Abstract
Functional cross talk between fatty acid biosynthesis and secondary metabolism has been discovered in several cases in microorganisms; none of them, however, involves a modular biosynthetic enzyme. Previously, we reported a hybrid modular nonribosomal peptide synthetase (NRPS)-polyketide synthase (PKS) pathway for the biosynthesis of FK228 anticancer depsipeptide in Chromobacterium violaceum strain 968. This pathway contains two PKS modules on the DepBC enzymes that lack a functional acyltransferase (AT) domain, and no apparent AT-encoding gene exists within the gene cluster or its vicinity. We report here that, through reconstitution of the FK228 biosynthetic pathway in Escherichia coli cells, two essential genes, fabD1 and fabD2, both encoding a putative malonyl coenzyme A (CoA) acyltransferase component of the fatty acid synthase complex, are positively identified to be involved in FK228 biosynthesis. Either gene product appears sufficient to complement the AT-less PKS modules on DepBC for polyketide chain elongation. Concurrently, a gene (sfp) encoding a putative Sfp-type phosphopantetheinyltransferase was identified to be necessary for FK228 biosynthesis as well. Most interestingly, engineered E. coli strains carrying variable genetic components produced significant levels of FK228 under both aerobic and anaerobic cultivation conditions. Discovery of the trans complementation of modular PKSs by housekeeping ATs reveals natural product biosynthesis diversity. Moreover, demonstration of anaerobic production of FK228 by an engineered facultative bacterial strain validates our effort toward the engineering of novel tumor-targeting bioagents.
Polyketide synthases (PKSs) share many similarities with fatty acid synthases (FASs) and can be largely classified as type I, II, and III, according to their architecture and mode of catalysis, yet many variants of PKSs exist in nature (5, 27, 29). Type I PKSs are multifunctional enzymes organized into modules, each of which harbors a set of distinct domains responsible for the catalysis of one cycle of polyketide chain elongation. A prototypical type I PKS elongation module contains minimally three integral domains—a ketosynthase (KS), an acyltransferase (AT), and an acyl carrier protein (ACP)—that together catalyze one round of polyketide chain elongation. Optional domains (such as ketoreductase [KR], dehydratase [DH], and enoylreductase [ER]) are found between the AT and ACP domains, which carry out variable steps of reductive modifications on polyketide intermediates. Type II PKSs are multienzyme complexes that carry a single set of catalytic domains acting iteratively. Type III PKSs are chalcone synthase (CHS)-like enzymes that essentially are iteratively acting condensing enzymes. FASs and type I and type II PKSs all use ACPs to tether acyl coenzyme A (CoA) substrates during fatty acid or polyketide biosynthesis, whereas type III PKSs directly condense acyl-CoA substrates without carrier proteins. Prior to biosynthesis, the apo form of ACPs must be activated to the holo form by attaching a 4′-phosphopantetheinyl moiety from CoA onto a conserved serine residue; this posttranslational modification reaction is catalyzed by phosphopantetheinyltransferases (PPTases) that can be largely classified into two groups, AcpS type and Sfp type (11, 17, 21).
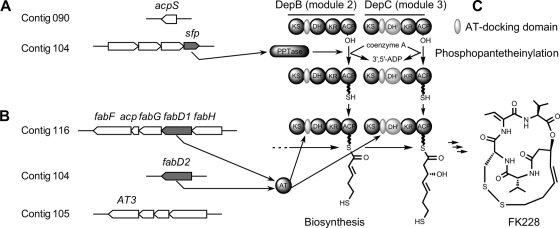
Integral AT domains in the prototypical type I PKSs were termed cognate ATs (8). A distinct variant of the type I PKSs contains no intact cognate ATs but a short segment of a remnant AT sequence in some or all modules. This subclass of type I PKSs was named the AT-less type I PKSs, and the remnant AT segment was named the AT docking domain (7, 8, 31). The essential AT activities are provided in trans by a discrete AT enzyme encoded by genes that are physically separated from the PKS genes but nevertheless within the same gene cluster.
Our studies of the biosynthesis of FK228, a potent histone deacetylase inhibitor recently approved by the FDA for the treatment of cutaneous T-cell lymphoma (CTCL) (14, 22), revealed that a 12-gene dep gene cluster encodes a hybrid modular nonribosomal peptide synthetase (NRPS)-PKS pathway (9, 23) and that a flavin adenine dinucleotide (FAD)-dependent oxidoreductase is responsible for a critical disulfide bond formation in FK228 biosynthesis in Chromobacterium violaceum no. 968 (32). Nevertheless, there are still several unanswered questions regarding the FK228 biosynthetic pathway. In particular, the two PKS modules on DepBC enzymes do not contain any cognate AT domain but a remnant AT docking domain (Fig. 1 C), and there is no AT-encoding gene anywhere in the defined dep gene cluster (9, 23). Where does the AT activity necessary for FK228 biosynthesis come from? In this work, we report the identification of two essential genes, fabD1 and fabD2, both encoding a putative malonyl CoA acyltransferase (MCAT; also known as FabD, encoded by the fabD gene) component of the FAS II complex, that are positively involved in FK228 biosynthesis. Concurrently, we identified a gene (sfp) encoding a putative Sfp-type PPTase necessary for FK228 biosynthesis as well. Furthermore, we show that, through reconstitution of the FK228 biosynthesis pathway, FK228 could be produced by recombinant Escherichia coli strains under both aerobic and anaerobic cultivation conditions. Our studies revealed, for the first time, that modular PKSs recruit FabD components of the primary metabolism for the biosynthesis of a secondary metabolite. Discovery of the trans complementation of modular PKSs by housekeeping ATs reveals natural product biosynthesis diversity. Moreover, demonstration of anaerobic production of FK228 by an engineered facultative bacterial strain validates our effort toward the engineering of novel tumor-targeting bioagents (Y.-Q. Cheng, unpublished data).
