Abstract
One emerging disease of grapevine in Europe is Bois noir (BN), a phytoplasmosis caused by “Candidatus Phytoplasma solani” and spread in vineyards by the planthopper Hyalesthes obsoletus (Hemiptera: Cixiidae). Here we present the first full characterization of the bacterial community of this important disease vector collected from BN-contaminated areas in Piedmont, Italy. Length heterogeneity PCR and denaturing gradient gel electrophoresis analysis targeting the 16S rRNA gene revealed the presence of a number of bacteria stably associated with the insect vector. In particular, symbiotic bacteria detected by PCR with high infection rates in adult individuals fell within the “Candidatus Sulcia muelleri” cluster in the Bacteroidetes and in the “Candidatus Purcelliella pentastirinorum” group in the Gammaproteobacteria, both previously identified in different leafhoppers and planthoppers. A high infection rate (81%) was also shown for another symbiont belonging to the Betaproteobacteria, designated the HO1-V symbiont. Because of the low level of 16S rRNA gene identity (80%) with the closest relative, an uncharacterized symbiont of the tick Haemaphysalis longicornis, we propose the new name “Candidatus Vidania fulgoroideae.” Other bacterial endosymbionts identified in H. obsoletus were related to the intracellular bacteria Wolbachia pipientis, Rickettsia sp., and “Candidatus Cardinium hertigii.” Fluorescent in situ hybridization coupled with confocal laser scanning microscopy and transmission electron microscopy showed that these bacteria are localized in the gut, testicles, and oocytes. As “Ca. Sulcia” is usually reported in association with other symbiotic bacteria, we propose that in H. obsoletus, it may occur in a bipartite or even tripartite relationship between “Ca. Sulcia” and “Ca. Purcelliella,” “Ca. Vidania,” or both.
Grapevine yellows is a severe insect-borne disease affecting grapes in many wine-producing countries. It is caused by phytoplasmas, cell wall-less bacteria belonging to the class Mollicutes that can multiply in the body of the insect vector and in phloem cells of the host plant (18, 32). An emerging grape yellows is “Bois noir” (BN), caused by a phytoplasma of the Stolbur group (16Sr-XII) recently proposed as “Candidatus Phytoplasma solani” (29). The insect vector of BN is Hyalesthes obsoletus, a polyphagous planthopper (Hemiptera: Cixiidae) that can occasionally feed on grapevine, although it is usually found on dicotyledonous weeds (1, 2). A direct approach for controlling BN is not available, but measures for limiting the spread of the disease are based on controlling the insect vector with insecticides and the management of weeds in the vineyard.
The use of biocontrol agents is of increasing interest in pest management (6, 7, 47, 51). One emerging strategy is “symbiotic control,” which applies the exploitation of microorganisms associated with the insect vector to provide antidisease strategies, such as a reduction of vector competence (6) or the manipulation of undesirable host traits (47).
For developing symbiotic control, the identification of dominant symbionts of the insect vector is necessary. In the case of H. obsoletus, despite the increasing relevance of BN in European vineyards, only few works describing the microbiota of this insect vector have been published: a preliminary characterization indicating an association with symbionts related to Wolbachia and the Bacteroidetes (23) and a symbiont screening of different planthoppers showing the affiliation of the bacteriome-restricted organisms “Candidatus Sulcia muelleri” and “Candidatus Purcelliella pentastirinorum” (12). However, that study did not provide details on the localization of symbionts in the insect body.
The present study examined, by means of molecular ecology techniques, the symbiont diversity residing in the body of H. obsoletus. We also provide information on the tissue localization of several endosymbionts. This study indicates that several symbionts cohabitate in the male and female gonads, suggesting that complex interactions between different vertically transmitted endosymbionts occur in the same insect host (20, 26).
MATERIALS AND METHODS
Insect material and DNA extraction.
H. obsoletus individuals were collected in 2005 to 2007 from wastelands close to vineyards affected by BN in Piedmont, North Italy. Ninety-nine individuals of H. obsoletus were killed with ethyl acetate and preserved frozen at −20°C or in ethanol until molecular analysis. Twenty H. obsoletus adult specimens were dissected to isolate salivary glands, gut, fat bodies, and ovaries. The total DNA of whole insects and dissected organs was extracted according to a method previously described by Doyle and Doyle (19).
Molecular techniques for characterizing the microflora of H. obsoletus.
Two different molecular methods were used to study the bacterial community associated with H. obsoletus. A length heterogeneity PCR (LH-PCR) (48, 53) was carried out to screen the diversity of the microbial population associated with H. obsoletus. The DNA extracted from insects was subjected to PCR amplification using eubacterial universal primers 27F and 338R (48); primer 27F was labeled with the fluorescent reporter dye 6-carboxyfluorescein (FAM) on the 5′ end. PCR conditions and sample preparation were previously described (35). LH-PCR fragments were loaded onto an ABI Prism 310 capillary electrophoresis system (Applied Biosystems) and run under denaturing conditions using the POP-4 running polymer (Applied Biosystems). The LH-PCR data were analyzed with Genescan 3.1.2 software (Applied Biosystems).
For DGGE (denaturing gradient gel electrophoresis) analysis, bacterial 16S rRNA genes were amplified by using forward primer GC357f, containing a 40-bp GC clamp, and reverse primer 907r, as previously described (35, 50). Polyacrylamide gels (7% of a 37:1 acrylamide-bisacrylamide mixture in 1× Tris-acetate-EDTA [TAE] buffer) with a gradient of 40 to 60% or 20 to 30% denaturant were used; 100% denaturant corresponds to 7 M urea and 40% formamide (45).
Sequencing of DGGE bands.
Selected DGGE bands were excised from the DNA eluted and used as a template in PCR reamplification reactions with primers 357F (without a GC clamp) and 907R, performed as previously described (35). The obtained PCR products were purified and sequenced (Primm, Milan, Italy), and the resulting sequences were compared with the those in the National Center for Biotechnology Information (NCBI) sequence database by using BLAST (http://www.ncbi.nlm.nih.gov/blast) (3).
Based on the sequences of DGGE bands corresponding to “Ca. Phytoplasma solani,” Wolbachia, “Ca. Cardinium hertigii,” “Ca. Sulcia muelleri,” “Ca. Purcelliella pentastiridorum,” and the HO1-V symbiont, additional sequences of the 16S rRNA genes of these microbes outside the 5′ and 3′ ends of the DGGE fragments were obtained by performing specific PCRs with primer pairs previously reported or designed for this work, as shown in Table 1. The six forward primers were used in combination with eubacterial reverse primer 1495R, while the six reverse primers were coupled with forward universal primer 27F (35). Therefore, the flanking regions at the 5′ and 3′ ends of the DGGE fragments of these bacteria were obtained.
TABLE 1.
Oligonucleotides adopted in this work to obtain almost the entire 16S rRNA gene sequences of the symbionts for prevalence screenings and for FISH analysesa
| Target organism | Primer pair (sequence [5′-3′] or reference) |
Probe (fluorochrome-sequence [5′-3′] or reference) | |
|---|---|---|---|
| Forward | Reverse | ||
| “Ca. Phytoplasma solani” | PhF (CTAAACAGTTTTCATAGCATCACAA) | PhR (TTGTGATGCTATGAAAACTGTTTAG) | ph1107 (TR-GATGGCAATTAACAACAAGGGT) |
| Wolbachia | WF (TTAAATATGGGAAGTTTACTTTCTGTATTAC) | WR (GTAATACAGAAAGTAAACTTCCCATATTTAA) | W1 (27) |
| W2 (27) | |||
| “Ca. Cardinium hertigii” | EndoF1 (35) | EndoR3 (35) | card172 (Cy3-ATCTTTCTAGCATGCGCTAA) |
| card1069 (Cy3-GCACCTTGTATTCCGTCC) | |||
| “Ca. Sulcia muelleri” | SF (ATMTAGACAKAAAATATTCAGTG) | SR (CACTGAATATTTTMTGTCTAKAT) | S1150 (Cy3-ACATTCCAGTTACTCCTATCT) |
| SF1 (AGATAGGAGTAACTGGAATGT) | |||
| “Ca. Purcelliella pentastiridorum” | PF (GTATTTTATTAATAATAAAATATG) | PR (CATATTTTATTATTAATAAAATAC) | P820 (HEX/6-JOE/ROX-AGAAAACACGGCAAAATCACC) |
| PR1 (AGAAAACACGGCAAAATCACC) | |||
| HO1-V symbiont | VF (GATGAAGGTTGATAAGATC) | VR (GATCTTATCAACCTTCATC) | V370 (HEX/6-JOE/ROX-GATCTTATCAACCTTCATC) |
| VF1 (TTTTAAATTCTTTATAAAGTT) | |||
| Mollicutes | MCP52 (55) | ||
| Bacteroidetes | CFB319 (42) | ||
| Eubacteria | 27F (9) | 1495R (9) | EUB338 (4) |
| “Ca. Baumannia cicadellinicola” | Pro319 (42) | ||
The sequences of oligonucleotides designed and reported in previous studies are not shown.
