Abstract
Lichens are commonly described as a mutualistic symbiosis between fungi and “algae” (Chlorophyta or Cyanobacteria); however, they also have internal bacterial communities. Recent research suggests that lichen-associated microbes are an integral component of lichen thalli and that the classical view of this symbiotic relationship should be expanded to include bacteria. However, we still have a limited understanding of the phylogenetic structure of these communities and their variability across lichen species. To address these knowledge gaps, we used bar-coded pyrosequencing to survey the bacterial communities associated with lichens. Bacterial sequences obtained from four lichen species at multiple locations on rock outcrops suggested that each lichen species harbored a distinct community and that all communities were dominated by Alphaproteobacteria. Across all samples, we recovered numerous bacterial phylotypes that were closely related to sequences isolated from lichens in prior investigations, including those from a lichen-associated Rhizobiales lineage (LAR1; putative N2 fixers). LAR1-related phylotypes were relatively abundant and were found in all four lichen species, and many sequences closely related to other known N2 fixers (e.g., Azospirillum, Bradyrhizobium, and Frankia) were recovered. Our findings confirm the presence of highly structured bacterial communities within lichens and provide additional evidence that these bacteria may serve distinct functional roles within lichen symbioses.
Marine sponges (31), the termite hindgut (34), mycorrhizal fungi (2, 13), and fungal endophytes (18) represent symbiotic niches where unique interactions between eukaryotes and rich, diverse bacterial populations occur. Lichens, a relationship between fungi (mycobionts, on which the species names and classifications are based) and green algae and/or cyanobacteria (photobionts), are another symbiotic niche that has received recent attention (15). Lichen thalli host diverse populations of organisms, such as lichenicolous (growing on the thallus) (21) and endolichenic (growing within the thallus) fungal species (1). While nonphotosynthetic bacteria have long been suspected to be associated with lichens (32; but also see reference 8), modern culture-independent studies are just beginning to reveal the diversity in bacteria inhabiting lichens, suggesting that lichens host diverse assemblages from several bacterial phyla (7, 8, 14, 16, 17, 23).
The few studies that have characterized the bacterial communities of lichens by using molecular fingerprinting techniques are limited in their phylogenetic resolution and have yielded conflicting results. Grube et al. (16) found that the community composition for certain lichen-associated bacterial groups (e.g., Alphaproteobacteria) exhibited species-specific patterns, and they advanced the idea that bacteria may be integral to the lichen symbiosis. Cardinale et al. (8), however, concluded that the structure of bacterial communities in lichens was not correlated with the host species and provided evidence that some lichen-associated bacteria may simply be opportunistic, perhaps as extensions of the immediate surrounding soil environment, rather than symbiotic partners.
Because the majority of lichens have green algal photobionts that are not capable of fixing atmospheric N2 like their cyanobacterial counterparts and because N acquisition is thought to occur exclusively via aerial deposition of inorganic N forms (29), work has also focused on the potential functional role for N2-fixing bacteria in the lichen symbiosis. For example, there have been reports of bacterial nitrogenase reductase (nifH) genes (responsible for N2 fixation) in bacteria isolated from lichens as well as within lichens themselves (16, 23). Bacteria found on internal and external surfaces of lichens may also help lichen symbionts to fulfill other nutritional requirements, including the acquisition of phosphorus and amino acids (16, 23).
Here we used bar-coded pyrosequencing with primers targeting the 16S rRNA gene to survey bacterial and archaeal diversity associated with surface-sterilized lichen thalli compared to that in nearby soils from a site in the western United States. Our objective was to determine if lichen-associated bacterial and archaeal communities are characteristic of lichen species, indicative of immediate spatial areas, or simply an extension of the surrounding soil environment. We also wanted to determine if the specific taxa found within lichens provide any clues regarding the functional roles of these microbes within the lichen symbiosis.
MATERIALS AND METHODS
Sampling design and site description.
Four different foliose green algal lichen species (Parmelia sulcata, Rhizoplaca chrysoleuca, Umbilicaria americana, and Umbilicaria phaea) (Fig. 1) were collected from granite rock outcrops at a site in northern Colorado (40.01°N, 105.47°W). The rocks were initially surveyed to ensure the presence of all four species; samples of individual lichen thalli of each species were then collected from each of three outcrops (one outcrop was sampled at two points within ∼3 m of one another) with a sterile knife and placed into individual sterile plastic collection bags. The thalli of the four different lichen species collected were generally within a few centimeters of one another and always within a 1-m2 area at each sampling location. A total of 16 samples were collected for the study (4 sampling locations, with 4 lichen species collected at each location). Each rock outcrop was located approximately 30 m apart from the others, and all were on the same southeast-facing slope. Eight soil samples (mollisols of the Typic Haplustoll subgroup) from sites adjacent to the rock outcrops were also collected and processed using previously described methods (12). All samples were transported back to the lab on ice and immediately processed.