FIG. 1.
Identification of missing genes necessary for FK228 biosynthesis in Chromobacterium violaceum no. 968. (A) Local genetic map of two candidate PPTase-encoding genes, with the sfp gene product postulated to participate in phosphopantetheinylation of carrier proteins of the FK228 biosynthetic pathway. (B) Local genetic map of three candidate AT-encoding genes, with the fabD1 and fabD2 gene products postulated to provide AT activities to complement the AT-less PKS modules on DepBC proteins for FK228 biosynthesis. (C) Scheme of two AT-less PKS modules on DepBC proteins that require a PPTase for carrier protein phosphopantetheinylation and a trans AT for polyketide chain elongation in FK228 biosynthesis. An inactive DH domain is shown in light gray and labeled as DHi. KS, AT, ACP, DH, KR, and PPTase are standard abbreviations of domain/enzyme names that have been described in the text.
MATERIALS AND METHODS
Bacterial strains, plasmids, culture conditions, and general molecular biological manipulations.
The bacterial strains and plasmids used in this study are listed in Table 1. Culture conditions and general molecular biological manipulations were performed as described previously (9, 32) or according to standard protocols (25).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Descriptiona | Source or reference |
|---|---|---|
| Chromobacterium violaceum | ||
| no. 968 (FERM BP-1968) | Wild-type strain, FK228 producing, Apr Thior | IPODb |
| Escherichia coli | ||
| DH5α | General cloning host | Lab stock |
| S17-1 | Host strain for interspecies conjugation | Lab stock |
| BL21(DE3) | Host strain for heterologous gene expression | Novagen |
| SW01 | BL21(DE3) harboring cosmid 18 | This work |
| SW02 | BL21(DE3) harboring cosmid 18 and pBMTL-3-depR | This work |
| SW03 | BL21(DE3) harboring cosmid 18, pBMTL-3-depR, and pCDFDuet-1-sfp | This work |
| SW04 | BL21(DE3) harboring cosmid 18, pBMTL-3-depR, and pCDFDuet-1-acpS | This work |
| SW05 | BL21(DE3) harboring cosmid 18, pBMTL-3-depR, and pCDFDuet-1-fabD1 | This work |
| SW06 | BL21(DE3) harboring cosmid 18, pBMTL-3-depR, and pCDFDuet-1-fabD2 | This work |
| SW07 | BL21(DE3) harboring cosmid 18, pBMTL-3-depR, and pCDFDuet-1-sfp-fabD1 | This work |
| SW08 | BL21(DE3) harboring cosmid 18, pBMTL-3-depR, and pCDFDuet-1-sfp-fabD2 | This work |
| SW09 | BL21(DE3) harboring cosmid 18, pBMTL-3-depR, and pCDFDuet-1-acpS-fabD1 | This work |
| SW10 | BL21(DE3) harboring cosmid 18, pBMTL-3-depR, and pCDFDuet-1-acpS-fabD2 | This work |
| Plasmids | ||
| pGEM-3Zf | Apr, general cloning vector | Promega |
| pGEM-T Easy | Apr, general cloning vector | Promega |
| Cosmid 18 | Apr Kanr, cosmid clone containing the FK228 biosynthetic gene cluster (dep) and flanking DNAs, shotgun sequenced | 9 |
| pBMTL-3 | Cmr, pBBR1 ori, broad-host-range vector | 19 |
| pBMTL-3-depR | Cmr, depR (with ribosome-binding site from pET29a) cloned into pBMTL-3 | 23 |
| pCDFDuet-1 | Smr, CDF ori, dual expression vector | Novagen |
| pCDFDuet-1-sfp | sfp cloned into the MCS1 region of pCDFDuet-1 | This study |
| pCDFDuet-1-acpS | acpS cloned into MCS1 region of pCDFDuet-1 | This study |
| pCDFDuet-1-fabD1 | fabD1 cloned into MCS2 region of pCDFDuet-1 | This study |
| pCDFDuet-1-fabD2 | fabD2 cloned into MCS2 region of pCDFDuet-1 | This study |
| pCDFDuet-1-sfp-fabD1 | sfp cloned into MCS1, fabD1 cloned into MCS2 of pCDFDuet-1 | This study |
| pCDFDuet-1-sfp-fabD2 | sfp cloned into MCS1, fabD2 cloned into MCS2 of pCDFDuet-1 | This study |
| pCDFDuet-1-acpS-fabD1 | acpS cloned into MCS1, fabD1 cloned into MCS2 of pCDFDuet-1 | This study |
| pCDFDuet-1-acpS-fabD2 | acpS cloned into MCS1, fabD2 cloned into MCS2 of pCDFDuet-1 | This study |
Apr, ampicillin resistance; Cmr, chloramphenicol resistance; Kanr, kanamycin resistance; Smr, streptomycin resistance; Thior, thiostrepton resistance; MCS, multiple cloning site.