After amplification and sequencing, all of the obtained 16S rRNA sequences were subjected to BLAST analysis and aligned with the corresponding 16S rRNA genes of close relatives and with other unrelated eubacterial sequences. Alignments were performed by using the software available at the Ribosomal Database Project (RDP) website (14). Phylogenetic analyses were performed by using Jukes and Cantor distance estimations with the TREECON 1.3b package (56). A 50% majority-rule bootstrap consensus tree (1,000 replicates) was generated. Gaps were treated as a fifth base.
Detection of the prevalence of H. obsoletus-associated microorganism populations by means of PCR.
By means of specific PCR screenings, we examined the abundances of six bacteria present in H. obsoletus (“Ca. Phytoplasma solani,” Wolbachia, “Ca. Cardinium hertigii,” “Ca. Sulcia muelleri,” “Ca. Purcelliella pentastiridorum,” and the HO1-V symbiont). Such microorganisms were considered of particular interest because either they were well known for their functions in other insect models or they appeared to be extremely abundant in the diversity screenings. The analyses were performed on 80 insect specimens, including those examined by DGGE and LH-PCR. Seventy individuals (28 females and 32 females) were used for whole-insect DNA extraction. Ten individuals (females and males, 5 each) were dissected, and DNA was extracted from the organs (fat bodies, gut, ovaries, testes, and salivary glands). To evaluate the prevalence of “Ca. Phytoplasma solani,” specific PCRs were performed by using primer pair M1-P8 (34) or the BN forward/reverse primer pair (5). The wsp gene of Wolbachia was amplified by using primers wsp81F and wsp691R as previously described (10).
The alignments of the “Ca. Sulcia,” “Ca. Purcelliella,” and HO1-V symbiont 16S rRNA sequences with related bacterial sequences were used to design primer pairs specifically targeting the symbionts (Table 1). Selected primers for “Ca. Sulcia” were SF1 (positions 656 to 677 of Escherichia coli strain K-12) and SR (positions 839 to 862 of E. coli strain K-12), and they amplified a 185-bp fragment. They did not match with any bacterial or invertebrate sequences in GenBank at the time of checking; moreover, they matched with the cixiid-associated “Ca. Sulcia” sequences (GenBank accession numbers FN428791 and FN428795). Selected primers for “Ca. Purcelliella” were PF (positions 472 to 496 of E. coli strain K-12) and PR1 (positions 855 to 876 of E. coli strain K-12), and they amplified a 404-bp fragment. They matched with the described H. obsoletus-associated “Ca. Purcelliella pentastiridorum” sequence (accession number FN428799) but not with other cixiid-associated “Ca. Purcelliella” sequences (accession number FN428803); furthermore, they did not correspond to any bacterial or invertebrate sequences in GenBank at the time of checking. Selected primers for the HO1-V symbiont were VF1 (positions 161 to 182 of E. coli strain K-12) and VR (positions 427 to 446 of E. coli strain K-12), and they amplified a 285-bp fragment. They did not coincide with any bacterial or invertebrate sequences in GenBank at the time of checking. Each PCR assay included a cloned amplicon sample specific for each microorganism as a positive control and a water sample as a negative control. Amplifications were performed under the following conditions: an initial denaturation step of 4 min at 94°C, followed by 35 cycles of 1 min at 94°C, 1 min at 54°C (when using primer pair SF1-SR) or 55°C (when using primer pair PF-PR1 or VF1-VR), and 1 min at 72°C and a final extension step of 7 min at 72°C. As a control, a sample of the PCR products obtained from each specific PCR was sequenced.
A PCR screening with primer pair VF-VR was also carried out with DNA samples of whole-body insects of the species Hyalesthes luteipes, Reptalus cuspidatus, and Reptalus melanochetus in order to assess the distribution of these symbionts among other cixiids.
Localization of symbionts in H. obsoletus by means of TEM and FISH.
Twenty-three individuals (5 females, 5 males, and 13 nymphs) were dissected and prepared to be studied by transmission electron microscopy (TEM), as previously reported (8). Thin sections (80 nm) were examined under a Zeiss EM900 transmission electron microscope.
Fluorescent in situ hybridization (FISH) was performed on 25 H. obsoletus individuals (10 females, 10 males, and 5 nymphs) to observe the distribution of phytoplasmas, “Ca. Cardinium,” Wolbachia, “Ca. Sulcia,” “Ca. Purcelliella,” and the HO1-V symbiont within the insect body. Specific fluorescent probes targeting the 16S rRNA gene were used (Table 1). The hybridization of Wolbachia was performed by using the probes W1 and W2 (27), while for the specific hybridization of the other bacteria, we designed the following probes: ph1107 for phytoplasmas, card172 and card1069 for “Ca. Cardinium,” S1150 for “Ca. Sulcia,” P820 for “Ca. Purcelliella,” and V370 for the HO1-V symbiont. We also used the probes MCP52 (55), matching with portions of 16S rRNA genes of different Mollicutes; CFB319 (42), targeting the 16S rRNA genes of the Bacteroidetes; and EUB338 (4), matching with 16S rRNA genes of all members of the Eubacteria. Probes card172, card1069, S1150, and W1 were labeled at their 5′ ends with the fluorochrome Cy3 (indocarbocyanine) (absorption and emission at 550 nm and 570 nm, respectively) or Cy5 (indodicarbocyanine) (absorption and emission at 650 nm and 670 nm, respectively); probe ph1107 was labeled with Texas Red (TR) (absorption and emission at 595 nm and 620 nm, respectively); probes P820 and V370 were labeled with HEX (4,7,2′,4′,5′,7′-hexachloro-6-carboxyfluorescein) (absorption and emission at 535 nm and 556 nm, respectively), 6-JOE (6-carboxy-4′,5′-dichloro-2′,7′-dimethoxy fluorescein) (absorption and emission at 520 nm and 548 nm, respectively), or ROX (carboxy-X-rhodamine) (absorption and emission at 580 nm and 600 nm, respectively); and probes W2, MCP52, CFB319, and EUB338 were labeled with fluorescein isothiocyanate (FITC) (absorption and emission at 494 nm and 520 nm, respectively). Insects were dissected to collect salivary glands, guts, and gonads. Paraformaldehyde-fixed insect dissection samples were hybridized according to a method described previously by Crotti et al. (15).
Nucleotide sequence accession numbers.
The nucleotide sequences of “Ca. Sulcia muelleri,” “Ca. Purcelliella pentastiridorum,” and the HO1-V symbiont's 16S rRNA genes were deposited in the GenBank/EMBL/DDBJ nucleotide sequence database under the following accession numbers: FM992371 for “Ca. Sulcia,” FR686933 for “Ca. Purcelliella,” FR686932 for the HO1-V symbiont of H. obsoletus, and FR733652 for the betaproteobacterial symbiont of R. melanochetus.
RESULTS AND DISCUSSION
Characterization of the bacterial community associated with H. obsoletus.
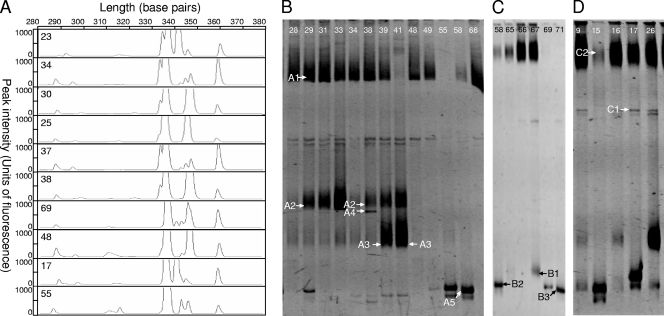
The bacterial community associated to H. obsoletus from BN-contaminated areas was studied by means of LH-PCR. The screened insects showed some dominant peaks (e.g., peaks at 338, 343, and 361 bp) that were conserved in almost all tested individuals, suggesting that certain bacterial species have a stable association with H. obsoletus (Fig. 1 A). Other peaks (e.g., peaks at 333, 342, and 349 bp) were found only for a few insects, indicating an occasional association. To identify the taxonomic affiliation, we amplified a portion of about 600 bp of the 16S rRNA gene from the total DNA of the insects and separated the amplified fragments by means of DGGE (Fig. 1B). Although the community profiles of different individuals showed some variability, certain bands were rather conserved in the individuals. DGGE experiments performed under different denaturing gradient conditions permitted us to recover some other bands associated with a few insects (Fig. 1C).