FIG. 1.
Species of foliose lichens containing green algal photobionts collected on rock outcrops at the sampling site. (A) Parmelia sulcata; (B) Rhizoplaca chrysoleuca (image courtesy of T. Wheeler); (C) Umbilicaria americana; (D) Umbilicaria phaea (image courtesy of F. Bungartz).
Surface sterilization and isolation of community DNA.
To focus on bacteria likely to be integral components of lichens (e.g., those found on internal surfaces of the thallus), we removed as many external surface-borne microbes from our samples as possible. Each lichen thallus was surface sterilized according to the most stringent protocol outlined by Arnold et al. (1): samples were washed for ∼30 s in sterile ultrapure laboratory-grade (Milli-Q) water to remove dirt or debris from surfaces and then were immersed and agitated separately in 96% ethanol for 10 s, followed by 0.5% NaOCl (bleach) for 2 min and 70% ethanol for 4 min. Immediately after surface sterilization, a small portion of material (∼2 cm2) was removed from the thallus of each lichen sample for genomic DNA extraction.
We followed a standard extraction protocol, using a commercially available PowerSoil DNA isolation kit (MoBio Laboratories, Carlsbad, CA), after an initial treatment optimized to enhance DNA yield. This initial treatment included grinding each sample separately with a sterile mortar and pestle under liquid N2, placing the macerated samples into individual 2-ml bead-beating tubes with kit buffer, and immersing them in a 65°C water bath for 10 min. After heating, the samples were processed according to the extraction kit protocol. Community DNA was extracted from soil samples in the same manner as described above, but with the elimination of the surface sterilization steps.
PCR amplification of 16S rRNA genes and bar-coded pyrosequencing.
Preparation of extracted DNA for pyrosequencing followed the protocol outlined by Fierer et al. (11). Briefly, the method includes targeted PCR amplification of a portion of the 16S small-subunit rRNA gene, triplicate PCR product pooling (per sample) to mitigate reaction-level PCR biases, and sequencing on a Roche GS-FLX 454 automated pyrosequencer. The primer set F515 (5′-GTGCCAGCMGCCGCGGTAA-3′) and R806 (5′-GGACTACVSGGGTATCTAAT-3′) was used on both lichen and soil samples. This primer set, generating ∼250-bp amplicons, is suited for community analysis of bacterial sequences (25) and has been used previously to amplify, with few biases, a wide range of bacterial and archaeal groups from soils (4). The F515 primer included a Roche 454-A pyrosequencing adapter and a 2-bp linker sequence (GT), and R806 incorporated 12-bp bar-coded sequences (each unique to an individual sample), a GG linker, and a Roche 454-B sequencing adapter.
The PCR was carried out in 25-μl reaction mixtures, which each contained 2 μl (15 μM [each]) of forward and reverse primers and 20 μl of 5Prime Hot master mix (Eppendorf-5Prime, Gaithersburg, MD). Each reaction mix received 1 μl of genomic community DNA as a template, and the following cycling parameters were used: 35 cycles (95°C for 30 s, 50°C for 1 min, and 72°C for 1 min) were performed after an initial denaturation at 95°C for 3 min. Triplicate reaction mixtures per sample were pooled, purified using an UltraClean PCR cleanup kit (MoBio), and quantified using the PicoGreen dsDNA assay (Invitrogen, Carlsbad, CA). The bar-coded PCR products from all samples (both lichen and soil) were normalized in equimolar amounts in a pooled sample, gel purified (precipitated with NaCl and ethanol) to remove any remaining residues, and sent for sequencing at EnGenCore at the University of South Carolina.
Sequence processing, assignment of taxonomic identity, diversity, and phylogeny.