IPOD, International Patent Organism Depositary, Tsukuba, Japan.
Rapid genome sequencing and gene identification.
Genomic DNA of wild-type C. violaceum no. 968 was prepared from an overnight culture with an UltraClean microbial DNA isolation kit (MO BIO Labs, Carlsbad, CA) and was submitted for single-end and paired-end shotgun sequencing on a G20 FLX platform (454 Life Sciences, Branford, CT) at the Research Technology Support Facility of Michigan State University (East Lansing, MI). De novo assembly of sequence reads by instrument software was performed and resulted in a quality draft genome sequence. Candidate genes were identified by using known protein sequences as bait to search the draft genome sequence of C. violaceum by the Blastx algorithm (2).
General strategies for targeted gene deletion in C. violaceum no. 968.
A multiplex PCR method, as described elsewhere (10, 23, 32), was used for all intended gene deletion experiments. This method utilized a broad-host-range Flp-FRT recombination system for site-specific gene replacement/deletion and subsequent marker removal (10, 15). Primers used for making gene deletion constructs and for the detection of genotypes are listed in Table S1 in the supplemental material.
Reconstitution of FK228 biosynthetic gene cluster in engineered E. coli strains.
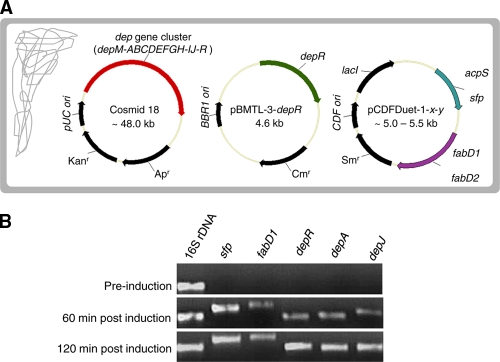
To probe whether individual candidate AT-encoding genes or PPTase-encoding genes are involved in FK228 biosynthesis, a three-plasmid system was utilized for gene cluster reconstitution and FK228 biosynthesis in bacterial strains derived from E. coli BL21(DE3) (Fig. 2A). First, all candidate genes were amplified by high-fidelity PCR from the genomic DNA of C. violaceum with primer sets carrying designed restriction sites. Second, two candidate PPTase-encoding gene amplicons were double digested with BamHI/HindIII and individually cloned into the first multiple cloning site (MCS) of pCDFDuet-1 (Novagen, Madison, WI) to create two intermediate constructs, pCDFDuet-1-acpS and pCDFDuet-1-sfp. Third, two candidate AT-encoding gene amplicons were double digested with NdeI/KpnI and individually cloned into the second multiple cloning site of pCDFDuet-1 to generate pCDFDuet-1-fabD1 and pCDFDuet-1-fabD2 or into the two previously made intermediate constructs to create a combination of four final constructs, pCDFDuet-1-acpS-fabD1, pCDFDuet-1-acpS-fabD2, pCDFDuet-1-sfp-fabD1, and pCDFDuet-1-sfp-fabD2 (Table 1). Finally, cosmid 18, which carries the original incomplete FK228 biosynthetic gene cluster that lacks any AT-encoding gene or PPTase-encoding gene (9), and pBMTL-3-depR, which was created to complement a depR deletion mutant of C. violaceum (23), were used in combination with the above expression constructs for the transformation of E. coli BL21(DE3) cells to create a series of bacterial strains (Table 1). Kanamycin at 25 μg/ml, chloramphenicol at 25 μg/ml, and streptomycin at 25 μg/ml were used individually or in combination for selection and maintenance of the respective E. coli strains.
FIG. 2.
Reconstitution of FK228 biosynthesis in E. coli cells. (A) Scheme of a three-plasmid approach for reconstitution of FK228 biosynthetic pathway in E. coli BL21(DE3) cells. Only one cell containing one copy of each plasmid is shown for simplicity. A series of engineered strains was generated with different combinations of plasmids or different genes on plasmids (Table 1). (B) Examination of gene expression by semiquantitative RT-PCR in engineered strain SW07 cultivated under aerobic conditions. The 16S rRNA gene was amplified as an internal control.
Bacterial fermentation and quantification of FK228 production by LC-MS.
