Abstract
A substantial proportion of infections caused by drug-resistant Gram-negative bacteria (GNB) in community and health care settings are recognized to be caused by evolutionarily related GNB strains. Their global spread has been suggested to occur due to human activities, such as food trade and travel. These multidrug-resistant GNB pathogens often harbor mobile drug resistance genes that are highly conserved in their sequences. Because they appear across different GNB species, these genes may have origins other than human pathogens. We hypothesized that saprophytes in common human food products may serve as a reservoir for such genes. Between July 2007 and April 2008, we examined 25 batches of prepackaged retail spinach for cultivatable GNB population structure by 16S rRNA gene sequencing and for antimicrobial drug susceptibility testing and the presence of extended-spectrum beta-lactamase (ESBL) genes. We found 20 recognized GNB species among 165 (71%) of 231 randomly selected colonies cultured from spinach. Twelve strains suspected to express ESBLs based on resistance to cefotaxime and ceftazidime were further examined for blaCTX-M and blaTEM genes. We found a 712-bp sequence in Pseudomonas teessidea that was 100% identical to positions 10 to 722 of an 876-bp blaCTX-M-15 gene of an E. coli strain. Additionally, we identified newly recognized ESBL blaRAHN-2 sequences from Rahnella aquatilis. These observations demonstrate that saprophytes in common fresh produce can harbor drug resistance genes that are also found in internationally circulating strains of GNB pathogens; such a source may thus serve as a reservoir for drug resistance genes that ultimately enter pathogens to affect human health.
Drug-resistant human infections caused by pathogenic Gram-negative bacteria (GNB) that contaminate fresh produce are becoming increasingly common in the United States (18, 28). One example is the large multistate epidemic of a strain of drug-resistant E. coli O157:H7 spread by contaminated spinach that occurred in 2006 (30). While food products are well recognized to disperse drug-resistant GNB pathogens that cause enteric illnesses, the modes of geographic dispersion of drug-resistant GNB pathogens that cause extraintestinal infections are less obvious. A sudden increase of extraintestinal infections caused by Enterobacteriaceae strains expressing New Delhi metallo-beta-lactamase (NDM-1) between 2008 and 2009 in the United Kingdom has been suggested to have occurred due to international travel to South Asia (15). The international spread of Escherichia coli serogroup O25 belonging to multilocus sequence type (MLST) ST131 has been suggested to occur due to food trade and travel; many of these ST131 strains express an extended-spectrum beta-lactamase (ESBL) encoded by blaCTX-M on a plasmid (21, 22, 26). A clonal group of E. coli strains belonging to ST69 that cause urinary tract infections (UTI), harboring the class 1 integron gene cassette configuration dfrA17-aadA5, has been isolated from clinical sources and food sources from all over the world (1, 13).
These so-called “international clones” of recognized pathogens all carry multiple drug resistance genes on mobile elements. These mobile elements are horizontally transferred to other pathogenic bacterial strains and even across bacterial species, which could cause a recipient strain to become multidrug resistant. Such a mechanism could suddenly increase the prevalence of drug-resistant infections in a community or institution without any selective pressures of antimicrobial drug use. However, all of these pathogens at some point acquire these mobile drug resistance genes horizontally from other sources. It has been suggested, for example, that blaCTX-M may have originated from an environmental saprophytic Kluyvera spp. (6). CTX-M-type genes have rapidly become the most common ESBL genes found in E. coli and Klebsiella pneumoniae isolates causing community- and hospital-acquired infections worldwide (23, 24). TEM-type ESBL genes are also found in a wide variety of environmental saprophytes (10).
Although it is recognized that saprophytes can harbor drug resistance genes, the extent to which they contribute to human drug-resistant infections is not evident. The observation that drug resistance genes with 100% identical nucleic acid sequences are found in different species of pathogenic GNB isolated from food sources all over the world suggests that they have common sources and are dispersed by the international food trade (22, 29). Therefore, the microbiota of common food products humans consume may serve as a reservoir for some of the drug resistance genes we observe in human pathogens. Because spinach is usually eaten uncooked in the United States and because it is increasingly implicated in outbreaks of drug-resistant GNB infections (28, 30), we sought to examine saprophytic bacteria of commercially obtained spinach for drug-resistant GNB and genes responsible for drug resistance.
MATERIALS AND METHODS
Spinach sampling.
Organic and nonorganic “baby” spinach packages were obtained from three local retail supermarkets in or around Berkeley, California. We obtained 6 brands of spinach, which were distributed by 6 different California producers; the source farms were not indicated on the packages. They are all sold in Bay Area supermarkets and are also distributed nationally. Organic spinach is defined by the USDA as spinach grown with no antibiotics or pesticides (12). The packages were purchased during two different seasons—summer (May 2007 to August 2007) and spring (March 2008). To avoid the effect of potential contamination from human handlers or consumers, we analyzed only prewashed and ready-to-eat packaged (boxes or bags) spinach. The spinach samples were stored at 4°C until processed, within 24 h of purchase.
Quantification and isolation of bacteria in spinach.