We used the QIIME software pipeline (6) for initial processing of sequence data, quality control, phylotype binning, taxonomic assignment of raw pyrosequence data, and sample grouping according to unique 12-bp bar codes. The data were also manually screened to remove sequences originating from chloroplast or other eukaryotic organisms. The phylogenetic placement of bacterial sequences obtained from our lichen samples was assessed by first assembling representative sequences of each bacterial phylotype (recovered sequences sharing ≥97% similarity) identified here, as well as representative sequences from other studies, and then aligning these using the NAST alignment function of the GreenGenes public database (http://greengenes.lbl.gov). Phylogenetic trees were created using the ARB software package (27) and were visualized using Interactive Tree of Life (iTOL) (22) software.
Microbial community and statistical analyses.
We used a taxonomic (Bray-Curtis) approach for community analysis, generating distance matrices for our samples in the QIIME pipeline. To control for different sequencing depths, we randomly sampled at an even depth (249 sequences per sample) before computing beta diversity. The matrices generated were then used in nonmetric multidimensional scaling (NMDS) and permutational multivariate analysis of variance (PERMANOVA), carried out using the PRIMER 6+ software package (9). Only phylotypes that were found in four or more samples were included in the distance matrix analyses to minimize the influence of low-frequency phylotypes that were restricted to individual samples.
RESULTS AND DISCUSSION
Phylotypes recovered from samples.
A total of 657 distinct bacterial phylotypes were recovered from the 11,572 high-quality sequences (∼250 bp, on average) generated by pyrosequencing of 16S rRNA genes from our 16 lichen samples. Each lichen sample yielded 723 sequences, on average (ranging from 257 to 1,082 sequences), which is a sufficient sampling effort for examining patterns of beta diversity across samples (20). Although the primer set should amplify bacterial as well as archaeal 16S rRNA genes (4), no archaeal sequences were recovered from these lichen samples. An additional 7,230 bacterial and archaeal sequences were generated from the soil samples, with an average of 1,033 sequences per soil sample (range, 922 to 1,144 sequences).
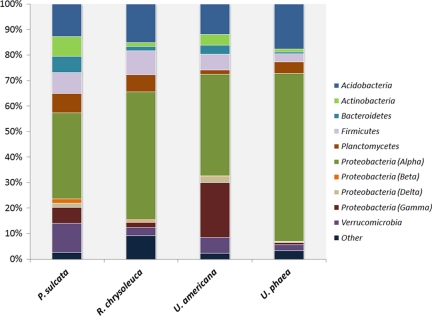
Lichen thalli appear to be associated with a broad spectrum of bacterial taxa, and phylotypes representing a number of phyla were recovered from our lichen samples (see Fig. S1 in the supplemental material). Some taxa that were ubiquitous across our lichen samples, namely, Actinobacteria, Firmicutes, and Gammaproteobacteria (Fig. 2), have also been observed in previous studies of lichen-associated bacteria (7, 8, 14, 16, 23). In addition, bacterial 16S rRNA gene sequences culled from a subset of these studies (8, 16, 23) were closely related to our phylotypes (see Fig. S1 in the supplemental material), which implies that particular bacterial taxa are widespread in lichens across very large geographic areas (Brazil, Europe, and the western to eastern United States). Our survey also recovered phylotypes representing abundant lichen-associated bacterial populations from the Bacteroidetes, Planctomycetes, and Verrucomicrobia (Fig. 2), which have not been reported previously from lichens. Finally, many of our phylotypes were not closely related to bacterial taxa that have been isolated previously and described formally, including some phylotypes representing bacterial lineages of high taxonomic ranks (see Fig. S1 in the supplemental material).
FIG. 2.
Relative abundances of various major bacterial lineages recovered from lichen species. Relative abundance was calculated as the percentage of sequences belonging to a particular lineage of all 16S rRNA gene sequences recovered from a given lichen species (4 samples per species).
Dominant bacterial taxa associated with lichens.
All lichen species were dominated by Alphaproteobacteria, and the lichens generally shared similar relative abundances of higher-level bacterial taxa (Fig. 2). Although not reflected in cultivation-based studies of lichen-associated bacteria, the prevalence of the Alphaproteobacteria in lichens has been observed previously via fluorescence in situ hybridization (FISH) studies of thallus surfaces (7, 16). Members of the Alphaproteobacteria commonly participate in symbiotic relationships (e.g., between Rhizobiales and vascular plants), and culture-independent studies have also pointed to the importance of the Alphaproteobacteria in microbial communities of belowground ectomycorrhizal fruiting bodies (3) as well as those coexisting with fungal species in decaying wood (33). Given the potentially expansive history of symbiotic relationships for the Alphaproteobacteria (19), their dominance in lichens is noteworthy, as lichens may represent an ancient symbiosis, perhaps dating to the Precambrian Era (28, 35).
