Abstract
2,4-Diacetylphloroglucinol (2,4-DAPG), an antibiotic produced by Pseudomonas fluorescens, has broad-spectrum antibiotic activity, inhibiting organisms ranging from viruses, bacteria, and fungi to higher plants and mammalian cells. The biosynthesis and regulation of 2,4-DAPG in P. fluorescens are well described, but the mode of action against target organisms is poorly understood. As a first step to elucidate the mechanism, we screened a deletion library of Saccharomyces cerevisiae in broth and agar medium supplemented with 2,4-DAPG. We identified 231 mutants that showed increased sensitivity to 2,4-DAPG under both conditions, including 22 multidrug resistance-related mutants. Three major physiological functions correlated with an increase in sensitivity to 2,4-DAPG: membrane function, reactive oxygen regulation, and cell homeostasis. Physiological studies with wild-type yeast validated the results of the mutant screens. The chemical-genetic fitness profile of 2,4-DAPG resembled those of menthol, sodium azide, and hydrogen peroxide determined in previous high-throughput screening studies. Collectively, these findings indicate that 2,4-DAPG acts on multiple basic cellular processes.
The polyketide antibiotic 2,4-diacetylphloroglucinol (2,4-DAPG), produced by certain Pseudomonas spp., plays a key role in microbial interactions and plant defense in a wide variety of natural and agricultural ecosystems. 2,4-DAPG is notable because it has antifungal, antibacterial, antiviral, antihelminthic, and phytotoxic properties (5, 11, 16, 25, 28, 29). It also induces systemic resistance in plants (24), promotes exudation of amino acids from roots (43), has toxicity to mammalian cells (30), and is a potent inhibitor of cyst reactivation of protists (27). Even Staphylococcus aureus, a multidrug-resistant human pathogen, is sensitive to 2,4-DAPG (28).
2,4-DAPG has been extensively studied as a key determinant in the biological control activity of Pseudomonas fluorescens against seedling and root diseases caused by plant-pathogenic fungi, bacteria, and nematodes (9, 11, 12, 13, 29, 44). Especially notable is that 2,4-DAPG producers are responsible for take-all decline (TAD), a spontaneous remission in take-all disease incidence and severity induced by continuous wheat monoculture after a severe outbreak of take-all disease (22, 54). In the United States alone, hundreds of thousands of hectares of wheat are protected from take-all disease by 2,4-DAPG-mediated TAD.
Plant pathogens defend themselves against toxins and pesticides by at least three types of mechanisms: degradation of the toxin by enzymes such as hydrolases or acetyltransferases (37), modifications of the target by mutation (53), and/or export of the toxin via membrane transporters (14, 38). However, very few studies have addressed either how plant pathogens respond to exposure to natural antibiotics like 2,4-DAPG or the modes of action of these antibiotics (12), even though antibiosis is a major mechanism of biological control. For example, upon exposure to sublethal concentrations of the biocontrol antibiotic 1-hydroxyphenazine produced by Pseudomonas, Mycosphaerella graminicola exhibited increased activity of reactive oxygen scavenging enzymes and melanin biosynthesis (33). A mutant of Caenorhabditis elegans, tolerant to oxidative stress, had increased tolerance to pyocyanin due to increased catalase and superoxide dismutase activity (34). In Botrytis cinerea, exposure to the antibiotic phenazine-1-carboxamide (PCN) activated ABC transporters that transported the antibiotic out of the cell (46), and degradation of 2,4-DAPG occurred through the enzyme laccase, which required tannic acid, a plant phenolic compound, as a mediator (48). Finally, Schouten et al. (47) showed that 18 of 117 isolates of Fusarium oxysporum were able to metabolize 2,4-DAPG.
Despite the diversity of roles played by 2,4-DAPG in microbe-microbe and plant-microbe interactions in nature, the molecular mechanisms underlying its inhibition of eukaryotic and prokaryotic cells are poorly understood. As a strategy for discovering the cellular pathways affected in eukaryotes by 2,4-DAPG, we screened a genome-wide deletion library of Saccharomyces cerevisiae for sensitivity to the antibiotic. Our results indicate that 2,4-DAPG affects multiple basic cellular processes, including membrane function, reactive oxygen regulation, and cell homeostasis.
MATERIALS AND METHODS
Yeast strains, media, and chemicals.
The S. cerevisiae deletion library (Open Biosystems) was generated from strain BY4741 (MATa haploid: his3Δ1 leu2 Δmet15 Δura3 Δ0) by the Saccharomyces Genome Deletion Project (55). The yeast deletion library (4,786 unique open reading frames [ORFs]) was stored at −80°C and grown in yeast extract (10 g liter−1)-peptone (20 g liter−1)-dextrose (20 g liter−1) (YPD) medium at 30°C or on YPD agar. 2,4-DAPG (Toronto Research Chemicals Inc.) was dissolved in methanol (1 mg ml−1). Monoacetylphloroglucinol (MAPG), propidium iodide (PI), 2,7-dichlorofluorescin diacetate (DCFH-DA), dihydroethidine (DHE), and acridine orange (AO) (Sigma-Aldrich Co.) were dissolved in methanol, distilled water, dimethyl sulfoxide (DMSO), methanol, and DMSO, respectively. The AO stock (5 mg ml−1) was diluted 1,000-fold before use.