FIG. 1.
Bacterial diversity associated with H. obsoletus. (A) Example of LH-PCR profiles of whole insects collected in 2005 to 2007 from uncultivated areas in Piedmont, Italy. Numbers refer to different individuals tested. (B and C) DGGE profiles, in 7% polyacrylamide gels with 20 to 40% (B) and 30 to 50% (C) denaturation gradients, of partial 16S rRNA bacterial genes amplified from DNA extracted from whole insects collected in 2005 to 2007 from wastelands in Piedmont, Italy. Numbers above the lanes refer to the numbers of tested individuals. The identities of sequences of bands marked with arrows are given in Table 1 according to the band identification (bands A1 to A5 and B1 to B3).
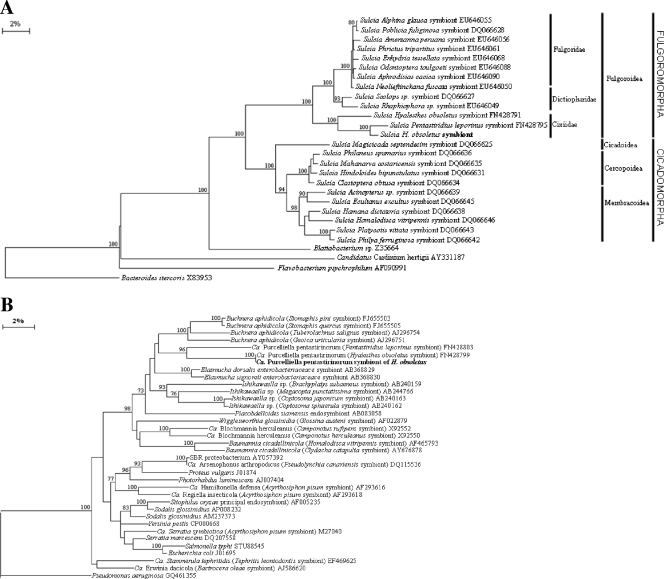
The sequences obtained from the bands isolated from DGGE gels are presented in Table 2, along with the closest relatives found in the RDP database. Band A1 was found for most of the tested individuals (83%) and showed 99% sequence identity with “Ca. Sulcia muelleri” of the Bacteroidetes. This bacterium was first reported as the “a-symbiont” of the Auchenorrhyncha by Müller (44), and it was later described by Moran et al. (42) as a novel clade of strap-shaped members of the Bacteroidetes that harbor a small genome and are associated with both the Cicadomorpha and Fulgoromorpha. “Ca. Sulcia muelleri” was recently reported in association with some cixiids (12). Almost the entire sequence of the 16S rRNA gene of this bacterium grouped into a branch of the neighbor-joining phylogenetic tree including “Ca. Sulcia” symbionts of Fulgoromorpha insect hosts belonging different families (Fig. 2 A). The phylogenetic analysis confirmed the strong congruency between the phylogeny of the symbiont and that of its host reported for all “Ca. Sulcia muelleri” isolates previously described (41, 54).
TABLE 2.
Identification of microorganisms associated to H. obsoletus according to DGGE profiles in Fig. 1
| Band | Most related species | GenBank accession no. | % nt identity (no. of identical bp/total no. of bp)b | Putative classification | No. of positive individuals/total no. of individualsa |
|---|---|---|---|---|---|
| A1 | “Candidatus Sulcia muelleri” | DQ066627 | 99 (525/528) | Bacteroidetes, Flavobacteriales | 16/18 |
| A2 | Wolbachia pipientis | DQ235291 | 100 (488/488) | Αlphaproteobacteria; Rickettsiales | 9/18 |
| A3 | Endosymbiont of Oppiella nova | AY279414 | 99 (515/520) | Bacteroidetes | 11/18 |
| A4 | “Candidatus Phytoplasma solani” | DQ222972 | 99 (505/506) | Mollicutes, Achioleplasmatales | 3/18 |
| A5 | Rickettsia limoniae | AF322443 | 99 (503/508) | Αlphaproteobacteria; Rickettsiales | 4/18 |
| B1 | Endosymbiont of Oppiella nova | AY279414 | 88 (362/410) | Bacteroidetes | 1/6 |
| B2 | Rickettsia limoniae | AF322443 | 99 (494/498) | Αlphaproteobacteria; Rickettsiales | 1/6 |
| B3 | Chryseobacterium joostei | AY466722 | 100 (529/529) | Bacteroidetes, Flavobacteriales | 1/6 |
| C1 | “Candidatus Purcelliella pentastirinorum” | FN428799 | 100 (543/543) | Gamma-3-proteobacteria | 15/18 |
| C2 | Hemaphysalis longicornis-associated microorganism | AB001520 | 79 (443/556) | Betaproteobacteria | 14/18 |
Number of individuals positive for the presence of the specific band in the DGGE analysis compared to the total number of individuals analyzed.
nt, nucleotide.
FIG. 2.
Phylogenetic affiliation of almost the entire 16S rRNA genes of bacteria associated with H. obsoletus. (A) “Ca. Sulcia muelleri.” Clades of insect hosts of “Ca. Sulcia” symbionts are shown. (B) “Ca. Purcelliella pentastirinorum.” Numbers at each node represent percentages of bootstrap replications calculated from 1,000 replicate trees. The scale bar represents the sequence divergence. SBR, syndrome “basses richesses.”
Band A2 was found for half of the tested individuals and showed 100% identity with Wolbachia pipientis, an intracellular reproductive manipulator previously described for different insect models, including leafhoppers (16, 20, 52, 59). Almost the entire 16S rRNA gene of this symbiont was obtained by combining the newly designed primers WF and WR, specific for Wolbachia, and bacterial universal primers in PCR experiments (data not shown). The sequence was phylogenetically affiliated within Wolbachia supergroup B.
Bands A3 and B1 were 99 and 88% similar, respectively, to the 16S rRNA gene of an endosymbiont of the mite Oppiella nova, affiliated with the genus “Ca. Cardinium” within the Bacteroidetes. “Ca. Cardinium hertigii” includes endosymbionts infecting numerous arthropods and able to induce multiple reproductive effects on their hosts (58, 59, 63, 64). The bands of “Ca. Cardinium” were detected in 50% (12 of 24) of the individuals examined by means of DGGE. “Ca. Cardinium hertigii” was detected in all the individuals that showed the presence of Wolbachia. To acquire the almost complete 16S rRNA gene sequence of this endosymbiont, “Ca. Cardinium”-specific primers (35) were combined with universal primers. The obtained sequence was affiliated with “Ca. Cardinium” endosymbionts of several mite and insect species (data not shown).
Band C1, observed for 75% of tested individuals, showed 100% sequence similarity with “Ca. Purcelliella pentastirinorum,” a gamma-3-proteobacterium recently described as one of the bacteriome-associated symbionts of several cixiid species (12). Evolutionary studies on this bacterium showed that it is restricted to the tribe Pentastirini, and it contributed to the diversification of this tribe within the Fulgoromorpha (12). Almost the entire 16S rRNA sequence of this bacterium, obtained by combining specific and universal primers, was incorporated into the branch of the Gammaproteobacteria phylogenetic tree that includes “Ca. Purcelliella” symbionts of the genus Hyalesthes and other cixiids (Fig. 2B), confirming the high level of congruency between symbiont and host.
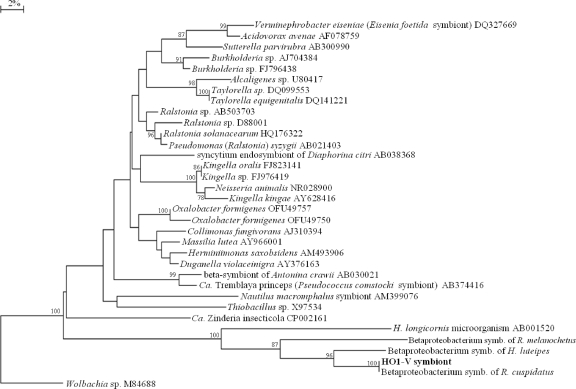
Furthermore, band C2, which was repeatedly found in the specimen tested by means of DGGE, did not show any significant affiliations based on sequence similarity and had an uncultured betaproteobacterium associated with the bush tick Haemaphysalis longicornis (46) as the closest relative, with 79% sequence similarity, while the nearest determined organism was Kingella kingae (GenBank accession number AY551998) of the Neisseriales. The genus Kingella includes human pathogens responsible for several pediatric infective diseases (62). Almost the entire sequence of the 16S rRNA gene, named HO1-V, grouped into a separate branch, together with its tick-associated closest relative, within the phylogenetic tree that includes different orders of symbiotic and free-living members of the Betaproteobacteria, neighboring the Neisseriales and the Burkholderiales (Fig. 3). The extremely low level of sequence identity even with the closest relatives suggests that the HO1-V symbiont is quite different from the nearest previously reported organisms. It is interesting that this bacterium is one of the few examples of major symbionts belonging to the beta subclass of the Proteobacteria, while primary symbionts fit more commonly into the Gammaproteobacteria or the Bacteroidetes group (43).