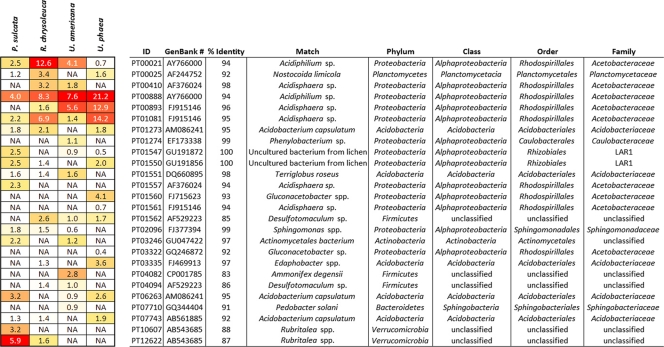
The dominant phylotypes of the lichens sampled here (those that were both abundant and widespread in samples of a given species) were members of the Acetobacteraceae within the Alphaproteobacteria (Fig. 3). Acetobacteraceae contains some N2-fixing species, such as Gluconacetobacter diazotrophicus, which shared 93% identity with one of our dominant phylotypes (PT001560). Since members of the Acetobacteraceae are also known to be chemoorganotrophs (5), our dominant phylotypes might also represent opportunistic organisms able to sustain growth on carbon sources present in the lichen thallus, such as mannitol (a common storage product of the mycobiont). However, the true identity and function within the lichen thallus for many of the dominant lichen-associated phylotypes listed in Fig. 3 remain unresolved, as many do not appear to be closely related to well-studied cultured isolates.
FIG. 3.
Heat map showing the 15 most dominant phylotypes in each of four lichen species sampled for this study and their average relative abundances (as percentages of all sample 16S rRNA gene sequences recovered). Phylotypes were considered dominant if they were both highly abundant and occurred frequently in samples of a given lichen species. Color tones moving from red to yellowish white indicate the highest to lowest relative abundance values (by species). “NA” indicates that the phylotype was not included within the 15 most dominant phylotypes for that species.
Potential for bacterium-mediated nutrient acquisition in lichens.
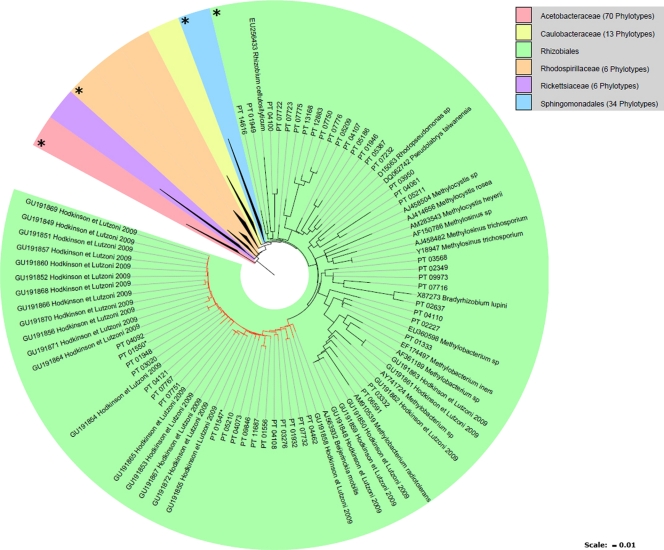
Of the dominant phylotypes recovered from our lichen samples, two (PT01547 and PT01550) directly matched previously identified bacteria within the putative N2-fixing Rhizobiales lineage (LAR1) recovered from lichen species collected in the eastern United States (17) (Fig. 3). These two dominant phylotypes each occurred, sometimes together, within three lichen species (with one or the other being found within all 4 species). Along with these dominant LAR1-affiliated phylotypes, many other LAR1 sequences (as well as others from the Rhizobiales) of Hodkinson and Lutzoni (17) also closely matched several of our less dominant phylotypes (Fig. 4), again suggesting that lichens harbor similar bacterial taxa regardless of the species or locality from which they were collected.
FIG. 4.