Screening for 2,4-DAPG sensitivity on solid medium.
All replications and inoculations were carried out with a 96-pin replicator (Fisher). Yeast knockout (YKO) mutants were transferred from 96-well master plates to single-well OMNI tray (128-mm by 86-mm) target plates (YPD agar) (Nunc) containing YPD agar without (control) or with (treatment) 2,4-DAPG at 200 μg ml−1. The replicator was sterilized between transfers by soaking the pins for 2 min in bleach (2% NaOCl) followed by 1 min in ethanol (70%) and then flaming. Each mutant was printed four times on the plate; a total of 384 spots were printed per target plate (see Fig. S1A in the supplemental material). Preliminary studies showed that the growth of wild-type BY4741 was inhibited by about 80% at this concentration. Plates were incubated for 48 h at 30°C, and then growth of each mutant was determined visually by the presence or absence of a colony on the treatment plate.
Screening for 2,4-DAPG sensitivity in liquid medium.
Yeast mutant cells were also tested in 96-well microtiter plates containing 100 μl of YPD per well. Plates were incubated for 5 h at 30°C without shaking; optical density at 600 nm (OD600) was then measured (initial OD) with a microtiter plate reader (Bio-Rad), and 2,4-DAPG was added to give a final concentration of 40 μg ml−1. Plates were incubated for an additional 16 h at 30°C, and the optical density was measured again (output OD). The sensitivity of mutants to 2,4-DAPG was determined by calculating the difference between the initial and output OD values (net OD) (see Fig. S1B in the supplemental material). The screening was performed in triplicate, and the average net OD reading of each mutant was compared with that of the wild-type strain. The t test (P = 0.01) was used to compare mutants to the control.
Bioinformatics analyses.
The Saccharomyces Genome Database (www.yeastgenome.org) provides information about the deleted ORF in each mutant. Analysis of gene ontology was performed at the MIPS site (mips.gsf.de) and the FUNspec site (funspec.med.utoronto.ca). The KEGG pathway database (www.genome.jp) was used for mutant pathway studies. STITCH (stitch.embl.de) and PubChem (pubchem.ncbi.nlm.nih.gov) were used for surveying chemical bioactivities and interactions, respectively.
Biochemical analysis.
To measure membrane permeability, cells of BY4741 from an overnight YPD culture were harvested by centrifugation and suspended in Hank's balanced salt solution (HBSS) to give ∼106 cells in 0.1 ml with 0 or 100 μg ml−1 of 2,4-DAPG. Propidium iodide (excitation at 540 nm and emission at 620 nm) (1 μg ml−1 in distilled water) was then added to the cell suspensions, and the suspensions were transferred to 1.5-ml tubes and incubated for 1 h at 30°C. Then, 0.1 ml of the cell suspensions was transferred onto a glass microscope slide, and images were taken by using an Olympus BX41TF fluorescence microscope (Olympus).
To measure reactive oxygen, cells of BY4741 from an overnight YPD culture were harvested by centrifugation, suspended in HBSS, and adjusted to ∼106 cells in 100 μl. Cells (0.1 ml) were transferred into 96-well plates, and 2,4-DAPG was added to give final concentrations of 0, 5, 10, 25, 50, or 100 μg ml−1. Each well also received the fluorescent reporter dye 2,7-dichlorofluorescin diacetate (excitation at 488 nm, emission at 520 nm) at 5 μM (final concentration) to measure hydrogen peroxide or dihydroethidine (DHE; excitation at 514 nm, emission at 560 nm) to measure superoxide (O2−). Plates were incubated at 30°C for 30 min (for superoxide) or 2 h (for H2O2) and monitored for fluorescence every minute (for O2−) or 2 min (for H2O2) with a SAFIRE microplate reader (Tecan Systems, Inc.). Each treatment was replicated four times.
To measure cell homeostasis, strain BY4741 was grown and harvested as described above to give ∼106 cells in 0.1 ml HBSS and then incubated in 96-well plates with 0, 100, or 200 μg ml−1 2,4-DAPG and acridine orange (5 μg ml−1) (excitation at 502 nm and emission at 525 nm) for 2 h at 30°C. Each treatment was replicated four times. Fluorescence was measured every 2 min, and after 2 h, a 0.1-ml aliquot was transferred to a glass microscope slide and images were taken with a fluorescence microscope. The slope of the curves showing changes in fluorescence was calculated using the linear regression function in SigmaPlot 8.0. Significant differences in slopes were determined by Tukey's honestly significant difference (HSD) test (P = 0.01).