For each spinach package, 25 g of spinach was weighed and placed in a UV-pretreated polyethylene bag containing 50 ml of phosphate-buffered saline (PBS; pH 7.4). The spinach was incubated in PBS for 30 to 60 min at room temperature after brief kneading. The PBS wash was then transferred to a 50-ml conical tube and centrifuged at 12,000 × g for 5 min at room temperature. The resulting pellet was resuspended in 2 ml of PBS, and 1 ml of it was saved in a 10% glycerol stock. The rest of the suspension was serially diluted in PBS by 10 logs and plated onto MacConkey agar plates to select for GNB. The plates were incubated for 24 to 48 h at 37°C. Plates containing 10 to 200 CFU were analyzed further. The number of CFU per gram of spinach was determined for each batch of spinach, and single isolated colonies with different morphologies were randomly selected from the plates for further analyses. Up to 12 colonies were picked from each plate, as determined by a rarefaction curve analysis generated in triplicate from 3 different spinach batches (see below). The colonies were saved on tryptic soy agar (TSA) slants and used for DNA extraction.
Bacterial DNA extraction.
We extracted DNA from the PBS bacterial suspensions for integron analysis and from individual bacterial colonies for 16S rRNA gene sequence and ESBL gene analyses. DNA was extracted from 650 μl of total PBS suspensions from all 25 batches. In addition, individual bacterial colonies were randomly picked from MacConkey agar plates. Each colony was suspended in 200 μl of sterile double-distilled water in a 1.5-ml Eppendorf tube and then vortexed. A freeze-thaw method of DNA extraction was used. The bacterial suspensions were boiled for 15 min and frozen overnight at −80°C. They were again boiled for 15 min before being used for PCR amplification.
PCR amplification of class 1 and 2 integron, 16S rRNA gene, and beta-lactamase gene (blaCTX-M and blaTEM) sequences.
PCR amplification was carried out with 2 μl of DNA template in 25 μl of PCR mixture. Each reaction mixture contained 0.2 mM deoxynucleoside triphosphates (dNTPs), 1 U of Taq polymerase (New England BioLabs [NEB]), 1× Taq reaction buffer, and 1 μM primers. The 16S rRNA gene PCRs were carried out with the primers 16s8F/16s806R18 as published previously (Table 1). Class 1 integron (intI1) and class 2 integron (intI2) genes were amplified from the total spinach batch DNA with primers Int1F/IntI1R19 and RB201/RB202 (Table 1), respectively, and the class 1 integron gene cassette was amplified with primers GC1_RLF/GC1_RLR (Table 1). Gram-negative bacterial colonies that were resistant to cefotaxime and ceftazidime were tested for beta-lactamase genes by PCR amplification of blaCTX-M, blaCTX-M-1, and blaTEM with primers CTX-M/F/CTX-M/R, CTX-M-1-MP-F/CTX-M-1-MP-R, and T1/T2, respectively (Table 1). The PCR conditions were modified from those previously published (3, 5, 11, 16, 19, 31, 32) (Table 1). The PCR products were electrophoresed on a 1 to 1.5% agarose gel. The gel was stained with ethidium bromide and visualized under UV transillumination. As a positive control for the 16S rRNA genes, we used E. coli ATCC 25922 DNA. PCR controls for blaCTX-M, blaCTX-M-1, and blaTEM DNA were obtained from a clinical E. coli strain (608.35) that contained the three genes, kindly provided by Satowa Suzuki of the National Institute of Infectious Diseases in Tokyo, Japan.
TABLE 1.
PCR primer sequences and conditions used in this study
| Target | Primers | Sequences (5′-3′) | PCR conditions | Expected amplicon size (bp) | Reference |
|---|---|---|---|---|---|
| 16S rRNA genes | 16s8F/16s806R | AGAGTTTGATCCTGGCTCAG/GGACTACCAGGGTATCTAATCC | 94°C, 5 min; 30 cycles of 94°C, 30 s, 62°C, 30 s, 72°C, 90 s | 800 | 19 |
| Integrase 1 gene (intl1) | Int1F/Int1R | CCTCCCGCACGATGATC/TCCACGCATCGTCAGGC | 94°C, 5 min; 33 cycles of 94°C, 45 s, 64°C, 30 s, 72°C, 1 min | 280 | 16 |
| Integrase 2 gene (intI2) | RB201/RB202 | GCAAACGCAAGCATTCATTA/ACGGATATGCGACAAAAAGG | 300 | 3 | |
| Class 1 gene cassette | GC1_RLF/GC1_RLR | GGCATCCAAGCAGCAAG/AAGCAGACTTGACCTGA | 95°C, 5 min; 35 cycles of 94°C, 1 min, 55°C, 1 min, 72°C, 1.5 min | Variable | 5 |
| CTX-M (blaCTX-M) | CTX-M/F/CTX-M/R | TTTGCGATGTGCAGTACCAGTAA/CTCCGCTGCCGGTTTTATC | 95°C, 5 min; 35 cycles of 94°C, 1 min, 55°C, 1 min, 72°C, 1.5 min | 500 | 11 |
| CTX-M-1 (blaCTX-M-1) | CTX-M-1-MP-F/CTX-M-1-MP-R | AAAAATCACTGCGCCAGTTC/AGCTTATTCATCGCCACGTT | 415 | 31 | |
| TEM (blaTEM) | T1/T2 | CCGTGTCGCCCTTATTCC/AGGCACCTATCTCAGCGA | 800 | 32 |
Sequencing analysis of 16S rRNA, blaCTX-M, and blaTEM genes.