ARB maximum parsimony constraint tree of Alphaproteobacteria sequences recovered from our lichen samples as well as closely related sequences obtained from GenBank (accession numbers are indicated, and those from previous lichen-bacterium studies show a citation). A large asterisk indicates that the lineage contains phylotypes closely related to sequences within genera containing known N2-fixing bacteria. PT, phylotype (those PT numbers with asterisks are found among the 15 most dominant phylotypes per species) (Fig. 3). Red branches indicate the LAR1 lineage. Bar = 1% sequence dissimilarity. The tree is rooted with Acinetobacter baumannii (GenBank accession no. AY847284).
Phylotypes were also recovered from our lichen thalli that were closely related to bacteria isolated from lichens with green algal photobionts by Liba et al. (23) and shown to be N2-fixing strains (e.g., Pseudomonas stutzeri). Furthermore, we found phylotypes (see Fig. S1 in the supplemental material) representing bacteria very closely related to several other known N2-fixing taxa, such as Frankia (Actinobacteria), Beijerinckia, Bradyrhizobium, and Azospirillum (Alphaproteobacteria), and others with less certain affiliations were found close to or within genera known to contain N2 fixers (e.g., Acinetobacter, Burkholderia, Gluconobacter, and Rhodospirillum). While this is by no means conclusive proof that these phylotypes are actually capable of N2 fixation, our results do suggest that lichens may harbor a variety of bacterial taxa capable of contributing to their nutritional requirements for N.
In addition to N, lichen-associated bacteria recovered in our survey may also play a role in the acquisition of phosphorus, confirming previous results demonstrating that many lichen-associated bacteria show phosphate-solubilizing activity (16, 23). Members of the Acetobacteraceae were dominant in our samples, and a number of strains from this family are known to solubilize phosphate (26, 30). One of our dominant phylotypes (PT01560) was closely related to Gluconobacter oxydans (see Fig. S1 in the supplemental material), an organism known to have genes coding for histidine acid phosphatase (HAP), an enzyme involved in the mineralization of phytate (a highly abundant organic phosphorus compound) (24). Although these connections are suggestive, we will need more directed and in-depth studies, such as specific functional gene analyses or comprehensive metagenomic studies, to more fully understand the functions of bacteria within the lichen symbiosis.
Structure of lichen-associated bacterial communities.
Lichen-associated bacterial communities were clearly distinct from those in nearby soils (see Fig. S2 in the supplemental material), suggesting that lichens harbor bacterial communities that are not simply an extension of those found in the surrounding soil environment. A lichen-specific structure would be expected if particular bacteria serve distinct functional roles within the lichen symbiosis; however, abiotic factors intrinsic to the lichen may also serve to shape these bacterial communities. For example, sizable populations of Acidobacteria were recovered from each of our lichen species (Fig. 2 and 3), perhaps representing acidophilic bacteria that can survive the conditions within the thalli of lichens, which are known to produce a wide range of organic acid secondary metabolites in abundance (10, 17).
As evident from Fig. 5, the overall beta diversity patterns exhibited by our lichen-associated bacterial communities were not a function of their rock outcrop of origin. Instead, bacterial communities clustered (and were statistically distinct) according to the lichen species from which they were isolated (Fig. 5). Species identity was clearly a much better predictor of lichen-associated bacterial communities than the spatial proximity of the lichen thalli collected.
FIG. 5.
Nonmetric multidimensional scaling plots (Bray-Curtis distance matrices) depicting patterns of beta diversity for bacterial communities associated with lichen thalli. Points that are closer together on the ordination have communities that are more similar, and dot- ted lines were added postanalysis to highlight species clusters. PERMANOVA indicated that differences between bacterial communities mapped according to the lichen species of origin (A) for the sample were highly significant (P = 0.001), while differences mapped according to rock outcrop of origin (B) were not significant (P = 0.65).
Conclusions.
Lichen-associated bacterial communities are not merely extensions of those found in surrounding soils; they are distinct in their structure. Different lichen species in close spatial proximity harbor dissimilar bacterial communities, and the lichen species itself appears to be the strongest predictor of community composition. Our findings lend credence to the notion that lichen-associated bacterial communities are highly structured, perhaps reflecting their functional role in the lichen symbiosis, and also suggest that some bacterial taxa are widespread in different lichen species, even across very large geographic areas. The Alphaproteobacteria appear to be the dominant bacterial group associated with lichens; however, a wide variety of bacteria from higher-order taxa, some undescribed, are also present in lichen thalli. Finally, our data suggest that assemblages of bacteria from divergent lineages may serve to provide lichens with N; however, the functional roles of lichen-associated bacteria still remain largely undetermined. Taken together, the results of this study build on previous work suggesting that bacteria are an additional, integral component of the lichen symbiosis.