The wild-type C. violaceum strain and recombinant E. coli strains were fermented aerobically for 4 days at 30°C under constant agitation (200 rpm) in 50 ml of LB medium supplemented with 1% (wt/vol) Diaion HP-20 resin (Sigma-Aldrich, St. Louis, MO) and appropriate antibiotics where necessary. Gene expression was induced with 0.5% (wt/vol) lactose and 0.1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) when bacterial culture reached an optical density at 600 nm (OD600) of 0.4. Strict anaerobic fermentation of bacterial strains was carried out similarly for 5 days at room temperature in a Coy anaerobic chamber (Grass Lake, MI) with occasional manual agitation, except that 0.05% (wt/vol) thioglycolate was added to the medium to capture any oxidative species generated during fermentation. Extraction of metabolites and detection and quantification of FK228 by liquid chromatography-mass spectrometry (LC-MS) were performed as described previously (23).
RNA extraction and reverse transcription (RT)-PCR.
Recombinant E. coli BL21(DE3) strains were grown in LB medium supplemented with appropriate antibiotics at 30°C under constant agitation (200 rpm) to an OD600 of 0.4. Five milliliters of each preinduction sample was collected, and the remaining cultures were induced with 0.5% (vol/vol) lactose and 0.1 mM IPTG. Aliquots of samples were collected at 60 min and again at 120 min postinduction. Preservation of sample aliquots, extraction of total RNA, and RT-PCR experiments were performed as described previously (23). Primers used for detection of individual gene expression are listed in Table S1 in the supplemental material.
Nucleotide sequence accession numbers.
The nucleotide sequences of C. violaceum no. 968 genes reported in this paper have been deposited in the GenBank database under accession numbers HM449690 for the 16S rRNA gene, HM449691 for fabD1, HM449692 for fabD2, HM449693 for AT3, HM449694 for acpS, and HM449695 for sfp.
RESULTS
Draft genome sequencing of Chromobacterium violaceum no. 968.
Shotgun sequencing of C. violaceum genomic DNA on a GS20 FLX sequencer generated a total of 163,954,650 input bases, which were assembled into 122 contigs; among them, 82 are large contigs (>500 nucleotides [nt]) with an average contig size of 59,514 bp. Those contigs were further aligned into 15 scaffolds with a total length of 4,909,141 bp. The sequence coverage for this draft bacterial genome was thus calculated at 33.4-fold, which exceeded the desired 30-fold oversampling of raw sequence for the 454 pyrosequencing and de novo assembly technology platform (12). Compared to the published 4.75-Mb complete genome of a type strain of C. violaceum ATCC 12472 with a G+C content of 64.8% (4), the 4.91-Mb draft genome sequence of C. violaceum no. 968 with an overall G+C content of 61.9% obtained in this study appears to be nearly complete. The quality of this draft genome sequence was assessed by the following analyses. First, a comparison of the 16S rRNA gene sequences of two C. violaceum strains revealed an overall 96% identity without a single gap (see Fig. S1 in supplemental material), indicating a high quality of the draft genome sequence and a taxonomical relatedness of the two strains. Second, a homology search by the Blastn algorithm of the draft genome sequence using our previously published FK228 biosynthetic gene cluster sequence (GenBank accession no. EF210776) (9) as bait identified two contigs that carry the gene cluster with a 100% sequence identity and with a 1,298-bp sequence gap (data not shown). This gap was artificially created due to two highly homologous regions within the gene cluster that were assembled into a single copy of sequence.
Identification and initial characterization of candidate genes.
Sequences of three bait proteins, FabD of E. coli K-12 (GenBank accession no. AAC74176), FabD of C. violaceum ATCC 12472 (GenBank accession no. NP_903085), and LnmG of Streptomyces atroolivaceus S-140 (GenBank accession no. AAN85520), were used to search the draft genome sequence and identified three candidate genes, fabD1, fabD2, and AT3, that encode putative AT enzymes (FabD, MCAT, or AT) (Table 2; Fig. 1B). Based on bioinformatic analysis, the fabD1 gene lies within an apparent FAS II gene cluster that also includes fabH, fabG, acp, and fabF, whereas fabD2 is a stand-alone gene whose function cannot be predicted a priori. The AT3 gene lies within a putative gene cluster that may be involved in cell surface O-antigen biosynthesis.
TABLE 2.
Candidate genes identified through rapid genome sequencing and their involvement in FK228 biosynthesis
| Genea | Deduced product and predicted function | Gene disposability | Involvement in FK228 biosynthesis |
|---|---|---|---|
| AT-encoding genes | |||
| fabD1 | FabD1, likely involved in fatty acid biosynthesis | No | Positive |
| fabD2 | FabD2, function unpredictable a priori | No | Positive |
| AT3 | AT3, likely involved in cell surface O-antigen biosynthesis | Yes | No |
| PPTase-encoding genes | |||
| acpS | AcpS-type PPTase, likely involved in primary metabolite biosynthesis | No | No |
| sfp | Sfp-type PPTase, likely involved in secondary metabolite biosynthesis | No | Yes |
AT, acyltransferase; PPTase, phosphopantetheinyltransferase.