Each PCR product that showed an electrophoretic band of an expected size was cleaned up by using ExoSAP-IT according to the manufacturer's instructions (USB Corporation). The purified PCR product was diluted 100 times and used for direct sequencing. Sequencing was done on an Applied Biosystems 3730 DNA analyzer (Applied Biosystems, Foster City, CA) at the University of California Berkeley DNA Sequencing Facility. DNA sequences of 600 bp or more for the 16S rRNA gene and 400 bp or more for blaCTX-M and blaTEM were visually inspected, aligned, and compared against sequences in GenBank by using BLAST (National Center for Biotechnology Information) or in the Greengenes server using the “Align” tool (greengenes.lbl.gov). Genus and species were determined by the criteria of >98% sequence identity for species and >95% sequence identity for genus (2). To identify all of the bacterial genera present in a batch of spinach, we randomly and sequentially picked individual colonies of distinct morphologies on MacConkey plates for 16S rRNA gene sequence analysis until no new genera could be identified (rarefaction curve analysis).
Antimicrobial susceptibility testing of bacterial colonies.
The drug susceptibility of a selected number of colonies on MacConkey agar plates was assessed with the Dade-Behring MicroScan Gram-negative breakpoint panel (Dade Behring MicroScan, Inc., West Sacramento, CA), following the manufacturer's protocol. More than 50% of the colonies from each species that we identified were randomly selected for drug susceptibility testing. These colonies were selected from growths from all 25 spinach batches. Resistance and susceptibility to the panel of antimicrobial drugs were determined with the Clinical and Laboratory Standards Institute standards (9). E. coli ATCC 29522 was used for quality control. Intermediate-resistant bacterial colonies were considered susceptible in this study.
Statistical analysis.
An independent sample t test was conducted to compare differences in mean CFU counts per gram of spinach.
Nucleotide sequence accession numbers.
Partial coding sequences of blaCTX-M PCR products that were less than 100% identical to previously identified genes have been deposited in GenBank under the following accession numbers: HQ339919, HQ339920, HQ339921, HQ339922, HQ339923, and HQ339924.
RESULTS
Sample collection and CFU counts in organic and nonorganic spinach.
Twenty-five spinach batches, purchased between July 2007 and April 2008, were examined. Among these, 12 (48%) were organic and 13 (52%) were nonorganic. The collection was spread out over two seasons, with 21 (84%) spinach batches collected in the summer and 5 (16%) in the spring. A mean of 1.39 × 107 CFU per gram of spinach (standard deviation [SD], ±1.44 × 107) was found for all samples. There was a significant difference in the CFU counts found in organic spinach (mean ± SD, 2.07 × 107 ± 1.53 × 107 CFU/g) and nonorganic spinach (7.51 × 106 ± 1.05 × 107) (P = 0.027, two-sample t test).
Analysis of 16S rRNA genes.
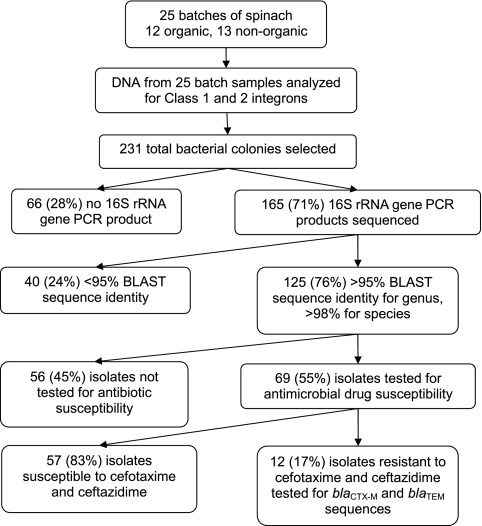
Rarefaction curve analyses showed that picking more than 12 colonies per spinach batch did not add to the number of bacterial genera found in the spinach samples. A total of 231 bacterial colonies were analyzed, and 165 (71%) were successfully sequenced for their 16S rRNA genes (Fig. 1). Of the 165 isolates sequenced, 125 (76%) could be identified to the species and genus level based on published criteria for 16S rRNA gene sequencing (2).
FIG. 1.
Flowchart of the spinach saprophyte analysis.
The majority of the colonies belonged to Pseudomonas spp. (43 [34%]) and Pantoea spp. (30 [24%]). The distribution by genus and species between organic and nonorganic spinach varied slightly (Table 2). Organic spinach contained 8 genera and 15 species, compared to 6 genera and 14 species in nonorganic spinach. Little difference in genus and species distribution was found between the two collection periods. The highest diversity in species was found in the collection from late August of 2007.
TABLE 2.