FIG. 3.
Phylogenetic positions of the nearly full-length 16S rRNA gene of the HO1-V symbiont. Orders within the Betaproteobacteria are indicated. Numbers at each node represent percentages of bootstrap replications calculated from 1,000 replicate trees. The scale bar represents the sequence divergence.
Other bands were found only for certain individuals with a low prevalence. For instance, band A4 showed 99% sequence identity with “Ca. Phytoplasma solani.” By combining primers specifically designed for sequence A4 with bacterial universal primers, the almost full 16S rRNA gene sequence of the pathogen was obtained and phylogenetically affiliated with that of the Bois noir phytoplasma (data not shown). The detection of the Bois noir phytoplasma by DGGE suggests that the titer of this pathogen is much higher than that of other phytoplasmas such as the Flavescence dorée phytoplasma, which has never been detected by means of PCR-DGGE in the total DNA extracted from insects (35).
For a few individuals, a sequence with 99% identity with Rickettsia limoniae, corresponding to bands A5 and B2 in the PCR DGGE gels in Fig. 1, was detected. Bacteria of the genus Rickettsia are human and animal pathogens vectored by blood-feeding insects. Nevertheless, members of this genus were found in association with the pea aphid Acyrthosiphon pisum (13) and with the whitefly Bemisia tabaci (25), and bacteria of the species R. limoniae have been identified in the cranefly Limonia chorea (GenBank accession number AF322443).
Band B3 in Fig. 1C, corresponding to a sequence strictly related to Chryseobacterium joostei of the Bacteroidetes, was detected for only one individual.
All the other bands detected by DGGE (Fig. 1B) did not give readable sequences.
Prevalence and localization of the main symbionts of H. obsoletus.
The prevalences of “Ca. Phytoplasma solani,” Wolbachia, “Ca. Cardinium hertigii,” “Ca. Sulcia muelleri,” “Ca. Purcelliella pentastiridorum,” and the HO1-V symbiont in H. obsoletus were studied by PCR assays targeting the 16S rRNA genes with symbiont-specific primers (Table 3).
TABLE 3.
Prevalence of symbionts in different organs or tissues of H. obsoletus determined with specific PCR assaysa
| Symbiont | No. of positive individuals/total no. of individuals testedb |
||||
|---|---|---|---|---|---|
| Whole insect | Gut | Ovaries | Testes | Salivary glands | |
| “Ca. Sulcia” | 60/70 (F, 28/31; M, 32/39) | 8/10 (F, 4/5; M, 3/5) | 4/5 | 2/5 | 2/10 (F, 1/5; M, 1/5) |
| Wolbachia | 43/70 (F, 23/31; M, 20/39) | 5/10 (F, 5/5; M, 0/5) | 3/5 | 0/5 | 2/10 (F, 2/5; M, 0/5) |
| “Ca. Cardinium” | 28/70 (F, 11/31; M, 17/39) | 3/10 (F, 2/5; M, 1/5) | 1/5 | 2/5 | 3/10 (F, 2/5; M, 1/5) |
| “Ca. Purcelliella” | 50/70 (F, 24/31; M, 26/39) | 3/10 (F, 2/5; M, 1/5) | 3/5 | 1/5 | 2/10 (F, 2/5, M, 0/5) |
| HO1-V | 61/70 (F, 28/31; M, 33/39) | 4/10 (F, 3/5; M, 1/5) | 4/5 | 1/5 | 3/10 (F, 1/5; M, 2/5) |
| “Ca. Phytoplasma solani” | 12/70 (F, 7/31; M, 5/39) | 2/10 (F, 1/10; M, 1/10) | 0/5 | 0/5 | 1/10 (F, 1/5; M, 0/5) |
A total of 80 individuals were used in the assays, including 36 females and 44 males. Seventy individuals were used as whole insects, while 10 were used for dissecting the different organs.
Number of individuals positive in specific PCR assays over the total number of tested individuals. M, males; F, females.
In whole insects, “Ca. Phytoplasma solani” showed an average infection rate of 17.5%, with 22.6% of females and 12.8% of males found to be positive. These values are in agreement with data from previous reports (2, 11, 33).
While the “Ca. Cardinium” symbiont was found in 38.8% of the checked insect population, with a slightly lower incidence in the whole-body females (35.5%) than in males (43.6%), the minimal infection rate of Wolbachia was on average 60%. The symbiont was found more frequently in females (74.2% positive insects) than in males (51.3% positive individuals). This alphaproteobacterium has been reported for all major orders of insects, with a variable infection rate at a specific level, from about 20% to more than 50% (30, 59, 60).
The minimal infection rates of “Ca. Sulcia” and of the HO1-V symbiont were similar, 85% and 83.8% of the samples, respectively. Also, the distributions of the two symbionts in males and females were comparable, with 90.3% “Ca. Sulcia”-infected and HO1-V-infected females and 82% “Ca. Sulcia”-infected and 84.6% HO1-V-infected males. Moreover, “Ca. Purcelliella” was present in 67.5% of tested specimens, with minimal infection rates of 74.4% for females and 66.7% for males.
The presence of the HO1-V symbiont was also detected in the cixiid species H. luteipes, R. cuspidatus, and R. melanochetus, with infection rates of 70% (7/10), 30% (3/10), and 40% (4/10), respectively.
A first insight into the localization of the symbionts was provided by specific PCR screenings of dissected body parts, as summarized in Table 3. All of the bacteria were found in the intestines and (with lower infection rates) in salivary glands; we detected almost all of the microbes in the gonads, with a few exceptions: we were not able to find the phytoplasmas in both male and female gonads, and Wolbachia was observed only in the ovaries and not in testes.
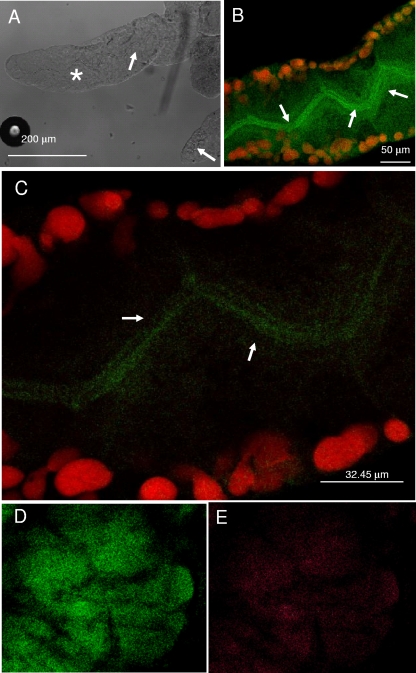
A more detailed localization of the bacteria associated with H. obsoletus was provided by FISH experiments. The localization of “Ca. Phytoplasma solani” was first explored by using a Mollicutes-specific probe. By dissecting salivary glands, it was possible to identify the different lobes and visualize the gland ducts that release saliva during feeding (Fig. 4 A and B). Positive hybridization signals were observed for one of five individuals tested (Fig. 4B). Signals were particularly concentrated in the duct of the salivary gland, suggesting that the phytoplasmas actively multiply in the salivary duct before injection into the plant (Fig. 4B and C). To confirm such results, a Stolbur-specific probe was designed and used on dissected salivary glands, where a neat amplification signal was observed for the whole gland lobe (Fig. 4D and E).
FIG. 4.