Similarly, sequences of five bait proteins, AcpS of E. coli K-12 (GenBank accession no. P24224), Sfp of Bacillus subtilis (GenBank accession no. P39135), AcpS of C. violaceum ATCC 12472 (GenBank accession no. NP_901742), EntD of C. violaceum ATCC 12472 (GenBank accession no. NP_902320), and PcpS of Pseudomonas aeruginosa PAO1 (GenBank accession no. AAG04554), were used to search the draft genome sequence and identified two candidate genes, acpS and sfp, that encode putative PPTase enzymes (AcpS; Sfp) (Table 2; Fig. 1A). The acpS gene appears to be a stand-alone gene likely involved in primary metabolite (e.g., fatty acid) biosynthesis. The sfp gene is located at the end of an apparent NRPS gene cluster; therefore, it is likely involved in secondary metabolite (e.g., nonribosomal peptide or hybrid molecule) biosynthesis.
Individual candidate genes were subjected to targeted gene deletion by a well-established multiplex PCR procedure to probe whether they play any roles in FK228 biosynthesis. Only AT3 was successfully mutated, and the mutant strain did not show any notable difference from the wild-type strain (data not shown), suggesting that AT3 is disposable and is independent of FK228 biosynthesis. Thus, AT3 was not further tested. In contrast, four other candidate genes, fabD1, fabD2, acpS, and sfp, could not be mutated despite numerous attempts, indicating that they are all essential to bacterial physiology and survival. They were then subjected to tests by a different strategy, described below.
Reconstitution of the FK228 biosynthetic pathway in E. coli.
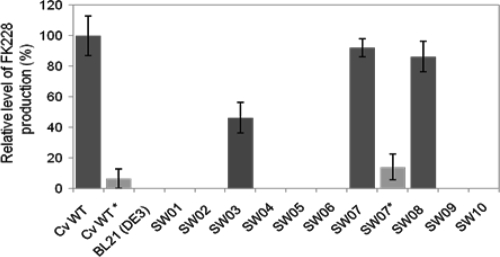
A series of recombinant E. coli strains were created herein (Table 1; Fig. 2A), and the relative levels of FK228 production by these strains were examined by LC-MS (Fig. 3; see also Table S2 and Fig. S2 in the supplemental material). When cosmid 18, the originally identified large construct that carries the dep gene cluster which definitely lacks an AT-encoding gene and a PPTase-encoding gene (9), was introduced into E. coli BL21(DE3) cells, the recombinant strain SW01 failed to produce FK228. When a newly defined pathway regulatory gene, depR (23), was introduced into SW01 via a broad-host-range construct, pBMTL-3-depR, the resulting strain SW02 still did not produce any detectable level of FK228. When the candidate sfp gene was introduced into SW02 via a compatible construct, pCDFDuet-1-sfp, the resulting strain, SW03, produced a moderate level of FK228. When either the candidate fabD1 or fabD2 gene was added in tandem with sfp on the pCDFDuet-1-sfp-fabD1 or pCDFDuet-1-sfp-fabD2 construct, the resulting strains SW07 and SW08 produced significant levels of FK228. All other strains that received a single candidate gene of either acpS (strain SW04), fabD1 (strain SW05), or fabD2 (strain SW06), or two genes with acpS in tandem with either fabD1 (strain SW9) or fabD2 (strain SW10), did not show production of a detectable level of FK228. Those observations led to the following conclusions: (i) the product of the sfp gene but not that of the acpS gene is capable of converting all ACPs (and presumably peptidyl carrier proteins [PCPs]) in the FK228 biosynthetic pathway from their inactive apo form to their active holo form; (ii) indigenous E. coli AcpS cannot promiscuously act on those heterologous carrier proteins from a secondary metabolic pathway; (iii) either gene product of the essential fabD1 or fabD2 gene of C. violaceum no. 968 is able to complement the AT-less PKS modules on DepBC for polyketide chain extension; (iv) even E. coli FabD (in the background in the case of strain SW03) is able to provide the necessary AT activities for FK228 biosynthesis, albeit at a lower rate; and (v) at this point, we believe that genes necessary for FK228 biosynthesis have all been identified.
FIG. 3.
Levels of FK228 produced by recombinant E. coli strains relative to the wild-type Chromobacterium violaceum no. 968 cultivated under aerobic conditions unless indicated by an asterisk, which indicates anaerobic conditions. Data are mean values of results from duplicate experiments, with error bars indicating standard deviation. Detailed strain information and FK228 levels are provided in Table 1 and in Table S2 in the supplemental material.
RT-PCR verification of gene expression.
To verify whether key biosynthetic and regulatory genes for FK228 biosynthesis were adequately expressed in the SW07 recombinant strain under normal aerobic conditions, aliquots of bacterial culture were collected at three time points, and total RNA samples were prepared and subjected to semiquantitative RT-PCR analysis (Fig. 2B). Prior to chemical induction, all examined genes, including two representative structural genes (depA and depJ) but excluding the 16S rRNA gene control, were not expressed. This suggests that the depR gene in its native position on cosmid 18 is not functioning in E. coli cells, likely due to a lack of proper external or internal stimulus or signal transduction pathway or other regulatory components; expression of the dep gene cluster carried by cosmid 18 in E. coli cells requires an ectopic copy of depR driven by a lactose-inducible promoter on a pBMTL-3 vector. Upon induction by lactose and IPTG for the expression of depR from pBMTL-3-depR and for the expression of sfp and fabD1 from pCDFDuet-1-sfp-fabD1, those genes and two representative structural genes (depA and depJ) were found to have been expressed at 60 min postinduction, and the gene expression reached higher levels at 120 min postinduction.