Prevalences of genera and speciesa found in organic and nonorganic spinach samples
| Genus | Species | No. (%) of isolates in spinach from indicated source |
No. (%) of isolates in total samples | |
|---|---|---|---|---|
| Organic | Nonorganic | |||
| Acinetobacter | rhizosphaerae | 1 (1.4) | 0 | 1 (0.8) |
| Enterobacter | aerogenes | 0 | 1 (1.8) | 1 (0.8) |
| amnigenus | 2 (2.8) | 2 (3.7) | 4 (3.2) | |
| kobei | 2 (2.8) | 0 | 2 (1.6) | |
| ludwigii | 1 (1.4) | 0 | 1 (0.8) | |
| ND | 3 (4.2) | 0 | 3 (2.4) | |
| Erwinia | persicina | 6 (8.4) | 11 (20.3) | 17 (13.6) |
| rhapontici | 0 | 2 (3.7) | 2 (1.6) | |
| Pantoea | agglomerans | 11 (6.6) | 15 (27.7) | 26 (20.8) |
| ananatis | 3 (4.2) | 1 (1.8) | 4 (3.2) | |
| Pseudomonas | fragi | 0 | 1 (1.8) | 1 (0.8) |
| libanensis | 1 (1.4) | 0 | 1 (0.8) | |
| orientalis | 2 (2.8) | 1 (1.8) | 3 (2.4) | |
| putida | 20 (28.1) | 8 (14.8) | 28 (22.4) | |
| reactans | 1 (1.4) | 0 | 1 (0.8) | |
| rhodesiae | 0 | 1 (1.8) | 1 (0.8) | |
| syringae | 1 (1.4) | 0 | 1 (0.8) | |
| teessidea | 1 (1.4) | 1 (1.8) | 2 (1.6) | |
| ND | 3 (4.2) | 2 (3.7) | 5 (4) | |
| Rahnella | aquatilis | 7 (9.8) | 3 (5.5) | 10 (8) |
| Rhizobium | ND | 1 (1.4) | 0 | 1 (0.8) |
| Serratia | fonticola | 5 (7.0) | 3 (5.5) | 8 (6.4) |
| proteamaculans | 0 | 2 (3.7) | 2 (1.6) | |
| Total | 71 (100.0) | 54 (100.0) | 125 (100.0) | |
The criteria for genus and species identification were BLAST identities above 95% and 98%, respectively. ND, not determined.
Antimicrobial drug resistance of Gram-negative bacteria in spinach.
Of 125 sequenced isolates, 69 (55%) were tested against 26 different antimicrobial agents (Fig. 1 and Tables 3 and 4). All of the individual colonies were resistant to more than one agent. All 27 Pseudomonas sp. isolates were resistant to 19 drugs. The 15 Pantoea agglomerans isolates showed resistance to 10 antimicrobial agents. Only 2 (3%) Pseudomonas isolates were resistant to kanamycin (MIC ≥ 4 μg/ml), and only 1 (1%) Rahnella aquatilis isolate was resistant to levofloxacin (MIC ≥ 4 μg/ml). Among the 41 (59%) isolates resistant to ampicillin (MIC ≥ 16 μg/ml), 28 (40%) were resistant to ampicillin-sulbactam (MIC ≥ 16/8 μg/ml) and to ticarcillin-K clavulanate (MIC ≥ 64/2 μg/ml). Similarly, 23 (33%) were resistant to piperacillin (MIC ≥ 64 μg/ml), and 9 (16%) were resistant to piperacillin-tazobactam (MIC ≥ 64/4 μg/ml). Six (8%) of the isolates were resistant to meropenem (MIC ≥ 8 μg/ml). Trimethoprim-sulfamethoxazole (MIC ≥ 2/38 μg/ml) resistance was seen in 18 (26%) isolates.
TABLE 3.
Frequency of resistance to various drugs among Gram-negative bacteria across genera and species isolated from organic and nonorganic spinach
| Organism (no. of isolates) | No. of isolates resistant toa: |
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Aminoglycoside |
Penicillin |
Penicillin/beta-lactamase inhibitor |
Fluoroquinolone |
Carbapenem |
Pol (Cl) | T/S | |||||||||||
| K | Ak | Gm | To | P | Am | Pi | A/S | P/T | Tim | Cp | Lvx | Mxf | Mer | Imp | |||
| Acinetobacter (1) | |||||||||||||||||
| rhizosphaerae (1) | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Enterobacter (5) | |||||||||||||||||
| aerogenes (1) | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 0 | 0 |
| amnigenus (2) | 0 | 0 | 0 | 0 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| kobei (1) | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ludwigii (1) | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Erwinia (7) | |||||||||||||||||
| persicina (6) | 0 | 0 | 0 | 0 | 6 | 2 | 2 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| rhapontici (1) | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Pantoea (18) | |||||||||||||||||
| agglomerans (15) | 0 | 0 | 0 | 0 | 15 | 4 | 6 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ananatis (3) | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 |
| Pseudomonas (27) | |||||||||||||||||
| fragi (1) | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| libanensis (1) | 1 | 0 | 0 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| orientalis (3) | 0 | 0 | 0 | 0 | 3 | 3 | 0 | 3 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| putida (16) | 0 | 0 | 0 | 0 | 16 | 15 | 3 | 15 | 2 | 15 | 0 | 0 | 0 | 2 | 1 | 0 | 11 |
| rhodesiae (1) | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| syringae (1) | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| teessidea (1) | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Species not determined (3) | 1 | 0 | 0 | 0 | 3 | 2 | 1 | 2 | 0 | 2 | 0 | 0 | 0 | 1 | 1 | 0 | 2 |
| Rahnella (7) | |||||||||||||||||
| aqualitis | 0 | 0 | 0 | 0 | 6 | 4 | 3 | 3 | 2 | 3 | 0 | 1 | 0 | 2 | 2 | 0 | 0 |
| Serratia (4) | |||||||||||||||||
| fonticola (3) | 0 | 0 | 0 | 0 | 3 | 3 | 2 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 |
| protemaculans (1) | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| TOTAL | 2 | 0 | 0 | 0 | 68 | 41 | 23 | 28 | 9 | 28 | 0 | 1 | 0 | 6 | 5 | 6 | 18 |
Pol, polymyxin; Cl, colistin; T/S, trimethoprim-sulfamethoxazole; K, kanamycin; Ak, amikacin; Gm, gentamicin; To, tobramycin; P, penicillin; Am, ampicillin; Pi, piperacillin; A/S, ampicillin/sulbactam; P/T, piperacillin-tazobactam; Tim, ticarcillin/K clavulanate; Cp, ciprofloxacin (expanded spectrum); Lvx, levofloxacin (broad spectrum); Mxf, moxifloxacin (broad spectrum); Mer, meropenem; Imp, imipenem.