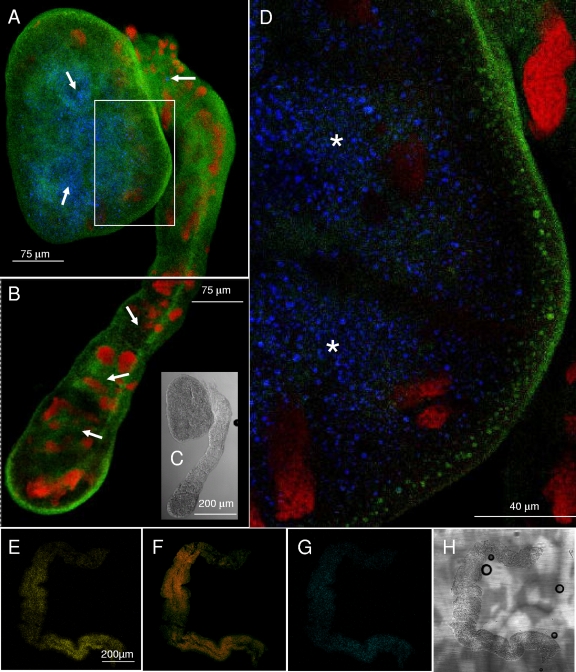
Localization of phytoplasma cells in the salivary glands of H. obsoletus. (A) Interferential contrast micrograph showing different lobes of the salivary gland. The salivary ducts, indicated by arrows, are visible in certain lobes. (B) Confocal laser scanning microscopy (CLSM) image of FISH of the salivary gland lobe, identified in A with an asterisk, hybridized with the Mollicutes-specific probe MCP-52. The image is reconstructed by overlapping 12 different focal planes. Epithelial cell nuclei stained with propidium iodide are marked by red spots. Mollicutes cells (green), presumably of “Ca. Phytoplasma solani,” are densely located within the salivary duct. Arrows indicate the salivary duct. (C) Magnification of a section of the lobe in B showing a single focal plane with a dense colonization by Mollicutes cells that are confined within the salivary duct (arrows). (D and E) CLSM image of FISH of salivary gland lobes with the eubacterial probe EUB338 (D) and with the Bois noir-specific probe ph1107 (E).
Although “Ca. Sulcia muelleri” has been observed in the typical position in the bacteriome in different leafhoppers and planthoppers, we were not able to observe and isolate the specific organ for any of the dissected specimens. On the other hand, the localization of “Ca. Sulcia” in the body of H. obsoletus was studied in the gut, the salivary glands, and the female and male gonads. All of these organs except salivary glands showed a massive presence of the symbiont. By using a specific probe, “Ca. Sulcia” appeared to be associated with the entire gut (Fig. 5 A to D), with a denser cell concentration in certain portions of the interior of the gut (Fig. 5A). When observed at higher magnifications (Fig. 5D), the symbiont appeared in clusters of strap-shaped cells previously described as being typical of “Ca. Sulcia muelleri” (42). In addition, close to the intestinal wall, cells of members of the Bacteroidetes other than “Ca. Sulcia” were found. It can be presumed that these bacteria are referable to “Ca. Cardinium hertigii,” the only other member of the Bacteroidetes massively represented in H. obsoletus.
FIG. 5.
Localization of symbionts in the gut of H. obsoletus. (A to D) FISH of the insect gut after hybridization with the Cy3-labeled “Ca. Sulcia”-specific probe S1150 (blue spots indicated by arrows) and the FITC-labeled CFB319 probe specific for the Bacteroidetes (green). Images in A an B allow us to analyze the differential distribution of bacteria in the gut. The images in A and B reconstruct the entire insect gut, shown in the interferential contrast micrograph (C). Epithelial cell nuclei are stained with propidium iodide (red). (D) Magnification of a portion of the gut (indicated by the white rectangle in A) shows the presence of several clusters of distinct “Ca. Sulcia” cells (indicated by asterisks). (E to G) FISH of the midgut of H. obsoletus with the probes specific for “Ca. Cardinium” (E), for the HO1-V symbiont (F), and for “Ca. Purcelliella” (G). (H) Intestine pictured by interferential contrast.
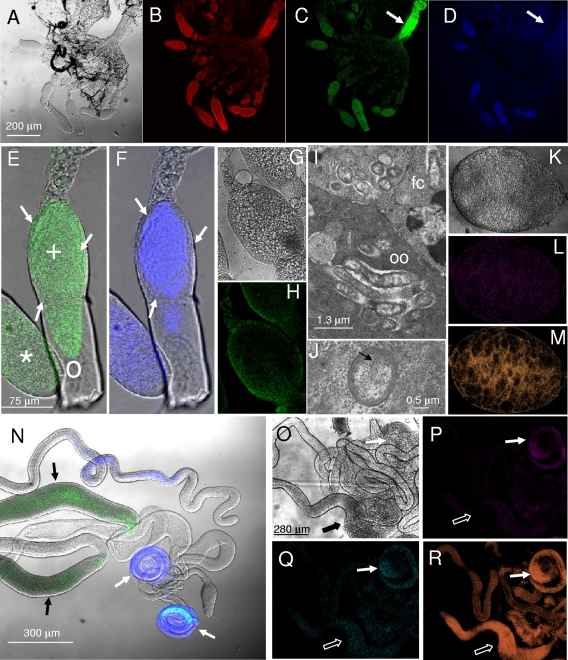
Examination of the gonads of H. obsoletus by means of FISH with the probe specific for “Ca. Sulcia” showed that the bacterium was associated with both the ovary (Fig. 6 A to F and L) and the testicles (Fig. 6N to R). By comparing FISH with the universal probe for bacteria and a specific probe for “Ca. Sulcia” in an entire ovary, it was possible to find signals for the symbiont in all the ovarioles (Fig. 6D) but not in the ovary duct, where other bacteria were resident (Fig. 6C). “Ca. Sulcia” appeared associated with the oocytes and the nurse cells (Fig. 6F) but not with the follicular cells of the ovariole, where other bacteria were detected by using a universal probe for bacteria (Fig. 6E). A more accurate analysis of the ovary by TEM showed at least three different cell morphologies associated with the oocyte and the follicular cells (Fig. 6I and J). While bacterial cells in the oocyte (Fig. 6I) showed the distinctive strap shape of “Ca. Sulcia muelleri” (42), some bacteria in the cytoplasm of the follicular cells are probably Wolbachia bacteria, confirming the hybridization signal observed for the follicles when using the Wolbachia-specific probe (Fig. 6G and H). In addition, other bacteria were also observed in the follicle cell cytoplasm with the brush-like structure of “Ca. Cardinium hertigii” (Fig. 6I and J) typical of these maternally transmitted endosymbionts (35, 64, 65). Within the male gonads, “Ca. Sulcia” was specifically associated with testicles but not with other organs (Fig. 6N to R). FISH with the universal probe EUB338 showed that bacteria other than “Ca. Sulcia” specifically colonize the accessory glands of the male gonads (Fig. 6N). These bacteria could be the HO1-V symbiont, as the hybridization of the male gonads of H. obsoletus with the specific probe for this microorganism gave a strongly positive result both for the testicles and for the accessory glands, while only a weak signal was obtained by FISH for organs other than the testicles with the “Ca. Sulcia”-specific and “Ca. Purcelliella”-specific probes (Fig. 6O to R).
FIG. 6.
Symbiont localization in the gonads of H. obsoletus. (A to D) Images of an insect ovary pictured by interferential contrast microscopy (A), CLSM after staining with propidium iodide (B), and FISH with the FITC-labeled EUB338 probe, specific for members of the Bacteria (C), and the Cy3-labeled probe S1150, specific for “Ca. Sulcia” (D). Arrows in C and D indicate the ovary duct densely colonized by bacteria other than “Ca. Sulcia.” (E and F) Magnification of an oocyte (labeled with a plus) and a nurse cell (labeled with an asterisk) present in A to D. Superpositions of the interferential contrast microscopy images and the FISH images are reported. Arrows indicate zones, corresponding to the follicular cells, with hybridization signals of the EUB338 probe but not of the S1150 probe. (G and H) Interferential contrast micrograph of a ovary portion (G) and CLSM image of FISH with the Wolbachia-specific probes W1 and W2 (H). The specific localization of these bacteria in the follicles is shown. (I) Transmission electron microscopy image of a follicle showing the interface between the oocyte (oo) and the follicular cell (fc). Different symbiont cell morphotypes are present in the oocyte and the follicular cell. Those in the follicular epithelium are probably Wolbachia, while bacterial cells in the oocyte showed the strap-like cell shape typical of “Ca. Sulcia.” (J) Detail of the follicular cell cytoplasm showing the typical brush-like structure (arrow) of “Ca. Cardinium hertigii.” (K to M) Image of an ovaric egg shown as an interferential contrast picture (K) and CLSM image of the hybridization with the “Ca. Sulcia”-specific probe S1150 (L) and the HO1-V-specific probe V370 (M). (N) Superposition of the FISH images over the interferential contrast microscopy image of a male reproductive system, hybridized with the FITC-labeled EUB338 probe, specific for members of the Bacteria (green), and the Cy3-labeled probe HOS1150, specific for the HO1-V symbiont (blue). (O to R) Interferential contrast (O) and CLSM images of a male reproductive system hybridized with the “Ca. Sulcia”-specific probe S1150 (P), with the “Ca. Purcelliella”-specific probe P820 (Q), or with the HO1-V-specific probe V370 (R). The different organs of the male reproductive system are indicated by arrows. In N, testes (white arrows) show the signal of the S1150 probe specific for “Ca. Sulcia,” while accessory glands (black arrows), hybridized with the bacterial probe EUB338, indicate the presence of bacteria other than the HO1-V symbiont. In P to R, while the testes (white arrows) hybridized with all of the probes, accessory glands showed a very weak signal after hybridization with both the “Ca. Sulcia”- and “Ca. Purcelliella”-specific probes (P and Q). On the other hand, FISH with the HO1-V probe showed a strong signal (R).