Production of FK228 under anaerobic fermentation conditions.
As a pilot study for the engineering of novel tumor-targeting bacterial agents that may effectively infiltrate, multiply, and continuously produce an anticancer drug inside the hypoxic core of solid tumors (Cheng, unpublished), we examined whether the engineered SW07 strain may produce FK228 under anaerobic fermentation conditions. To our delight, the SW07 strain produced ca. 14.1% as much FK228 as the wild-type C. violaceum strain or 15.3% as much as the SW07 strain under aerobic conditions (Fig. 3; see also Table S2 and Fig. S2 in the supplemental material); this relative level of FK228 production was translated into 0.40 mg/liter of actual yield or a ca. 741 nM concentration when normalized to the FK228 standard. As a control, the wild-type C. violaceum strain was also found to have produced a lower level of FK228 under anaerobic conditions. This Gram-negative bacterial species appeared to be able to survive and grow slightly during the first 24 h under the strict anaerobic conditions tested.
DISCUSSION
Organizational and functional variation of PKSs leads to a diverse structure of polyketide natural products (5, 27). The modular AT-less PKSs represent a severe deviation from the canonical type I PKSs (7, 8), and this phenomenon appears to be a transition state of complex enzyme evolution from FAS II to type II PKSs (28). Nevertheless, in most AT-less PKS pathways, the AT activities required for polyketide chain elongation are often encoded by discrete genes within the respective biosynthetic gene clusters. In the present work, for the first time, we uncovered an extreme case of the AT-less PKS system where the AT activity is encoded by fabD genes, the essential bacterial genes involved in fatty acid biosynthesis.
As more and more modular AT-less PKSs are identified (7), it was not so surprising to find that the two PKS modules on DepBC do not contain an intact cognate AT domain but a remnant of an AT-docking segment (Fig. 1C). However, a failure to identify any AT-encoding gene within or in the vicinity of the defined dep gene cluster was intriguing (9, 23). To address the mystery about where the necessary AT activities would come from, the genome of C. violaceum no. 968 was decoded by a rapid genome sequencing platform, revealing three candidate genes that may encode the AT activities for FK228 biosynthesis. However, attempts to identify the exact AT-encoding gene by a direct gene deletion approach was not successful, because two candidate genes (fabD1 and fabD2) could not be mutated due to their apparent essential roles in bacterial viability. A third gene (AT3) could be mutated, but the mutant did not show any obvious sign of physiological defect or decrease in FK228 production.
We then engineered a series of recombinant E. coli strains that harbor one to three compatible vectors that carry either the previously reported incomplete dep gene cluster or candidate genes identified in this work with three objectives in mind (Fig. 2A). First, we hoped to identify the missing AT-encoding gene and PPTase-encoding gene necessary for FK228 biosynthesis. Second, we hoped to reconstitute a functional FK228 biosynthetic pathway in a heterologous host, such as E. coli cells. Third, we hoped to demonstrate that FK228 could be produced by a recombinant E. coli strain under both aerobic conditions and particularly anaerobic conditions as a pilot experiment for genetic engineering of novel cancer-targeting bioagents (Cheng, unpublished).
Three engineered E. coli strains, SW03, SW07, and SW08, were subsequently found to produce variable amounts of FK228 under normal (aerobic) fermentation conditions. In addition to receiving cosmid 18 and the depR gene on a vector, all three of these strains received the sfp gene, while other E. coli strains, regardless of having the acpS gene or not, did not produce FK228. It was thus clear that the AcpS-type PPTase from either the E. coli host or from C. violaceum cannot activate the carrier proteins from the FK228 biosynthetic pathway. Therefore, it was concluded that the sfp gene, which was predicted to encode a broad-substrate-range Sfp-type PPTase, is involved in FK228 biosynthesis. This PPTase should have other essential functions as well; otherwise, the sfp gene could have been mutated in early experimental attempts. In addition, two of the three strains, SW07 and SW08, which also received either the fabD1 or fabD2 gene of C. violaceum, produced much higher levels of FK228 than SW03. Those observations suggested that either fabD1 or fabD2 of C. violaceum is involved in FK228 biosynthesis and their involvement is interchangeable; it is also possible that both genes are redundantly involved in FK228 biosynthesis. Surprisingly, the indigenous fabD of E. coli appeared to function as well for FK228 biosynthesis in the SW03 strain, which received the sfp gene but not fabD1 or fabD2.
Sequence alignment of C. violaceum FabD1 and FabD2 and E. coli FabD shows that FabD1 and FabD share a much higher amino acid sequence identity than the other two sets compared (FabD1/FabD2 and FabD2/FabD) (see Fig. S3 in the supplemental material), suggesting that FabD1 is very likely the housekeeping FAS II component for bacterial fatty acid biosynthesis. The role of FabD2 is less certain. Besides a positive role in FK228 biosynthesis, FabD2 should have other essential functions; otherwise, the fabD2 gene could have been mutated in early experimental attempts.