TABLE 4.
Antimicrobial drug resistance to cephalosporins and monobactam among Gram-negative bacteria across genera and species in spinach
| Organism (no. of isolates) | No. of isolates resistant toa: |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Cephalosporin of indicated class |
Monobactam (Azt) | ||||||||
| Narrow spectrum |
Expanded spectrum |
Broad spectrum |
|||||||
| Cfz | Cf | Ctn | Crm | Cft | Caz | Cax | Cpe | ||
| Acinetobacter (1) | |||||||||
| rhizosphaerae (1) | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 1 |
| Enterobacter (5) | |||||||||
| aerogenes (1) | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 0 | 0 |
| amnigenus (2) | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 2 |
| kobei (1) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ludwigii (1) | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 1 |
| Erwinia (7) | |||||||||
| persicina (6) | 3 | 4 | 0 | 2 | 0 | 0 | 0 | 0 | 2 |
| rhapontici (1) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Pantoea (18) | |||||||||
| agglomerans (15) | 8 | 11 | 0 | 4 | 1 | 1 | 0 | 0 | 3 |
| ananatis (3) | 1 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Pseudomonas (27) | |||||||||
| fragi (1) | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| libanensis (1) | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 |
| orientalis (3) | 3 | 3 | 3 | 3 | 3 | 0 | 1 | 0 | 3 |
| putida (16) | 16 | 15 | 15 | 15 | 8 | 5 | 4 | 5 | 14 |
| rhodesiae (1) | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 1 |
| syringae (1) | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| teessidea (1) | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 |
| Species not determined (3) | 3 | 2 | 2 | 2 | 2 | 0 | 1 | 1 | 2 |
| Rahnella (7) | |||||||||
| aqualitis | 5 | 6 | 1 | 4 | 3 | 3 | 0 | 2 | 2 |
| Serratia (4) | |||||||||
| fonticola (3) | 3 | 3 | 0 | 3 | 2 | 2 | 1 | 1 | 3 |
| protemaculans (1) | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 0 | 1 |
| Total | 50 | 58 | 26 | 41 | 23 | 14 | 10 | 10 | 38 |
Cfz, cefazolin; Cf, cephalothin; Ctn, cefotetan; Crm, cefuroxime; Cft, cefotaxime; Caz, ceftazidime; Cax, ceftriaxone; Cpe, cefepime; Azt, aztreonam.
Cephalosporin and aztreonam resistance was assessed separately (Table 4). Most isolates (58) were resistant to cephalothin (MIC ≥ 8 μg/ml), a narrow-spectrum cephalosporin, but fewer (26) were resistant to cefotetan (MIC ≥ 32 μg/ml), an expanded-spectrum cephalosporin. The resistance to broad-spectrum cephalosporins varied: 14 (20%) isolates were resistant to ceftazidime (MIC ≥ 16 μg/ml), 23 (33%) to cefotaxime (MIC ≥ 32 μg/ml), and 10 (14%) to ceftriaxone (MIC ≥ 32 μg/ml). Resistance to cefepime (MIC ≥ 32 μg/ml) was observed in 10 (14%) isolates. Finally, 38 (55%) isolates were resistant to aztreonam (MIC ≥ 16 μg/ml).
For all of the drugs, the difference in frequency of resistance between Gram-negative bacteria of organic versus nonorganic spinach source was not statistically significant. There was no statistically significant difference in antimicrobial resistance in the isolates collected during summer 2007 or spring 2008 as well.
Class 1 and class 2 integrons and ESBL genes blaCTX-M, blaCTX-M-1, and blaTEM.
Class 1 integron was detected in DNA extracts from 3 spinach batch samples. However, only one of these contained an insert DNA, which was 98 bp in length. This sequence did not match any recognized drug-resistance gene sequences. Class 2 integrons were not found in any of the 25 batches. To rule out the possible effect of PCR inhibitors in the total spinach wash, we spiked the wash with integron DNA at a concentration equivalent to 1 bacterium. The PCR assay was able to amplify the DNA.