Supplementary Material
Acknowledgments
Members of the Fierer lab, particularly Chris Lauber and Donna Berg-Lyons, helped in the laboratory and with analyses and sample processing. We also thank Albert Barberán, who helped with sequence analyses. Joe Jones, from EnGenCore, supervised the pyrosequencing. Tim Wheeler (http://waysofenlichenment.net/) and Frank Bungartz graciously allowed permission for use of their images of Rhizoplaca chrysoleuca and Umbilicaria phaea, respectively. Constructive comments from two anonymous reviewers greatly improved the quality of the manuscript.
This research was funded by grants to R.K. and N.F. from the National Institutes of Health, the National Science Foundation, and the U.S. Department of Agriculture.
Footnotes
Published ahead of print on 17 December 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Arnold, A. E., et al. 2009. A phylogenetic estimation of trophic transition networks for ascomycetous fungi: are lichens cradles of symbiotrophic fungal diversification? Syst. Biol. 58:283-297. [DOI] [PubMed] [Google Scholar]
- 2.Artursson, V., R. D. Finlay, and J. K. Jansson. 2006. Interactions between arbuscular mycorrhizal fungi and bacteria and their potential for stimulating plant growth. Environ. Microbiol. 8:1-10. [DOI] [PubMed] [Google Scholar]
- 3.Barbieri, E., et al. 2005. New evidence for bacterial diversity in the ascoma of the ectomycorrhizal fungus Tuber borchii Vittad. FEMS Microbiol. Lett. 247:23-35. [DOI] [PubMed] [Google Scholar]
- 4.Bates, S. T., et al. 18 November 2010. Examining the global distribution of dominant archaeal populations in soil. ISME J. doi: 10.1038/ismej.2010.171. [DOI] [PMC free article] [PubMed]
- 5.Brenner, D. J., N. R. Krieg, and J. T. Staley. 2005. Bergey's manual of systematic bacteriology, 2nd ed., vol. 2. The proteobacteria. Part C. Alpha-, beta-, delta-, and epsilonproteobacteria. Springer, New York, NY.
- 6.Caporaso, J. G., et al. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7:335-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cardinale, M., et al. 2008. In situ analysis of the bacterial community associated with the reindeer lichen Cladonia arbuscula reveals predominance of Alphaproteobacteria. FEMS Microbiol. Ecol. 66:63-71. [DOI] [PubMed] [Google Scholar]
- 8.Cardinale, M., A. M. Puglia, and M. Grube. 2006. Molecular analysis of lichen-associated bacterial communities. FEMS Microbiol. Ecol. 57:484-495. [DOI] [PubMed] [Google Scholar]
- 9.Clark, K. R., and R. N. Gorley. 2006. PRIMER v6: user manual/tutorial. PRIMER-E, Plymouth, United Kingdom.
- 10.Elix, J. A., and E. Stocker-Wörgötten. 2008. Biochemistry and secondary metabolites, p. 104-133. In T. H. Nash III (ed.), Lichen biology, 2nd ed. Cambridge University Press, Cambridge, United Kingdom.