Now it is proven that an MCAT component of FAS II is recruited by modular type I PKS modules on DepBC for FK228 biosynthesis, which establishes a unique functional cross talk between bacterial primary metabolism and secondary metabolism and adds new evidence to the phenomenon of complex enzyme evolution from FAS II to type II PKSs (28). Previously, two type II PKS systems responsible for the biosynthesis of actinorhodin (24) or tetracenomycin (3, 13, 30) were found to recruit FabD of FAS II. Additional variants of cross talk between bacterial primary metabolism and secondary metabolism have also been reported. For example, in the biosynthesis of quinoxaline antibiotics in two Streptomyces strains, an ACP from the primary FAS II is recruited by a stand-alone condensation domain of type II NRPS to form an initiation module of the biosynthetic pathway (26). Furthermore, cross talk between a type I FAS (FAS I) for primary metabolism and a secondary FAS I for HC-toxin biosynthesis had also been postulated in the fungal species Cochliobolus carbonum, where only one gene encoding the β-subunit of FAS I was found in the secondary metabolic gene cluster, while the α-subunit of FAS I necessary for HC-toxin biosynthesis might be recruited from the primary FAS I (1). Recently, an opposite example was reported in the apicidin biosynthetic pathway in Fusarium semitectum, where a gene encoding the α-subunit of FAS I is present in the apicidin biosynthetic gene cluster, while the β-subunit of FAS I necessary for apicidin biosynthesis is speculated to be recruited from the primary FAS I (16).
Lastly, the ability by which the engineered SW07 strain produced 0.4 mg/liter of actual yield or a ca. 741 nM concentration of FK228 under strict anaerobic conditions is worth highlighting. This accomplishment represents an important milestone toward the goal of engineering of tumor-targeting bioagents (Cheng, unpublished). The next step will be undertaken to introduce the FK228 biosynthetic capacity into one or more selected nonpathogenic anaerobic bacterial species, such as Bifidobacterium longum or Clostridium oncolyticum (20, 33), so that when the engineered bacteria infiltrate and multiply inside the necrotic region of solid tumors, FK228 will be produced in situ. Because FK228 is a potent histone deacetylase inhibitor and a new anticancer drug effective in the lower nanomolar range (18, 34), the synergistic efforts of bacterial oncolysis of tumor tissue and the anticancer activity of FK228 could potentially be clinically effective against many types of cancer.
Supplementary Material
Acknowledgments
We thank the Research Technology Support Facility of Michigan State University for providing quality shotgun genome sequencing and assembly services. We also thank Daad Saffarini for assistance with bacterial anaerobic fermentation and Patrick Anderson for assistance with LC-MS.
This work was supported in part by a Research Growth Initiative Award from the University of Wisconsin—Milwaukee and by an Idea Award BC073985 from the U.S. Department of Defense Breast Cancer Research Program.
Footnotes
Published ahead of print on 23 December 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Ahn, J. H., and J. D. Walton. 1997. A fatty acid synthase gene in Cochliobolus carbonum required for production of HC-toxin, cyclo(d-prolyl-l-alanyl-d-alanyl-l-2-amino-9, 10-epoxi-8-oxodecanoyl). Mol. Plant Microbe Interact. 10:207-214. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Bao, W., E. Wendt-Pienkowski, and C. R. Hutchinson. 1998. Reconstitution of the iterative type II polyketide synthase for tetracenomycin F2 biosynthesis. Biochemistry 37:8132-8138. [DOI] [PubMed] [Google Scholar]
- 4.Brazilian National Genome Project Consortium. 2003. The complete genome sequence of Chromobacterium violaceum reveals remarkable and exploitable bacterial adaptability. Proc. Natl. Acad. Sci. U. S. A. 100:11660-11665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chan, Y. A., A. M. Podevels, B. M. Kevany, and M. G. Thomas. 2009. Biosynthesis of polyketide synthase extender units. Nat. Prod. Rep. 26:90-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reference deleted.