Chemical-genetic profiles of 2,4-DAPG in S. cerevisiae genome-wide mutants.
Results of the screen of the mutant library against 2,4-DAPG were compared with published (18, 41) yeast fitness profiles. Table S1 in the supplemental material shows two main public chip-based data sets used for comparison. The protocol is described in greater detail in Methods in the supplemental material.
RESULTS
Screening for 2,4-DAPG sensitivity.
In YPD agar with 200 μg ml−1 of 2,4-DAPG and YPD broth with 40 μg ml−1 2,4-DAPG, 324 and 438 mutants, respectively, exhibited increased sensitivity to the antibiotic. A total of 231 mutants with greater sensitivity were identified by both screening methods (see Fig. S2 in the supplemental material). Ninety-three mutants with increased sensitivity to 2,4-DAPG were identified only on agar, and 207 were detected only in broth (see Fig. S2). Mutants with greater tolerance to 2,4-DAPG were not detected by either screening method. Among the commonly selected 231 mutants, 22 were classified in the multidrug resistance (MDR) category (data not shown). Based on gene ontology, 112 mutants were related to the mitochondrion (data not shown).
Alterations in membrane structure increased 2,4-DAPG sensitivity.
Ergosterol, the primary membrane sterol in the yeast cell, serves a structural role and functions in vesicular trafficking. Ergosterol mutants have critical problems with cellular homeostasis, resulting from reduced membrane fluidity, process signal transduction, and ion channel activity (36). Among the 22 MDR mutants, seven were annotated as functioning in specific biosynthesis pathways. Four of the seven were related to membrane structure and biosynthesis, three (the ERG2, ERG4, and ERG5 mutants) were defective in terminal enzymes of the ergosterol biosynthesis pathway, and one (FEN1 mutant) was related to the biosynthesis of unsaturated fatty acids (see Fig. S3 and S4 in the supplemental material). The ERG3 mutant was selected from the list of 231 commonly selected mutants as not belonging to the MDR category. Glycerolphospholipid metabolism (CHO1) and sphingolipid metabolism (LSB5) mutants also were selected because of increased sensitivity to 2,4-DAPG (see Fig. S5 in the supplemental material).
Mutants in the oxidative phosphorylation pathway had increased sensitivity to 2,4-DAPG.
Among the 231 commonly selected mutants, 22 were related to the oxidative phosphorylation pathway, including nine vacuolar ATPase (V-ATPase) assembly and biosynthesis mutants (VMA2, VMA5, VMA6, VMA7, VMA8, VMA9, VMA13, CUP5, and PPA1 mutants) (see Fig. S5 and S6 in the supplemental material). Five mitochondrial proton-transporting ATP synthase-related mutants (ATP4, ATP5, ATP7, ATP15, and INH1 mutants), four cytochrome c oxidase mutants (COX6, COX7, COX9, and COX11 mutants), and four cytochrome c reductase activity-related mutants (CYT1, COR1, QCR7, and QCR8 mutants) exhibited increased sensitivity to 2,4-DAPG. The pathway study was performed using the KEGG pathway database.
Confirmation of selected pathways with physiological assays.
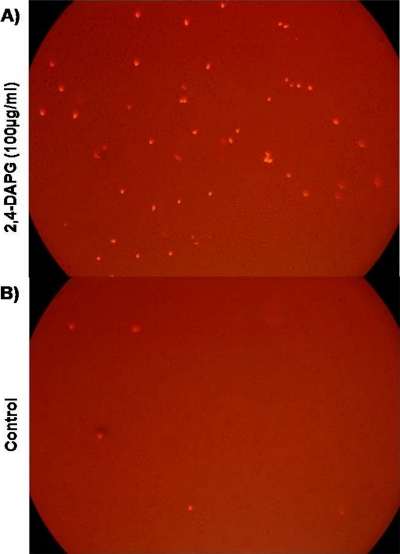
We measured the effect of 2,4-DAPG on membrane permeability by treating wild-type yeast cells with propidium iodide (1 μg ml−1) in the presence or absence of 2,4-DAPG. Fluorescence microscopy revealed that a larger number of cells exposed to 2,4-DAPG (100 μg ml−1) were permeable to the dye than were cells not exposed to the antibiotic (Fig. 1).
FIG. 1.
Interruption of membrane permeability in wild-type strain BY4741 exposed to 2,4-DAPG. Propidium iodide (1 μg ml−1) was added to a cell suspension treated with 100 μg ml−1 2,4-DAPG. The image was taken with a fluorescence microscope (excitation at 540 nm and emission at 620 nm). (A) Cells treated with 100 μg ml−1 2,4-DAPG; (B) control, no 2,4-DAPG added.