Of 12 isolates (4 Pseudomonas putida, 1 Pseudomonas teessidea, 3 Rahnella aquatilis, 2 Serratia fonticola, 1 Serratia proteamaculans, and 1 Pantoea agglomerans) that showed resistance to both cefotaxime and ceftazidime, 10 tested positive by PCR primers designed to amplify CTX-M-type beta-lactamase genes. None of the 12 samples showed a PCR product for blaTEM. The PCR-amplified sequences were compared against the NCBI database (Table 5). Of the 10 isolates, 2 gave PCR products with the blaCTX-M primers whose amplicon sequences did not match with any entry in the database (Serratia proteamaculans and Pantoea agglomerans). A 479-bp sequence from one Pseudomonas putida strain (S24M) and a 444-bp sequence from another P. putida strain (MS40-16) were 100% identical to animal and human E. coli blaCTX-M-15 sequences deposited in the NCBI database (Table 5). One P. teessidea strain had a 712-bp sequence that was 100% identical to positions 10 to 722 of an 876-bp blaCTX-M-15 sequence (GenBank sequence accession no. HQ157357.1) from an E. coli strain. The other strains, 3 Rahnella aquatilis isolates and 2 Serratia fonticola isolates, contained sequences that were 99% and 98% identical to a newly found extended-spectrum beta-lactamase gene, blaRAHN-2, reported by others to be found in vegetables (27), and beta-lactamase FONA-5 from an unidentified source (GenBank sequence accession no. AJ251243.1), respectively (Table 5).
TABLE 5.
Genetic analysis of putative ESBL-producing spinach saprophytes
| Strain | Spinach type | Size (bp) of sequenced DNA fragment (accession no.) | Position (bp) of sequence on matched sequence | Matched sequence(s) in NCBI database (organism of origin, sequence length [bp], accession no.), % match |
|---|---|---|---|---|
| Pseudomonas putida | ||||
| S24-M | Organic | 479 | 47-525 | CTX-M-15 (Salmonella enterica, 619, HQ589352.1), 100 |
| MS40-16 | Nonorganic | 444 | 87-526 | CTX-M-15 (Salmonella enterica, 619, HQ589352.1), 100 |
| Pseudomonas teessidea | ||||
| MS40-13 | Nonorganic | 712 | 10-722 | CTX-M-15 (E. coli, 876, HQ157357.1), 100 |
| Rahnella aquatilis | ||||
| MS32-6A | Organic | 523 (HQ339921) | 67-552 | RAHN-2 (Rahnella sp. 366, 655, GU584921.1), 99 |
| MS32-6C | Organic | 523 (HQ339922) | 67-552 | RAHN-2 (Rahnella sp. 366, 655, GU584921.1), 99 |
| MS37-1 | Organic | 489 (HQ339923) | 250-737 | RAHN-2 (Rahnella sp. 344, 887, GU584905.1), 99 |
| Serratia fonticola | ||||
| MS23-3 | Nonorganic | 522 (HQ339919) | 217-738 | FONR-5 (transcriptional regulator) and FONA-5 (beta-lactamase) (Serratia fonticola, 888, AJ251243.1), 98 |
| MS26-1 | Nonorganic | 523 (HQ339920) | 216-738 | FONR-5 (transcriptional regulator) and FONA-5 (beta-lactamase) (Serratia fonticola, 888, AJ251243.1), 98 |
DISCUSSION
In this study, we sought to characterize the population structure, drug susceptibility profile, and drug resistance genes of GNB saprophytes in spinach. To our surprise, we found several well-recognized and common CTX-M sequences as well as new ESBL gene sequences that have been previously reported from GNB pathogens from human clinical sources and GNB pathogens from food products from other parts of the world. We found partial blaCTX-M sequences in strains of Pseudomonas putida and Pseudomonas teessidea that were identical to the CTX-M-15 gene sequences deposited in the NCBI database (Table 5). These NCBI sequences were identified in E. coli strains from human clinical samples and animal food products. We found no other blaCTX-M-15 sequences deposited in NCBI that were obtained from saprophytes in fresh produce items. One study of fresh produce items in France found blaRAHN-2, a new ESBL gene, in Rahnella aquatilis from fruits and vegetables (27). In our study, we also found new beta-lactamase genes similar to blaRAHN-2 in 3 Rahnella aquatilis samples, as well as genes similar to fonA-5 in two Serratia fonticola strains.
The design of this study may have underestimated the total number of GNB species represented among the spinach microbiota because we focused on analyzing only bacteria cultivated on MacConkey agar plates. Our original rationale for using MacConkey agar plates was that we wanted to select for E. coli in spinach since there had been a national epidemic of E. coli O157:H7 shortly before this project was undertaken. Nevertheless, the main findings—the identification of ESBL gene sequences in saprophytes—are not affected by this study limitation.