- 7.Cheng, Y.-Q., J. M. Coughlin, S. K. Lim, and B. Shen. 2009. Type I polyketide synthases that require discrete acyltransferases. Methods Enzymol. 459:165-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheng, Y.-Q., G. L. Tang, and B. Shen. 2003. Type I polyketide synthase requiring a discrete acyltransferase for polyketide biosynthesis. Proc. Natl. Acad. Sci. U. S. A. 100:3149-3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng, Y.-Q., M. Yang, and A. M. Matter. 2007. Characterization of a gene cluster responsible for the biosynthesis of anticancer agent FK228 in Chromobacterium violaceum no. 968. Appl. Environ. Microbiol. 73:3460-3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Choi, K. H., and H. P. Schweizer. 2005. An improved method for rapid generation of unmarked Pseudomonas aeruginosa deletion mutants. BMC Microbiol. 5:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Copp, J. N., and B. A. Neilan. 2006. The phosphopantetheinyl transferase superfamily: phylogenetic analysis and functional implications in cyanobacteria. Appl. Environ. Microbiol. 72:2298-2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Droege, M., and B. Hill. 2008. The genome sequencer FLX system—longer reads, more applications, straightforward bioinformatics and more complete data sets. J. Biotechnol. 136:3-10. [DOI] [PubMed] [Google Scholar]
- 13.Florova, G., G. Kazanina, and K. A. Reynolds. 2002. Enzymes involved in fatty acid and polyketide biosynthesis in Streptomyces glaucescens: role of FabH and FabD and their acyl carrier protein specificity. Biochemistry 41:10462-10471. [DOI] [PubMed] [Google Scholar]
- 14.Furumai, R., et al. 2002. FK228 (depsipeptide) as a natural prodrug that inhibits class I histone deacetylases. Cancer Res. 62:4916-4921. [PubMed] [Google Scholar]
- 15.Hoang, T. T., R. R. Karkhoff-Schweizer, A. J. Kutchma, and H. P. Schweizer. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77-86. [DOI] [PubMed] [Google Scholar]
- 16.Jin, J. M., et al. 2010. Functional characterization and manipulation of the apicidin biosynthetic pathway in Fusarium semitectum. Mol. Microbiol. 76:456-466. [DOI] [PubMed] [Google Scholar]
- 17.Lambalot, R. H., et al. 1996. A new enzyme superfamily—the phosphopantetheinyl transferases. Chem. Biol. 3:923-936. [DOI] [PubMed] [Google Scholar]
- 18.Lane, A. A., and B. A. Chabner. 2009. Histone deacetylase inhibitors in cancer therapy. J. Clin. Oncol. 27:5459-5468. [DOI] [PubMed] [Google Scholar]
- 19.Lynch, M. D., and R. T. Gill. 2006. Broad host range vectors for stable genomic library construction. Biotechnol. Bioeng. 94:151-158. [DOI] [PubMed] [Google Scholar]
- 20.Minton, N. P. 2003. Clostridia in cancer therapy. Nat. Rev. Microbiol. 1:237-242. [DOI] [PubMed] [Google Scholar]
- 21.Mootz, H. D., R. Finking, and M. A. Marahiel. 2001. 4′-Phosphopantetheine transfer in primary and secondary metabolism of Bacillus subtilis. J. Biol. Chem. 276:37289-37298. [DOI] [PubMed] [Google Scholar]
- 22.National Cancer Institute. 2010. StatBite: FDA oncology drug product approvals in 2009. J. Natl. Cancer Inst. 102:219. [DOI] [PubMed] [Google Scholar]
- 23.Potharla, V. Y., S. R. Wesener, and Y.-Q. Cheng. 2011. New insights into the genetic organization of the FK228 biosynthetic gene cluster in Chromobacterium violaceum strain 968. Appl. Environ. Microbiol. 77:1508-1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Revill, W. P., M. J. Bibb, and D. A. Hopwood. 1995. Purification of a malonyltransferase from Streptomyces coelicolor A3(2) and analysis of its genetic determinant. J. Bacteriol. 177:3946-3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sambrook, J., and D. W. Russell. 2000. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 26.Schmoock, G., et al. 2005. Functional cross-talk between fatty acid synthesis and nonribosomal peptide synthesis in quinoxaline antibiotic-producing streptomycetes. J. Biol. Chem. 280:4339-4349. [DOI] [PubMed] [Google Scholar]
- 27.Shen, B. 2003. Polyketide biosynthesis beyond the type I, II and III polyketide synthase paradigms. Curr. Opin. Chem. Biol. 7:285-295. [DOI] [PubMed] [Google Scholar]
- 28.Smith, J. L., and D. H. Sherman. 2008. Biochemistry: an enzyme assembly line. Science 321:1304-1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Staunton, J., and K. J. Weissman. 2001. Polyketide biosynthesis: a millennium review. Nat. Prod. Rep. 18:380-416. [DOI] [PubMed] [Google Scholar]
- 30.Summers, R. G., A. Ali, B. Shen, W. A. Wessel, and C. R. Hutchinson. 1995. Malonyl-coenzyme A:acyl carrier protein acyltransferase of Streptomyces glaucescens: a possible link between fatty acid and polyketide biosynthesis. Biochemistry 34:9389-9402. [DOI] [PubMed] [Google Scholar]
- 31.Tang, G. L., Y.-Q. Cheng, and B. Shen. 2004. Leinamycin biosynthesis revealing unprecedented architectural complexity for a hybrid polyketide synthase and nonribosomal peptide synthetase. Chem. Biol. 11:33-45. [DOI] [PubMed] [Google Scholar]
- 32.Wang, C., S. R. Wesener, H. Zhang, and Y.-Q. Cheng. 2009. An FAD-dependent pyridine nucleotide-disulfide oxidoreductase is involved in disulfide bond formation in FK228 anticancer depsipeptide. Chem. Biol. 16:585-593. [DOI] [PubMed] [Google Scholar]
- 33.Wei, M. Q., et al. 2007. Facultative or obligate anaerobic bacteria have the potential for multimodality therapy of solid tumours. Eur. J. Cancer 43:490-496. [DOI] [PubMed] [Google Scholar]
- 34.Yoo, C. B., and P. A. Jones. 2006. Epigenetic therapy of cancer: past, present and future. Nat. Rev. Drug Discov. 5:37-50. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.