“Ca. Sulcia muelleri” is typically associated with another bacterial symbiont that varies among insect groups: in sharpshooters, it is coresident with the gammaproteobacterium “Ca. Baumannia cicadellinicola” (41); in cicadas, it is associated with the alphaproteobacterium “Ca. Hodkinia cicadicola” (38); and in spittlebugs, its cosymbiont is the betaproteobacterium “Ca. Zinderia insetticola” (37). In all of these systems, the symbionts both provide essential nutrients to the host and are nutritionally interdependent on each other (36, 37, 39). We cannot exclude the possibility that the HO1-V symbiont is the complementary symbiont of “Ca. Sulcia.” Indeed, although we do not have knowledge of the possible colocalization of “Ca. Sulcia” and the HO1-V symbiont within the same bacteriome, we observed members of both the Bacteriodetes and the Betaproteobacteria within the ovaries or eggs, implying that they are maternally transmitted together. This suggests that the two symbionts could have undergone coevolution, with the possible development of complementarity.
The distributions of “Ca. Purcelliella,” previously known to be in the bacteriome, as well as “Ca. Sulcia” in H. obsoletus were studied for salivary glands, guts, and male and female reproductive systems. A positive hybridization signal was present in the salivary glands (data not shown) and in the gut (Fig. 5G and H). Nevertheless, we were not able to observe any detectable fluorescence either in ovaries or in ovaric eggs, while a weak hybridization signal was present in the male gonads (Fig. 6Q).
Hybridization with the probe specific for the HO1-V symbiont was first performed on the insect gut, where a heavy signal was detected, indicating a considerable amount of bacterial cells residing in this organ (Fig. 5F). Also, ovaric tissues and oocytes (Fig. 6K and L), together with male gonads (Fig. 6R), were observed to host the HO1-V symbiont. On the contrary, no hybridization signal was visible in H. obsoletus salivary glands.
“Ca. Cardinium hertigii” is known to be associated with several reproductive disorders, including parthenogenesis in parasitoid wasps of the genus Encarsia (64), feminization in the mite Brevipalpus phoenicis (57), and cytoplasmic incompatibility in Encarsia pergandiella (28). It localizes in different organs and tissues of insect hosts (31, 49, 65), including follicle cells of ovaries as well as oocytes and nurse cells (65), as observed for Encarsia spp. To evaluate the localization of this member of the Bacteroidetes in the body of H. obsoletus, hybridization with the specific probes was first carried out on the salivary glands and gut. No successful hybridization was obtained with the first hybridization, while a massive signal was detected in the digestive tube (Fig. 5E). This symbiont was also detected in the ovaries, with a specific localization in the follicle area, confirming what was observed by means of TEM, and in the male gonads (data not shown). These data suggest a peculiar localization pattern for “Ca. Cardinium” and “Ca. Sulcia” in H. obsoletus, with “Ca. Sulcia” in the oocytes and “Ca. Cardinium” in the follicle cells; nevertheless, we were not able to define a precise localization within the gonad tissues of the HO1-V symbiont.
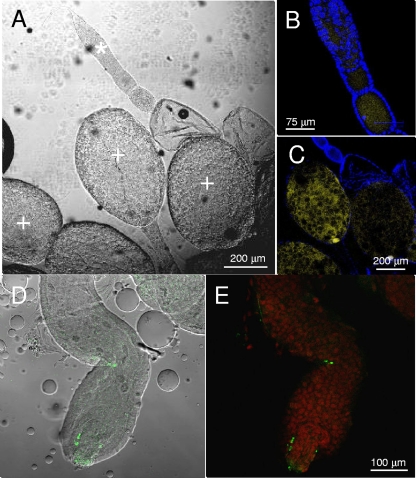
FISH using a Wolbachia-specific probe showed the bacterium associated with the female oocytes and the mature eggs (Fig. 6G and H and 7 A to C). Hybridization signals were also found in the gut of nymphs (Fig. 7D and E). The localization of Wolbachia in different tissues of female gonads of H. obsoletus also suggests for the planthopper the vertical transmission pattern reported for insect hosts of this bacterium.
FIG. 7.
Visualization by CSLM of the gut of nymphs and female gonads of H. obsoletus. (A) Interferential contrast microscopy image of a female gonad. (B and C) DAPI (4′,6-diamidino-2-phenylindole) staining and FISH with the Cy5-labeled W1 probe specific for Wolbachia (yellow). Insect cell nuclei stained with DAPI are blue. Magnifications of an immature ovariole (asterisk) (B) and of a mature egg (plus) (C) are shown. (D) Interferential contrast microscopy image of a nymphal gut overlapped with an FITC-labeled W2 probe specific for Wolbachia (green). (E) The same image after propidium iodide staining and FISH using the FITC-labeled probe W2 specific for Wolbachia (green).
As reported previously for several mite and hymenopteran species, we observed a double infection of both the sexual manipulators “Ca. Cardinium hertigii” and Wolbachia in H. obsoletus (21, 24, 58, 64). The presence of both of these potential sexual manipulators in gonads opens up new perspectives for the investigation of possible reproductive abnormalities such as sex ratio alterations and ways of action and interference between sexual symbionts.
Overall considerations.
Our investigations of the bacterial diversity associated with H. obsoletus indicated that several bacterial species inhabit the insect body, revealing a complex symbiotic organization. Some of the bacterial symbionts were related to bacteria previously described to be reproductive manipulators, such as Wolbachia and “Ca. Cardinium hertigii”; others, like “Ca. Sulcia” and “Ca. Purcelliella,” were proven to be primary symbionts of different members of the Auchenorrhyncha, often involved in the host's nutrients supply (12, 36, 39, 61). Indeed, such bacteria were found in almost all of the individuals, suggesting that they could play important—if not essential—roles in the host. The high infection rate of the HO1-V symbiont also suggests that this bacterium has a strict association with its host. Although we do not have knowledge of the possible role of this microorganism in insect biology, we can suppose a major function.
All of these bacteria were widely distributed within the insect body, massively colonizing different organs, especially the gut and male and female gonads. Interestingly, in the gonads the symbionts were detected in both oocytes and testicles; this suggests a venereal transmission from male to female, as reported previously for beneficial symbionts in aphids (40) and for the acetic acid bacterium Asaia sp. in Anopheles stephensi (17, 22).
Potential interactions between bacteria colocalized in the host tissues, particularly in the gonads, should be deeply investigated in the future. The elucidation of the role of these microorganisms in the host could be useful for a symbiotic approach to controlling phytoplasmoses either with the expression of antagonistic factors by microorganisms cross-living with the phytoplasmas or by means of reproductive manipulators helping to drive the establishment of antagonistic symbionts or to imbalance natural populations of the planthopper, with the final aim of limiting BN diffusion.
The low level of 16S rRNA gene identity of the HO1-V symbiont with the closest relative (80%), which moreover is an uncultured organism, supports the proposal of a novel clade of symbionts of cixiids. Indeed, the HO1-V symbiont is strongly associated with H. obsoletus; moreover, at least 3 other cixiid species (one of the genus Hyalesthes and two of the genus Reptalus) were shown to host this bacterium, for which we propose the new name “Candidatus Vidania fulgoroideae.” The generic name honors Carlo Vidano, an Italian auchenorrhynchologist of the University of Turin who first described and studied the biology of phytoplasma vectors in Italy. The species name refers to the superfamily Fulgoroidea, which includes the family of H. obsoletus harboring the symbiont. Distinctive features of “Ca. Vidania fulgoroideae” are the following unique 16S rRNA gene sequences (positions according to homologous E. coli positions): ACA ATC AAA TAT GCC TTT TGA AAA GGG ATT TTA AAT TCT TTA TAA AGT TAT ATT TAA AAA TAT AAT AAA ATG GAC TTA TTA AAT AAA TTA TGT TTT AA (positions 133 to 231), GAT GAA GGT TGA TAA GAT CGT AAA ACA CTT TTT TTA ATT AAT AAA AAC TTG TAT AAA (positions 427 to 484), AGT TTT TAA CTT ATC ATA AAA GGA CCG CTA AAA ATA TAA AAA (positions 1139 to 1181), and TTT TTA CAG CGA GTA AAT AAG CTG A (positions 1254 to 1279).