CTX-M beta-lactamase was first identified in Japan in 1986 (20). Since 2000, CTX-M-producing E. coli strains have began to emerge in different regions of the world, often associated with community-acquired UTI (8, 23). More than 80 CTX-M beta-lactamases have been described to date (23). Among these, the CTX-M-15 type has been reported from all continents except Antarctica (22). It was first described in an E. coli strain from India (14). An international E. coli clone belonging to serotype O25 and MLST type ST131 carrying blaCTX-M-15 is now reported from many regions of the world, from clinical sources, companion animals, and food-producing animals (17, 21, 22, 25, 26, 29). However, blaCTX-M-15 has not been previously identified from any saprophytic organisms from any food source. It may not have been sought in saprophytes from such sources before.
Integrons play an important role in the dissemination of drug-resistant genes across bacterial organisms (7). Class 1 integrons are found in up to 59% of Gram-negative bacteria causing clinical infections (7). A recent study of drug resistance in oxidase-positive organisms in fresh produce in Canada found class 1, 2, and 3 integrase genes in Pseudomonas fluorescens from alfalfa sprouts (4). In our study, we found Class 1 integrase sequences in 3 of the spinach batches, and only one had an insert, which did not encode any drug resistance.
We did not identify any recognized pathogens in the sampled spinach, but saprophytes on food can certainly get introduced into hospitals. If they are resistant, they could conceivably spread their mobile drug resistance determinants to pathogens prevalent in hospital settings. Our findings provide evidence that microbiota found in fresh produce—organic or nonorganic—could serve as a potential reservoir for mobile drug resistance genes. The identification of saprophytes encoding ESBLs is particularly worrisome, given the worldwide dispersion of GNB pathogens that harbor ESBL genes.
Acknowledgments
We thank Satowa Suzuki of the National Institute of Infectious Diseases, Japan, for kindly providing the universal CTX-M and TEM primers as well as the positive-control strains for the CTX-M and TEM genes. We thank Remi Ajiboye, Owen Solberg, Olivera Marjanovic, Kate Williams, Camila Barcia, and Sheila Adams-Sapper for their technical support and their input in discussions.
This study was funded in part by a grant from the National Institutes of Health (grant AI059523).
Footnotes
Published ahead of print on 7 January 2011.
REFERENCES
- 1.Ajiboye, R. M., et al. 2009. Global spread of mobile antimicrobial drug resistance determinants in human and animal Escherichia coli and Salmonella strains causing community-acquired infections. Clin. Infect. Dis. 49:365-371. [DOI] [PubMed] [Google Scholar]
- 2.Backhed, F., R. E. Ley, J. L. Sonnenburg, D. A. Peterson, and J. I. Gordon. 2005. Host-bacterial mutualism in the human intestine. Science 307:1915-1920. [DOI] [PubMed] [Google Scholar]
- 3.Barlow, R. S., J. M. Pemberton, P. M. Desmarchelier, and K. S. Gobius. 2004. Isolation and characterization of integron-containing bacteria without antibiotic selection. Antimicrob. Agents Chemother. 48:838-842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bezanson, G. S., R. MacInnis, G. Potter, and T. Hughes. 2008. Presence and potential for horizontal transfer of antibiotic resistance in oxidase-positive bacteria populating raw salad vegetables. Int. J. Food Microbiol. 127:37-42. [DOI] [PubMed] [Google Scholar]
- 5.Bissonnette, L., and P. H. Roy. 1992. Characterization of In0 of Pseudomonas aeruginosa plasmid pVS1, an ancestor of integrons of multiresistance plasmids and transposons of gram-negative bacteria. J. Bacteriol. 174:1248-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonnet, R. 2004. Growing group of extended-spectrum beta-lactamases: the CTX-M enzymes. Antimicrob. Agents Chemother. 48:1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cambray, G., A. M. Guerout, and D. Mazel. 2010. Integrons. Annu. Rev. Genet. 44:141-166. [DOI] [PubMed] [Google Scholar]
- 8.Carattoli, A. 2008. Animal reservoirs for extended spectrum beta-lactamase producers. Clin. Microbiol. Infect. 14(Suppl. 1):117-123. [DOI] [PubMed] [Google Scholar]
- 9.Clinical and Laboratory Standards Institute. 2008. Performance standards for antimicrobial susceptibility testing; 18th informational supplement. CLSI M100-S18. CLSI, Wayne, PA.
- 10.D'Costa, V. M., E. Griffiths, and G. D. Wright. 2007. Expanding the soil antibiotic resistome: exploring environmental diversity. Curr. Opin. Microbiol. 10:481-489. [DOI] [PubMed] [Google Scholar]
- 11.Edelstein, M., M. Pimkin, I. Palagin, I. Edelstein, and L. Stratchounski. 2003. Prevalence and molecular epidemiology of CTX-M extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae in Russian hospitals. Antimicrob. Agents Chemother. 47:3724-3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.FACT: Food, Agriculture, Conservation, and Trade Act of 1990. Organic Foods Production Act of 1990, Title XXI. Public Law 109-97; 25:1-21. (Amended in 2005.) [Google Scholar]
- 13.Johnson, J. R., et al. 2005. Distribution and characteristics of Escherichia coli clonal group A. Emerg. Infect. Dis. 11:141-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karim, A., L. Poirel, S. Nagarajan, and P. Nordmann. 2001. Plasmid-mediated extended-spectrum beta-lactamase (CTX-M-3 like) from India and gene association with insertion sequence ISEcp1. FEMS Microbiol. Lett. 201:237-241. [DOI] [PubMed] [Google Scholar]
- 15.Kumarasamy, K. K., et al. 2010. Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect. Dis. 10:597-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lévesque, C., and P. H. Roy. 1993. PCR analysis of integrons, p. 590-594. In D. H. Persing, T. F. Smith, F. C. Tenover, and T. J. White (ed.), Diagnostic molecular microbiology: principles and applications. ASM Press, Washington, DC.
