiotropic effects of favoured alleles. Proc. R. Soc. Lond. B Biol. Sci. 271:705-714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peakall, R., and P. E. Smouse. 2006. GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 6:288-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Postius, C., and A. Ernst. 1999. Mechanisms of dominance: coexistence of picocyanobacterial genotypes in a freshwater ecosystem. Arch. Microbiol. 172:69-75. [DOI] [PubMed] [Google Scholar]
- 33.Rasconi, S., M. Jobard, L. Jouve, and T. Sime-Ngando. 2009. Use of Calcofluor White for detection, identification, and quantification of phytoplanktonic fungal parasites. Appl. Environ. Microbiol. 75:2545-2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rohrlack, T., et al. 2005. Ingestion of microcystins by Daphnia: intestinal uptake and toxic effects. Limnol. Oceanogr. 50:440-448. [Google Scholar]
- 35.Rohrlack, T., et al. 2008. Oligopeptide chemotypes of the toxic freshwater cyanobacterium Planktothrix can form subpopulations with dissimilar ecological traits. Limnol. Oceanogr. 53:1279-1293. [Google Scholar]
- 36.Rohrlack, T., R. Skulberg, and O. M. Skulberg. 2009. Distribution of oligopeptide chemotypes of the cyanobacterium Planktothrix and their persistence in selected lakes in Fennoscandia. J. Phycol. 45:1259-1265. [DOI] [PubMed] [Google Scholar]
- 37.Rouhiainen, L., et al. 2004. Genes coding for hepatotoxic heptapeptides (microcystins) in the cyanobacterium Anabaena strain 90. Appl. Environ. Microbiol. 70:686-692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rounge, T. B., et al. 2010. Subpopulation differentiation associated with nonribosomal peptide synthetase gene cluster dynamics in the cyanobacterium Planktothrix spp. J. Phycol. 46:645-652. [Google Scholar]
- 39.Rounge, T. B., T. Rohrlack, T. Kristensen, and K. S. Jakobsen. 2008. Recombination and selectional forces in cyanopeptolin NRPS operons from highly similar, but geographically remote Planktothrix strains. BMC Microbiol. 8:e141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sigee, D. C., A. Selwyn, P. Gallois, and A. P. Dean. 2007. Patterns of cell death in freshwater colonial cyanobacteria during the late summer bloom. Phycologia 46:284-292. [Google Scholar]
- 41.Sparrow, F. K. 1960. Aquatic phycomycetes, 2nd ed. The University of Michigan Press, Ann Arbor, MI.
- 42.Strom, S. L. 2008. Microbial ecology of ocean biogeochemistry: a community perspective. Science 320:1043-1045. [DOI] [PubMed] [Google Scholar]
- 43.Suda, S., et al. 2002. Taxonomic revision of water-bloom-forming species of oscillatorioid cyanobacteria. Int. J. Syst. Evol. Microbiol. 52:1577-1595. [DOI] [PubMed] [Google Scholar]
- 44.Symonds, E. P., D. J. Trott, P. S. Bird, and P. Mills. 2008. Growth characteristics and enzyme activity in Batrachochytrium dendrobatidis isolates. Mycopathologia 166:143-147. [DOI] [PubMed] [Google Scholar]
- 45.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596-1599. [DOI] [PubMed] [Google Scholar]
- 46.Thingstad, T. F., and R. Lignell. 1997. Theoretical models for the control of bacterial growth rate, abundance, diversity and carbon demand. Aquat. Microb. Ecol. 13:19-27. [Google Scholar]
- 47.Vilgalys, R., and M. Hester. 1990. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 172:4238-4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Welker, M., G. Christiansen, and H. von Döhren. 2004. Diversity of coexisting Planktothrix (cyanobacteria) chemotypes deduced by mass spectral analysis of microystins and other oligopeptides. Arch. Microbiol. 182:288-298. [DOI] [PubMed] [Google Scholar]
- 49.Welker, M., and H. von Döhren. 2006. Cyanobacterial peptides—nature's own combinatorial biosynthesis. FEMS Microbiol. Rev. 30:530-563. [DOI] [PubMed] [Google Scholar]
- 50.White, T. J., T. Bruns, S. Lee, and J. W. Taylor. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics, p. 315-322. In M. A. Innis, D. H. Gelfand, J. J. Sninsky, and T. J. White (ed.), PCR protocols: a guide to methods and applications. Academic Press, Inc., New York, NY.
- 51.Young, F. M., C. Thomson, J. S. Metcalf, J. M. Lucocq, and G. A. Codd. 2005. Immunogold localisation of microcystins in cryosectioned cells of Microcystis. J. Struct. Biol. 151:208-214. [DOI] [PubMed] [Google Scholar]
- 52.Yourth, C. P., and P. Schmid-Hempel. 2006. Serial passage of the parasite Crithidia bombi within a colony of its host, Bombus terrestris, reduces success in unrelated hosts. Proc. R. Soc. Lond. B Biol. Sci. 273:655-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.