Acknowledgments
We are grateful to the Centro Interdipartimentale Grandi Strumenti of the University of Modena and Reggio Emilia for confocal microscopy analysis.
Partial financial contribution comes from the Italian Ministry for Research (MIUR), within the project PRIN 2007, Caratterizzazione del Microbiota Associato a Scaphoideus titanus e Hyalesthes obsoletus, Cicaline Vettrici di Fitoplasmi nella Vite ed Isolamento e Studio della Localizzazione di Batteri Acetici Simbionti, and the European Union in the ambit of project BIODESERT (European Community's Seventh Framework Programme CSA-SA REGPOT-2008-2 under grant agreement no. 245746). E.C., C.B., and D.D. benefited from travel grants from Cost Action FA0701, Arthropod Symbiosis: from Fundamental Studies to Pest and Disease Management.
Footnotes
Published ahead of print on 23 December 2010.
REFERENCES
- 1.Alma, A., C. Arnò, A. Arzone, and C. Vidano. 1988. New biological reports on auchenorrhyncha in vineyards, p. 509-516. In C. Vidano and A. Arzone (ed.), Proceedings of the 6th Auchenorrhyncha Meeting, Turin, Italy. Consiglio Nazionale delle Ricerche, Rome, Italy.
- 2.Alma, A., G. Soldi, R. Tedeschi, and C. Marzachì. 2002. Role of Hyalesthes obsoletus Signoret (Homoptera Cixiidae) in the transmission of grapevine Bois noir in Italy. Petria 12:411-412. [Google Scholar]
- 3.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 4.Amann, R. I., et al. 1990. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 56:1919-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Angelini, E., G. L. Bianchi, L. Filippin, C. Morassutti, and M. Borgo. 2007. A new TaqMan method for the identification of phytoplasmas associated with grapevine yellows by real-time PCR assay. J. Microbiol. Methods 68:613-622. [DOI] [PubMed] [Google Scholar]
- 6.Beard, C. B., R. V. Durvasula, and F. F. Richards. 1998. Bacterial symbiosis in arthropods and the control of disease transmission. Emerg. Infect. Dis. 4:581-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beard, C. G., C. Cordon-Rosales, and R. V. Durvasula. 2002. Bacterial symbionts of the triatominae and their potential use in control of Chagas disease transmission. Annu. Rev. Entomol. 47:123-141. [DOI] [PubMed] [Google Scholar]
- 8.Beninati, T., et al. 2004. A novel alpha-proteobacterium resides in the mitochondria of ovarian cells of the tick Ixodes ricinus. Appl. Environ. Microbiol. 70:2596-2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bianciotto, V., et al. 1996. An obligately endosymbiotic mycorrhizalfungus itself harbors obligately intracellular bacteria. Appl. Environ. Microbiol. 62:3005-3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Braig, H. R., W. Zhou, S. Dobson, and S. L. O'Neill. 1998. Cloning and characterization of a gene encoding the major surface protein of the bacterial endosymbiont Wolbachia. J. Bacteriol. 180:2373-2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bressan, A., R. Turata, S. Spiazzi, E. Boudon-Padieu, and V. Girolami. 2006. Hyalesthes obsoletus: dispersal from nettle and transmission efficiency of stolbur phytoplasma to grapevine, p. 173-175. Extended Abstr. 15th Meet. International Council for the Study of Virus and Virus-like Diseases of the Grapevine (ICVG), Stellenbosch, South Africa, 3 to 7 April 2006.
- 12.Bressan, A., J. Arneodo, M. Simonato, W. P. Haines, and E. Boudon-Padieu. 2009. Characterization and evolution of two bacteriome-inhabiting symbionts in cixiid planthoppers (Hemiptera: Fulgoromorpha: Pentastirini). Environ. Microbiol. 11:3265-3279. [DOI] [PubMed] [Google Scholar]
- 13.Chen, D. Q., D. C. Campbell, and A. H. Purcell. 1996. A new Rickettsia from a herbivorous insect, the pea aphid Acyrthosiphon pisum (Harris). Curr. Microbiol. 33:123-128. [DOI] [PubMed] [Google Scholar]
- 14.Cole, J. R., et al. 2003. The Ribosomal Database Project (RDP-II): previewing a new autoaligner that allows regular updates and the new prokaryotic taxonomy. Nucleic Acids Res. 31:442-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crotti, E., et al. 2009. Asaia, a versatile acetic acid bacterial symbiont, capable of cross-colonizing insects of phylogenetically distant genera and orders. Environ. Microbiol. 11:3252-3264. [DOI] [PubMed] [Google Scholar]
- 16.Curley, C. M., E. L. Brodie, M. G. Lechner, and A. H. Purcell. 2007. Exploration for facultative endosymbionts of glassy-winged sharpshooter (Hemiptera: Cicadellidae). Ann. Entomol. Soc. Am. 100:345-349. [Google Scholar]
- 17.Damiani, C., et al. 2008. Paternal transmission of symbiotic bacteria in malaria vectors. Curr. Biol. 18:R1087-R1088. [DOI] [PubMed] [Google Scholar]
- 18.Doi, Y. M., M. Teranaka, K. Yora, and H. Asuyama. 1967. Mycoplasma or PLT-group-like microorganisms found in the phloem elements of plants infected with mulberry dwarf, potato witches' broom, aster yellows, or paulonia witches' broom. Ann. Phytopathol. Soc. Jpn. 33:259-266. [Google Scholar]
- 19.Doyle, J. J., and J. L. Doyle. 1990. Isolation of plant DNA from fresh tissues. Focus 12:13-15. [Google Scholar]
- 20.Duron, O., G. D. D. Hurst, E. A. Hornett, J. A. Josling, and J. Engelstädter. 2008. High incidence of the maternally inherited bacterium Cardinium in spiders. Mol. Ecol. 17:1427-1437. [DOI] [PubMed] [Google Scholar]
- 21.Enigl, M., and P. Schausberger. 2007. Incidence of the endosymbionts Wolbachia, Cardinium and Spiroplasma in phytoseiid mites and associated prey. Exp. Appl. Acarol. 42:75-85. [DOI] [PubMed] [Google Scholar]
- 22.Favia, G., et al. 2007. Bacteria of the genus Asaia stably associate with Anopheles stephensi, an Asian malarial mosquito vector. Proc. Natl. Acad. Sci. U. S. A. 104:9047-9051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gonella, E., et al. 2008. Study of the bacterial community affiliated to Hyalesthes obsoletus, the insect vector of “bois noir” phytoplasma of grape. Bull. Insectol. 61:221-222. [Google Scholar]
- 24.Gotoh, T., H. Noda, and S. Ito. 2007. Cardinium symbionts cause cytoplasmic incompatibility in spider mites. Heredity 98:13-20. [DOI] [PubMed] [Google Scholar]
- 25.Gottlieb, Y., et al. 2006. Identification and localization of a Rickettsia sp. in Bemisia tabaci (Homoptera: Aleyrodidae). Appl. Environ. Microbiol. 72:3646-3652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gottlieb, Y., et al. 2008. Inherited intracellular ecosystem: symbiotic bacteria share bacteriocytes in whiteflies. FASEB J. 22:2591-2599. [DOI] [PubMed] [Google Scholar]
- 27.Heddi, A., A. M. Grenier, C. Khatchadourian, H. Charles, and P. Nardon. 1999. Four intracellular genomes direct weevil biology: nuclear, mitochondrial, principal endosymbiont, and Wolbachia. Proc. Natl. Acad. Sci. U. S. A. 96:6814-6819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hunter, M. S., S. J. Perlman, and S. E. Kelly. 2003. A bacterial symbiont in the Bacteroidetes induces cytoplasmic incompatibility in the parasitoid wasp Encarsia pergandiella. Proc. R. Soc. Lond. B Biol. Sci. 270:2185-2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.IRPCM Phytoplasma/Spiroplasma Working Team—Phytoplasma Taxonomy Group. 2004. ‘Candidatus Phytoplasma’, a taxon for the wall-less, non-helical prokaryotes that colonize plant phloem and insects. Int. J. Syst. Evol. Microbiol. 54:1243-1255. [DOI] [PubMed] [Google Scholar]
- 30.Jeyaprakash, A., and M. A. Hoy. 2000. Long PCR improves Wolbachia DNA amplification: wsp sequences found in 76% of 63 arthropod species. Insect Mol. Biol. 4:393-405. [DOI] [PubMed] [Google Scholar]