- 17.Li, J., et al. 2010. Dissemination of cefotaxime-M-producing Escherichia coli isolates in poultry farms, but not swine farms, in China. Foodborne Pathog. Dis. 7:1387-1392. [DOI] [PubMed] [Google Scholar]
- 18.Lynch, M. F., R. V. Tauxe, and C. W. Hedberg. 2009. The growing burden of foodborne outbreaks due to contaminated fresh produce: risks and opportunities. Epidemiol. Infect. 137:307-315. [DOI] [PubMed] [Google Scholar]
- 19.Martinez-Freijo, P., et al. 1998. Class I integrons in Gram-negative isolates from different European hospitals and association with decreased susceptibility to multiple antibiotic compounds. J. Antimicrob. Chemother. 42:689-696. [DOI] [PubMed] [Google Scholar]
- 20.Matsumoto, Y., F. Ikeda, T. Kamimura, Y. Yokota, and Y. Mine. 1988. Novel plasmid-mediated beta-lactamase from Escherichia coli that inactivates oxyimino-cephalosporins. Antimicrob. Agents Chemother. 32:1243-1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oteo, J., M. Perez-Vazquez, and J. Campos. 2010. Extended-spectrum β-lactamase producing Escherichia coli: changing epidemiology and clinical impact. Curr. Opin. Infect. Dis. 23:320-326. [DOI] [PubMed] [Google Scholar]
- 22.Peirano, G., and J. D. Pitout. 2010. Molecular epidemiology of Escherichia coli producing CTX-M beta-lactamases: the worldwide emergence of clone ST131 O25:H4. Int. J. Antimicrob. Agents 35:316-321. [DOI] [PubMed] [Google Scholar]
- 23.Pitout, J. D. 2010. Infections with extended-spectrum beta-lactamase-producing Enterobacteriaceae: changing epidemiology and drug treatment choices. Drugs 70:313-333. [DOI] [PubMed] [Google Scholar]
- 24.Pitout, J. D., and K. B. Laupland. 2008. Extended-spectrum beta-lactamase-producing Enterobacteriaceae: an emerging public-health concern. Lancet Infect. Dis. 8:159-166. [DOI] [PubMed] [Google Scholar]
- 25.Rodriguez, I., et al. 2009. Extended-spectrum {beta}-lactamases and AmpC {beta}-lactamases in ceftiofur-resistant Salmonella enterica isolates from food and livestock obtained in Germany during 2003-07. J. Antimicrob. Chemother. 64:301-309. [DOI] [PubMed] [Google Scholar]
- 26.Rogers, B. A., H. E. Sidjabat, and D. L. Paterson. 2011. Escherichia coli O25b-ST131: a pandemic, multiresistant, community-associated strain. J. Antimicrob. Chemother. 66:1-14. [DOI] [PubMed] [Google Scholar]
- 27.Ruimy, R., D. Meziane-Cherif, S. Momcilovic, G. Arlet, A. Andremont, and P. Courvalin. 2010. RAHN-2, a chromosomal extended-spectrum class A beta-lactamase from Rahnella aquatilis. J. Antimicrob. Chemother. 65:1619-1623. [DOI] [PubMed] [Google Scholar]
- 28.Sivapalasingam, S., C. R. Friedman, L. Cohen, and R. V. Tauxe. 2004. Fresh produce: a growing cause of outbreaks of foodborne illness in the United States, 1973 through 1997. J. Food Prot. 67:2342-2353. [DOI] [PubMed] [Google Scholar]
- 29.Vincent, C., et al. 2010. Food reservoir for Escherichia coli causing urinary tract infections. Emerg. Infect. Dis. 16:88-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wendel, A. M., et al. 2009. Multistate outbreak of Escherichia coli O157:H7 infection associated with consumption of packaged spinach, August-September 2006: the Wisconsin investigation. Clin. Infect. Dis. 48:1079-1086. [DOI] [PubMed] [Google Scholar]
- 31.Woodford, N., E. J. Fagan, and M. J. Ellington. 2006. Multiplex PCR for rapid detection of genes encoding CTX-M extended-spectrum (beta)-lactamases. J. Antimicrob. Chemother. 57:154-155. [DOI] [PubMed] [Google Scholar]
- 32.Yagi, T., H. Kurokawa, N. Shibata, K. Shibayama, and Y. Arakawa. 2000. A preliminary survey of extended-spectrum beta-lactamases (ESBLs) in clinical isolates of Klebsiella pneumoniae and Escherichia coli in Japan. FEMS Microbiol. Lett. 184:53-56. [DOI] [PubMed] [Google Scholar]