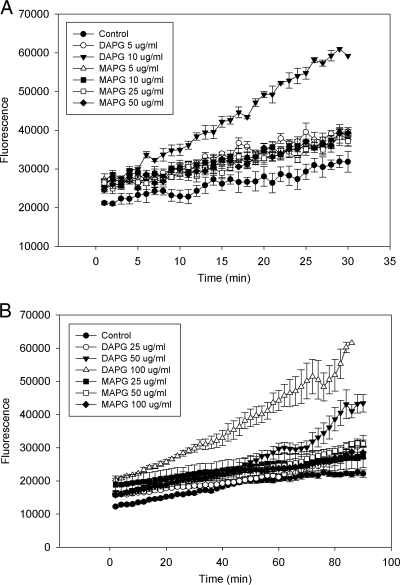
Dihydroethidine (DHE) was used to assess the level of the superoxide radical (O2−) after exposure to 2,4-DAPG. Five μg ml−1 of 2,4-DAPG increased the superoxide level only slightly compared with that of the control. Ten micrograms per milliliter increased superoxide generation further but did not inhibit cell growth, and at 20 min, the fluorescence intensity was twice that of the control (Fig. 2 A). The effect of 2,4-DAPG at 25 μg ml−1 was even greater (data not shown). MAPG, which has substantially less antimicrobial activity than 2,4-DAPG, had essentially no effect on superoxide generation at 5, 10, 25, or 50 μg ml−1 (Fig. 2A).
FIG. 2.
Oxidative burst in wild-type strain BY4741 exposed to 2,4-DAPG or MAPG. (A) Production of O2− was determined by measuring fluorescence of oxidation of DHE. (B) Production of H2O2 was determined by measuring DCFH-DA fluorescence. Each point of each treatment is the mean of four replicates. Fluorescence was measured every minute for DHE and every 2 min for DCFH-DA by a SAFIRE microplate reader.
DCFH-DA was used to measure the effect of 2,4-DAPG on hydrogen peroxide production. Yeast exposed to 2,4-DAPG at 25 and 50 μg ml−1 showed minimal and increased fluorescence, respectively, compared to that of nontreated cells (Fig. 2B). 2,4-DAPG at 100 μg ml−1 increased fluorescence intensity further, and a difference relative to the control was apparent after 10 min. At 90 min, 100 μg ml−1 of 2,4-DAPG caused a 3-fold increase in fluorescence intensity compared to that of nontreated cells. Concentrations of MAPG up to 100 μg ml−1 had no effect on fluorescence (Fig. 2B).
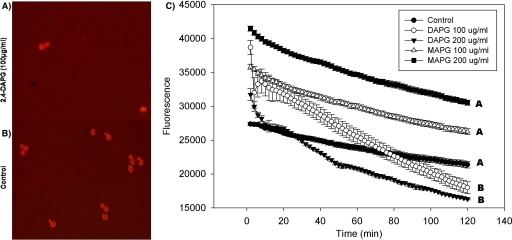
Acridine orange is commonly used to detect intracellular pH gradients and to measure proton pump activity (10). Yeast incubated in the presence of 2,4-DAPG and acridine orange showed a more rapid decrease in fluorescence than did cells treated only with acridine orange (Fig. 3 A and B), which is consistent with interrupted cell homeostasis. 2,4-DAPG at 200 μg ml−1 caused fluorescence to decrease significantly (P = 0.01) faster than at 100 μg ml−1 (Fig. 3C). MAPG had no effect on fluorescence.
FIG. 3.
Interrupted cell homeostasis in wild-type strain BY4741 by 2,4-DAPG. Cells treated with 2,4-DAPG or MAPG at 100 or 200 μg ml−1 were exposed to acridine orange for 2 h at 30°C. (A) Cells treated with 100 μg ml−1 2,4-DAPG; (B) control, no 2,4-DAPG added; (C) acridine orange fluorescence intensities. Each point of each treatment is the mean of results from four replicates. Fluorescence was measured every 2 min with a SAFIRE microplate reader (excitation at 502 nm, emission at 525 nm). Slopes of curves were calculated using the linear regression function in SigmaPlot 8.0. Lines with the same letters are not significantly different according to Tukey's HSD test (P = 0.01).
2,4-DAPG shows a similar mechanism of action to menthol, sodium azide, and hydrogen peroxide.
Chemical-genetic profiling is a well-established technique that seeks to organize small molecules and yeast genes into functionally relevant groups and to identify sets of compounds with biological effects and genes whose deletion leads to sensitivity to similar compound sets (41). The fitness of the 231 mutants with increased sensitivity to 2,4-DAPG was compared to other public profiles generated by screening 5,000 yeast haploid mutants exposed to approximately 1,200 chemical or environmental stress conditions (18, 41). By using the FitSearch algorithm recently developed for robust fitness profile comparisons (see Methods in the supplemental material), compounds with fitness profiles most similar to that of 2,4-DAPG were identified, and they included menthol, H2O2, and sodium azide; all are directly or indirectly related to membrane permeability, oxidative stress, and cellular homeostasis (Table 1).