- 31.Kitajima, E. W., et al. 2007. In situ observation of the Cardinium symbionts of Brevipalpus (Acari: Tenuipalpidae) by electron microscopy. Exp. Appl. Acarol. 42:263-271. [DOI] [PubMed] [Google Scholar]
- 32.Lee, I. M., R. E. Davis, and D. E. Gundersen. 2000. Phytoplasma: phytopathogenic mollicutes. Annu. Rev. Microbiol. 54:221-255. [DOI] [PubMed] [Google Scholar]
- 33.Maixner, M., H. Darimont, and H. D. Mohr. 2001. Studies on the transmission of Bois noir to weeds and potential ground-cover plants by Hyalesthes obsoletus Signoret (Auchenorrhyncha: Cixiidae). IOBC/WPRS Bull. 24:249-251. [Google Scholar]
- 34.Marzachì, C., F. Veratti, M. d'Aquilio, A. Vischi, M. Conti, and G. Boccardo. 2000. Molecular hybridization and PCR amplification of non-ribosomal DNA to detect and differentiate Stolbur phytoplasma isolates from Italy. J. Plant Pathol. 82:201-212. [Google Scholar]
- 35.Marzorati, M., et al. 2006. A novel Bacteroidetes symbiont is localized in Scaphoideus titanus, the insect vector of Flavescence dorée in Vitis vinifera. Appl. Environ. Microbiol. 72:1467-1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McCutcheon, J. P., and N. A. Moran. 2007. Parallel genomic evolution and metabolic interdependence in an ancient symbiosis. Proc. Natl. Acad. Sci. U. S. A. 104:19392-19397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McCutcheon, J. P., and N. A. Moran. 2010. Functional convergence in reduced genomes of bacterial symbionts spanning 200 My of evolution. Genome Biol. Evol. 2:708-718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McCutcheon, J. P., B. R. McDonald, and N. A. Moran. 2009. Origin of an alternative genetic code in the extremely small and GC-rich genome of a bacterial symbiont. PLoS Genet. 5:1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moran, N. A. 2007. Symbiosis as an adaptive process and source of phenotypic complexity. Proc. Natl. Acad. Sci. U. S. A. 104:8627-8633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moran, N. A., and H. E. Dunbar. 2006. Sexual acquisition of beneficial symbionts in aphids. Proc. Natl. Acad. Sci. U. S. A. 103:12803-12806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moran, N. A., C. Dale, H. E. Dunbar, W. A. Smith, and H. Ochman. 2003. Intracellular symbionts of sharpshooters (Insecta: Hemiptera: Cicadellinae) form a distinct clade with a small genome. Environ. Microbiol. 5:116-126. [DOI] [PubMed] [Google Scholar]
- 42.Moran, N. A., L. P. Tran, and M. N. Gerardo. 2005. Symbiosis and insect diversification: an ancient symbiont of sap-feeding insects from the bacterial phylum Bacteroidetes. Appl. Environ. Microbiol. 71:8802-8810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moran, N. A., J. P. McCutcheon, and A. Nakabachi. 2008. Genomics and evolution of heritable bacterial symbionts. Annu. Rev. Genet. 42:165-190. [DOI] [PubMed] [Google Scholar]
- 44.Müller, H. J. 1962. Neure Vorstellungen uber Verbreitung und Phylogenie der Endosymbiosen der Zikaden. Z. Morphol. Okol. Tiere. 51:190-210. [Google Scholar]
- 45.Muyzer, G. E., C. de Waal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Noda, H., U. G. Munderloh, and T. J. Kurtti. 1997. Endosymbionts of ticks and their relationship to Wolbachia spp. and tick-borne pathogens of humans and animals. Appl. Environ. Microbiol. 63:3926-3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rio, R. V. M., Y. Hu, and S. Aksoy. 2004. Strategies for the home team: symbioses exploited for vector-borne disease control. Trends Microbiol. 12:325-336. [DOI] [PubMed] [Google Scholar]
- 48.Ritchie, N. J., M. E. Schutter, R. P. Dick, and D. D. Myrold. 2000. Use of length heterogeneity PCR and fatty acid methyl ester profiles to characterize microbial communities in soil. Appl. Environ. Microbiol. 66:1668-1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sacchi, L., et al. 2008. Multiple symbiosis in the leafhopper Scaphoideus titanus (Hemiptera: Cicadellidae): details of transovarial transmission of Cardinium sp. and yeast-like endosymbionts. Tissue Cell 40:231-242. [DOI] [PubMed] [Google Scholar]
- 50.Sass, A. M., H. Sass, M. J. Coolen, H. Cypionka, and J. Overmann. 2001. Microbial communities in the chemocline of a hypersaline deep-sea basin (Urania basin, Mediterranean Sea). Appl. Environ. Microbiol. 67:5392-5402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schnepf, E., et al. 1998. Bacillus thuringiensis and its pesticidal crystal proteins. Microbiol. Mol. Biol. Rev. 62:775-806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stouthamer, R., J. A. Breeuwer, and G. D. Hurst. 1999. Wolbachia pipientis: microbial manipulator of arthropod reproduction. Annu. Rev. Microbiol. 53:71-102. [DOI] [PubMed] [Google Scholar]
- 53.Suzuki, M., M. S. Rappé, and S. J. Giovannoni. 1998. Kinetic bias in estimates of coastal picoplankton community structure obtained by measurements of small-subunit rRNA gene PCR amplicon length heterogeneity. Appl. Environ. Microbiol. 64:4522-4529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Takiya, D. M., L. P. Tran, C. H. Dietrich, and N. A. Moran. 2006. Co-cladogenesis spanning three phyla: leafhoppers (Insecta: Hemiptera: Cicadellidae) and their dual bacterial symbionts. Mol. Ecol. 15:4175-4191. [DOI] [PubMed] [Google Scholar]
- 55.Tanaka, R., M. Ootsubo, T. Sawabe, Y. Ezura, and K. Tajima. 2004. Biodiversity and in situ abundance of gut microflora of abalone (Haliotis discus hannai) determined by culture-independent techniques. Acquaculture 21:453-463. [Google Scholar]
- 56.Van de Peer, Y., and R. De Wachter. 1994. TREECON for Windows: a software package for the construction and drawing of evolutionary trees for the Microsoft Windows environment. Comput. Appl. Biosci. 10:569-570. [DOI] [PubMed] [Google Scholar]
- 57.Weeks, A. R., F. Marec, and J. A. J. Breeuwer. 2001. A mite species that consists entirely of haploid females. Science 292:2479-2482. [DOI] [PubMed] [Google Scholar]
- 58.Weeks, A. R., R. Velten, and R. Stouthamer. 2003. Incidence of a new sex-ratio-distorting endosymbiotic bacterium among arthropods. Proc. R. Soc. Lond. B Biol. Sci. 270:1857-1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Werren, J. H., and D. Windsor. 2000. Wolbachia infection frequencies in insects: evidence of a global equilibrium? Proc. Biol. Sci. 267:1277-1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Werren, J. H., D. Windsor, and L. R. Guo. 1995. Distribution of Wolbachia among neotropical arthropods. Proc. R. Soc. Lond. B Biol. Sci. 267:1277-1285. [Google Scholar]
- 61.Wu, D., et al. 2006. Metabolic complementarity and genomics of the dual bacterial symbiosis of sharpshooters. PLoS Biol. 4:1079-1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yagupsky, P. 2004. Kingella kingae: from medical rarity to an emerging paediatric pathogen. Lancet Infect. Dis. 4:358-367. [DOI] [PubMed] [Google Scholar]
- 63.Zchori-Fein, E., and S. J. Perlman. 2004. Distribution of the bacterial symbiont Cardinium in arthropods. Mol. Ecol. 13:2009-2016. [DOI] [PubMed] [Google Scholar]
- 64.Zchori-Fein, E., et al. 2001. A newly discovered bacterium associated with parthenogenesis and a change in host selection behavior in parasitoid wasps. Proc. Natl. Acad. Sci. U. S. A. 98:12555-12560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zchori-Fein, E., S. J. Perlman, S. E. Kelly, N. Katzir, and M. S. Hunter. 2004. Characterization of a ‘Bacteroidetes’ symbiont in Encarsia wasps (Hymenoptera: Aphelinidae): proposal of ‘Candidatus Cardinium hertigii’. Int. J. Syst. Evol. Microbiol. 54:961-968. [DOI] [PubMed] [Google Scholar]