- 11.Fierer, N., M. Hamady, C. L. Lauber, and R. Knight. 2008. The influence of sex, handedness, and washing on the diversity of hand surface bacteria. Proc. Natl. Acad. Sci. U. S. A. 105:17994-17999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fierer, N., and R. B. Jackson. 2006. The diversity and biogeography of soil bacterial communities. Proc. Natl. Acad. Sci. U. S. A. 103:626-631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Founoune, H., et al. 2002. Mycorrhiza helper bacteria stimulate ectomycorrhizal symbiosis of Acacia holosericea with Pisolithus alba. New Phytol. 153:81-89. [Google Scholar]
- 14.González, I., A. Ayuso-Sacido, A. Anderson, and O. Genilloud. 2005. Actinomycetes isolated from lichens: evaluation of their diversity and detection of biosynthetic gene sequences. FEMS Microbiol. Ecol. 54:401-415. [DOI] [PubMed] [Google Scholar]
- 15.Grube, M., and G. Berg. 2009. Microbial consortia of bacteria and fungi with focus on the lichen symbiosis. Fungal Biol. Rev. 23:72-85. [Google Scholar]
- 16.Grube, M., et al. 2009. Species-specific structural and functional diversity of bacterial communities in lichen symbioses. ISME J. 3:1105-1115. [DOI] [PubMed] [Google Scholar]
- 17.Hodkinson, B., and F. Lutzoni. 2009. A microbiotic survey of lichen-associated bacteria reveals a new lineage from the Rhizobiales. Symbiosis 49:163-180. [Google Scholar]
- 18.Hoffman, M. T., and A. E. Arnold. Diverse bacteria inhabit living hyphae of phylogenetically diverse fungal endophytes. Appl. Environ. Microbiol. 76:4063-4075. [DOI] [PMC free article] [PubMed]
- 19.Horner, D. S., R. P. Hirt, S. Kilvington, D. Lloyd, and T. M. Embley. 1996. Molecular data suggest an early acquisition of the mitochondrion endosymbiont. Proc. R. Soc. Lond. B Biol. Sci. 263:1053-1059. [DOI] [PubMed] [Google Scholar]
- 20.Kuczynski, J., et al. 2010. Microbial community resemblance methods differ in their ability to detect biologically relevant patterns. Nat. Methods 7:813-819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lawrey, J. D., and P. Diederich. 2003. Lichenicolous fungi: interactions, evolution, and biodiversity. Bryologist 106:80-120. [Google Scholar]
- 22.Letunic, I., and P. Bork. 2007. Interactive Tree of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics 23:127-128. [DOI] [PubMed] [Google Scholar]
- 23.Liba, C., et al. 2006. Nitrogen-fixing chemo-organotrophic bacteria isolated from cyanobacteria-deprived lichens and their ability to solubilize phosphate and to release amino acids and phytohormones. J. Appl. Microbiol. 101:1076-1086. [DOI] [PubMed] [Google Scholar]
- 24.Lim, B. L., P. Yeung, C. Cheng, and J. E. Hill. 2007. Distribution and diversity of phytate-mineralizing bacteria. ISME J. 1:321-330. [DOI] [PubMed] [Google Scholar]
- 25.Liu, Z., C. Lozupone, M. Hamady, F. D. Bushman, and R. Knight. 2007. Short pyrosequencing reads suffice for accurate microbial community analysis. Nucleic Acids Res. 35:e120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Loganathan, P., and S. Nair. 2004. Swaminathania salitolerans gen. nov., sp. nov., a salt-tolerant, nitrogen-fixing and phosphate-solubilizing bacterium from wild rice (Porteresia coarctata Tateoka). Int. J. Syst. Evol. Microbiol. 54:1185-1190. [DOI] [PubMed] [Google Scholar]
- 27.Ludwig, W., et al. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lutzoni, F., M. Pagel, and V. Reeb. 2001. Major fungal lineages are derived from lichen symbiotic ancestors. Nature 411:937-940. [DOI] [PubMed] [Google Scholar]
- 29.Nash, T. H., III. 2008. Nitrogen, its metabolism and potential contribution to ecosystems, p. 216-233. In Lichen biology, 2nd ed. Cambridge University Press, Cambridge, United Kingdom.
- 30.Silva, L., et al. 2002. Microorganisms with capacity for phosphate solubilization in Dão red wine (Portugal). Dev. Plant Soil Sci. 102:245-248. [Google Scholar]
- 31.Taylor, M. W., R. Radax, D. Steger, and M. Wagner. 2007. Sponge-associated microorganisms: evolution, ecology, and biotechnological potential. Microbiol. Mol. Biol. Rev. 71:295-347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Uphof, J. C. 1925. Purple bacteria as symbionts of a lichen. Science 61:67. [DOI] [PubMed] [Google Scholar]
- 33.Valášková, V., W. de Boer, P. J. A. Klein Gunnewiek, M. Pospíšek, and P. Baldrian. 2009. Phylogenetic composition and properties of bacteria coexisting with the fungus Hypholoma fasciculare in decaying wood. ISME J. 3:1218-1221. [DOI] [PubMed] [Google Scholar]
- 34.Warnecke, F., et al. 2007. Metagenomic and functional analysis of hindgut microbiota of a wood-feeding higher termite. Nature 450:560-565. [DOI] [PubMed] [Google Scholar]
- 35.Yuan, X., S. Xiao, and T. N. Taylor. 2005. Lichen-like symbiosis 600 million years ago. Science 308:1017-1020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.