TABLE 1.
Sensitivity of yeast mutants to 2,4-DAPG compared with results of published chemical screens (Tc > 0.2 and Hp < 1.00e−30)
| Compound | Overlapping scorea | Overlapping significanceb | Reference |
|---|---|---|---|
| Menthol | 0.25 | <1.00e−30 | 41 |
| Sodium azide | 0.23 | <1.00e−30 | 41 |
| Hydrogen peroxide | 0.21 | <1.00e−30 | 41 |
Tanimoto coefficient (Tc) is used as the overlapping score between the query profile (2,4-DAPG) and the target profiles (published fitness profiles). When two profiles are perfectly overlapped, Tc is 1.
Hypergeometric P value (Hp) means the overlapping significance between query and target profiles compared with random overlapping chance. Lower Hp tends to indicate more significant overlapping between fitness profiles of two compounds, which are then likely to have more similar mechanisms of action.
DISCUSSION
The antibiotic 2,4-DAPG is active against a broad range of organisms, including viruses, bacteria, protists, fungi, mammalian cells, and plants. 2,4-DAPG is produced by P. fluorescens in the rhizosphere, and it plays an important role in the biological control of soilborne plant pathogens. However, despite its role in plant defense in many agroecosystems, virtually nothing is known about the mode(s) of action of 2,4-DAPG against fungal plant pathogens. We selected the S. cerevisiae genome-wide mutant library as a tool to provide insight into those cellular pathways that are interrupted by 2,4-DAPG in plant-pathogenic fungi. S. cerevisiae is ideal for this type of study because it has a short life cycle and is amenable to genetic studies, and whole genome-wide mutants are readily available and fairly inexpensive. In this study, we focused on the 231 mutants that showed greater sensitivity to 2,4-DAPG in both agar- and broth-based screening assays. Yeasts have frequently served as model organisms for elucidating the mechanism(s) of action of antimicrobial compounds and pharmaceuticals (45).
The function of the plasma membrane is to separate the cytoplasm from the environment. Ergosterol is the predominant sterol in the fungal plasma membrane and is critical to proper membrane fluidity, permeability, vesicle trafficking, homeostasis, and the activity of membrane-bound proteins (17, 49). It is similar in structure and functions to cholesterol in mammalian cells. We found that ergosterol biosynthesis mutants had increased sensitivity to 2,4-DAPG (see Fig. S3 and S4 in the supplemental material), and four of these mutants occurred in sequence at the very end of the ergosterol pathway (ERG2, ERG3, ERG5, and ERG4 mutants), indicating that intact membrane function is important for protection against damage by 2,4-DAPG. However, 2,4-DAPG does not directly target ergosterol like amphotericin B, a polyene-type antifungal agent that binds hydrophobically to ergosterol and extends pore size across the cell membrane. Eventually, altered cell permeability and leakage of cytoplasm components occur, and the cell finally dies (8). ERG6, ERG2, ERG5, and ERG4 ergosterol mutants showed tolerance to amphotericin B due to the lack of binding to the targets (26), but these same mutants had dramatically increased sensitivity to cycloheximide, which interferes with the translocation step in protein synthesis but has no effect on membrane structure. The ergosterol mutants were supersensitive to cycloheximide due to increased membrane permeability (26). Our assay using propidium iodide demonstrated that 2,4-DAPG-treated yeast cells had altered plasma membrane permeability (Fig. 1), which is consistent with increased sensitivity of the ergosterol mutants to 2,4-DAPG. Sphingolipid biosynthesis (FEN1, LCB5) and phospholipid biosynthesis (CHO1) mutants also were selected by both the agar and broth screening methods for increased sensitivity to 2,4-DAPG. These two lipids function similarly to ergosterol in the targeting and assembly of membrane proteins, in having integral structural and transport roles, and responding to stress (6). Collectively, our results indicate that although ergosterol or other lipids are not direct targets of 2,4-DAPG, any factor that compromises normal membrane structure and function will increase the sensitivity of the fungal cell to 2,4-DAPG.
2,4-DAPG sensitivity was also increased in mutants involved in coenzyme Q biosynthesis (COQ2 mutant), cytochrome c oxidase (COX6, COX7, COX9, and COX11 mutants), and cytochrome c reductase (CYT1, COR1, QCR7, and QCR8 mutants). COQ2 is an enzyme of the ubiquinone biosynthetic pathway and transports electrons between the mitochondrial respiration complexes (1). Cytochrome c oxidase is involved in the terminal step in the electron transport chain in cellular respiration. Cytochrome c reductase is an essential complex for the energy-generating process of oxidative phosphorylation (23). Apparently, disruption of the electron transport chain in these mutants resulted in free radical leakage and the production of reactive oxygen species. Our physiological tests provided support for this conclusion, because 2,4-DAPG altered superoxide radical and hydrogen peroxide levels in wild-type S. cerevisiae. Among the 231 mutants, 112 mutants were related to the mitochondria based upon their cellular component category of gene ontology (data not shown). In total, these localization results indicate that a normal mitochondrial electron transport system and oxidoreduction function are important to reduce 2,4-DAPG toxicity.
The vacuole contains high concentrations of basic amino acids, carbohydrates, inorganic phosphate, and critical ions such as Ca2+ (4). V-ATPase is a large, multicomplex enzyme that hydrolyzes ATP to pump protons across membranes and is responsible for the acidification of cellular compartments. V-ATPase also contributes to maintaining the level of cytosolic Ca2+ and generating a pH gradient. Activity of V-ATPase is inhibited by oxidizing agents, by dissociation of the V1 sector, and by increasing the formation of disulfide bonds (4). Nine V-ATPase assembly and biosynthesis-related mutants (the VMA2, VMA5, VMA6, VMA7, VMA8, VMA9, VMA13, CUP5, and PPA1 mutants) had increased sensitivity to 2,4-DAPG (see Fig. S5 in the supplemental material). VMA2 plays a critical role in ATP binding (39). VMA5 is required for assembling the V1 domain, and the VMA5 mutant is sensitive to calcium level (21). VMA6 has a critical role in complex assembly and stabilization and is also sensitive to calcium level (2). VMA7 assists in the assembly of the complex on the vacuolar membrane and has a specific interaction with VMA8 (40). VMA8 plays a role in coupling of proton transport (56). VMA9 is a subunit of the V0 subcomplex and interacts with V-ATPase assembly factor VMA21p. VMA9 is involved in V0 biogenesis (7). VMA13 is required for the activity of the complex but not its assembly and stability (20). CUP5, alias of VMA3, is a highly hydrophobic integral membrane proteolipid and is essential to maintaining iron homeostasis (52). PPA1, alias of VMA16, is involved in the assembly of the subunits onto the vacuolar membrane and proton transport (19). Based on our screening results and biochemical assays, we suggest that 2,4-DAPG inactivates V-ATPase in the yeast cell. V-ATPase previously has been reported to be inactivated by reactive oxygen in mammalian and fungal cells (4). Collectively, our results indicate that V-ATPase activity is inhibited by reactive oxygen, which is triggered by 2,4-DAPG. Following this inhibition, the cell begins losing control of homeostasis and suffers incremental damage.
Chemical-genetic profiles can be generated from gene deletion libraries exposed to different chemical or environmental stresses, with responses detected by a variety of techniques (i.e., DNA chip-based parallel measurement, high-density colony measurements on an agar plate, high-density optical density or fluorescence responses in liquid culture). Similarly, there are many ways to express the relative fitness scores (i.e., fold ratio, z-score, P value, ranks, or simple ±binary value). In large-scale experiments using the same setup, profiles in a data set can be compared, thus making it possible to use data-mining algorithms, such as clustering, to discover similar modes of action among bioactive compounds. In small-scale experiments, especially those based on a nonchip platform like our data set, such data mining is not effective. Recently, FitSearch (a data-mining algorithm [see Methods in the supplemental material]) was developed, and it enables comparison of a compound's fitness profile with public chip-based yeast fitness data profiles and nonchip-based single-compound fitness profiles. More importantly, FitSearch can provide the possible mode(s) of action for a compound of interest based upon comparisons to similar compounds with known mode(s) of action. As a result of fitness profile comparisons of 2,4-DAPG with compounds in databases, (1R, 2S, 5R)-2-isopropyl-5-methylcyclohexanol (menthol), sodium azide, and hydrogen peroxide were ranked as having the best match with the profile of 2,4-DAPG. Menthol is well known as an antipruritic drug (42) through activation of the cold-sensitive transient receptor potential cation channel subfamily M member 8 (TRPM8), an ion channel regulating Na+ and Ca2+ ions. Menthol also has antimicrobial activity, and the inhibitory effect of this phenolic compound could be explained by its hydrophobicity and interaction with the membrane (3). Trombetta et al. (51) demonstrated that menthol caused a perturbation in the lipid fraction of the membrane, altered membrane permeability, and caused leakage of intracellular materials. 2,4-DAPG is also a phenolic compound and has an assigned antimicrobial action mediated via surface interactions (http://pubchem.ncbi.nlm.nih.gov/).
Sodium azide is well known as a rapid and reversible inhibitor of the cytochrome c oxidase-respiratory chain complex IV due to enhanced cytochrome c holoenzyme dissociation (32). Membrane-associated protein kinase C activity can also be altered by sodium azide. Marino et al. (35) demonstrated that sodium azide increased intracellular calcium in mammalian systems, causing azide neurotoxicity. In our study, mutants in cytochrome c oxidase (COX6, COX7, COX9, and COX11 mutants) and cytochrome c reductase (CYT1, COR1, QCR7, and QCR8 mutants) showed increased sensitivity to 2,4-DAPG. Recently, Gleeson et al. (15) demonstrated that 2,4-DAPG can cause impairment of mitochondrial function in S. cerevisiae.
Hydrogen peroxide (H2O2) can damage proteins, lipids, and DNA. The primary source of reactive oxygen species is free leakage of electrons generated by the mitochondrial respiratory system. Thorpe et al. (50) screened a homozygous diploid deletion yeast library for sensitivity to H2O2. Among 121 H2O2-sensitive mutants, 58 were related to mitochondrial functions, including respiratory chain, mitochondrial genome maintenance, and protein synthesis.
Many vacuolar function and ergosterol biosynthesis mutants have been shown to be extremely sensitive to diamide, a superoxide-generating agent. Among the 231 mutants we focused on, ergosterol and V-ATPase function mutants showed increased sensitivity to 2,4-DAPG. Biochemical assays demonstrated clearly that 2,4-DAPG triggered a reactive oxygen burst, producing both superoxide and hydrogen peroxide.
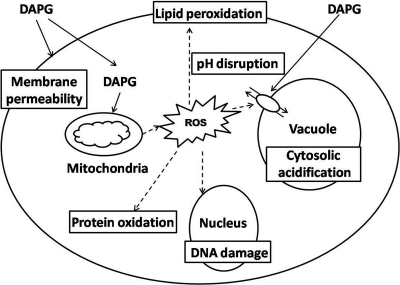
To our knowledge, this is the first systematic approach for identifying 2,4-DAPG targets and pathways in yeast. In conclusion, our study indicates that the antibiotic 2,4-DAPG disturbs cell membrane permeability, triggers reactive oxygen burst, and interrupts cell homeostasis as its mechanism of action (Fig. 4).
FIG. 4.
A possible model for the mechanisms of action of 2,4-DAPG in S. cerevisiae. 2,4-DAPG causes membrane damage, reactive oxygen burst, cytosolic acidification, and disturbance of cell homeostasis.
The antibiotic 2,4-DAPG is the key mechanism of take-all decline (TAD), one of the most successful and widely used natural biological controls in wheat production. 2,4-DAPG, produced in the rhizosphere by P. fluorescens, does not eliminate the pathogen Gaeumannomyces graminis var. tritici but significantly inhibits its growth, thus reducing the incidence and severity of take-all disease. As a result, the pathogen can be exposed to the antibiotic throughout the growing season and during continuous wheat monoculture for years or decades. A practical outcome of our study is a potential explanation for why resistance to 2,4-DAPG has not emerged in G. graminis var. tritici and take-all suppressiveness has not been lost during long-term wheat monoculture (up to >40 years) in a field (31). We propose that because 2,4-DAPG attacks multiple basic cellular pathways, emergence of antibiotic resistance in the take-all pathogen in the field is unlikely.
Supplementary Material
Acknowledgments
Sangjo Han was supported by a National Research Foundation of Korea grant funded by the Korean Government (KRF-2009-352-C00140).
Footnotes
Published ahead of print on 30 December 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Ashby, M. N., S. Y. Kutsunai, S. Ackerman, A. Tzagoloff, and P. A. Edwards. 1992. COQ2 is a candidate for the structural gene encoding para-hydroxybenzoate:polyprenyltransferase. J. Biol. Chem. 267:4128-4136. [PubMed] [Google Scholar]
- 2.Bauerle, C., M. N. Ho, M. A. Lindorfer, and T. H. Stevens. 1993. The Saccharomyces cerevisiae VMA6 gene encodes the 36-kDa subunit of the vacuolar H+-ATPase membrane sector. J. Biol. Chem. 268:12749-12757. [PubMed] [Google Scholar]
- 3.Ben Arfa, A., S. Combes, L. Preziosi-Belloy, N. Gontard, and P. Chalier. 2006. Antimicrobial activity of carvacrol related to its chemical structure. Lett. Appl. Microbiol. 43:149-154. [DOI] [PubMed] [Google Scholar]
- 4.Bowman, E. J., and B. J. Bowman. 2000. Cellular role of the V-ATPase in Neurospora crassa: analysis of mutants resistant to concanamycin or lacking the catalytic subunit A. J. Exp. Biol. 203:97-106. [DOI] [PubMed] [Google Scholar]
- 5.Brazelton, J. N., E. E. Pfeufer, T. A. Sweat, B. B. McSpadden Gardener, and C. Coenen. 2008. 2,4-Diacetylphloroglucinol alters plant root development. Mol. Plant Microbe Interact. 21:1349-1358. [DOI] [PubMed] [Google Scholar]
- 6.Carman, G. 2005. Regulation of phospholipid synthesis in yeast by zinc. Biochem. Soc. Trans. 33:1150-1153. [DOI] [PubMed] [Google Scholar]
- 7.Compton, M. A., L. A. Graham, and T. H. Stevens. 2006. Vma9p (subunit e) is an integral membrane V0 subunit of the yeast V-ATPase. J. Biol. Chem. 281:15312-15319. [DOI] [PubMed] [Google Scholar]
- 8.Cowen, L. E. 2008. The evolution of fungal drug resistance: modulating the trajectory from genotype to phenotype. Nat. Rev. Microbiol. 6:187-198. [DOI] [PubMed] [Google Scholar]
- 9.Cronin, D., et al. 1997. Ecological interaction of a biocontrol Pseudomonas fluorescens strain producing 2,4-diacetylphloroglucinol with the soft rot potato pathogen Erwinia carotovora subsp. atroseptica. FEMS Microbiol. Ecol. 23:95-106. [Google Scholar]
- 10.Darzynkiewicz, Z., et al. 1992. Features of apoptotic cells measured by flow cytometry. Cytometry 13:795-808. [DOI] [PubMed] [Google Scholar]
- 11.de Souza, J. T., D. M. Weller, and J. M. Raaijmakers. 2003. Frequency, diversity, and activity of 2,4-diacetylphloroglucinol-producing fluorescent Pseudomonas spp. in Dutch take-all decline soils. Phytopathology 93:54-63. [DOI] [PubMed] [Google Scholar]
- 12.Duffy, B., A. Schouten, and J. M. Raaijmakers. 2003. Pathogen self-defense: mechanisms to counteract microbial antagonism. Annu. Rev. Phytopathol. 41:501-538. [DOI] [PubMed] [Google Scholar]
- 13.Duffy, B. K., and G. Défago. 1997. Zinc improves biocontrol of Fusarium crown and root rot of tomato by Pseudomonas fluorescens and represses the production of pathogen metabolites inhibitory to bacterial antibiotic biosynthesis. Phytopathology 87:1250-1257. [DOI] [PubMed] [Google Scholar]
- 14.Fleissner, A., C. Sopalla, and K. M. Weltring. 2002. An ATP-binding cassette multidrug-resistance transporter is necessary for tolerance of Gibberella pulicaris to phytoalexins and virulence on potato tubers. Mol. Plant Microbe Interact. 15:102-108. [DOI] [PubMed] [Google Scholar]
- 15.Gleeson, O., F. O'Gara, and J. P. Morrissey. 2010. The Pseudomonas fluorescens secondary metabolite 2,4-diacetylphloroglucinol impairs mitochondrial function in Saccharomyces cerevisiae. Antonie Van Leeuwenhoek 97:261-273. [DOI] [PubMed] [Google Scholar]
- 16.Haas, D., and C. Keel. 2003. Regulation of antibiotic production in root-colonizing Pseudomonas spp. and relevance for biological control of plant disease. Annu. Rev. Phytopathol. 41:117-153. [DOI] [PubMed] [Google Scholar]
- 17.Henneberry, A. L., and S. L. Sturley. 2005. Sterol homeostasis in the budding yeast, Saccharomyces cerevisiae. Semin. Cell Dev. Biol. 16:155-161. [DOI] [PubMed] [Google Scholar]
- 18.Hillenmeyer, M. E., et al. 2008. The chemical genomic portrait of yeast: uncovering a phenotype for all genes. Science 320:362-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hirata, R., L. A. Graham, A. Takatsuki, T. H. Stevens, and Y. Anraku. 1997. VMA11 and VMA16 encode second and third proteolipid subunits of the Saccharomyces cerevisiae vacuolar membrane H+-ATPase. J. Biol. Chem. 272:4795-4803. [DOI] [PubMed] [Google Scholar]
- 20.Ho, M. N., et al. 1993. VMA13 encodes a 54-kDa vacuolar H+-ATPase subunit required for activity but not assembly of the enzyme complex in Saccharomyces cerevisiae. J. Biol. Chem. 268:18286-18292. [PubMed] [Google Scholar]
- 21.Ho, M. N., K. J. Hill, M. A. Lindorfer, and T. H. Stevens. 1993. Isolation of vacuolar membrane H-ATPase-deficient yeast mutants; the VMA5 and VMA4 genes are essential for assembly and activity of the vacuolar H+-ATPase. J. Biol. Chem. 268:221-227. [PubMed] [Google Scholar]
- 22.Hornby, D. 1998. Take-all disease of cereals: a regional perspective. CAB International, Wallingford, United Kingdom.
- 23.Hunte, C., H. Palsdottir, and B. L. Trumpower. 2003. Protonmotive pathways and mechanisms in the cytochrome bc1 complex. FEBS Lett. 545:39-46. [DOI] [PubMed] [Google Scholar]
- 24.Iavicoli, A., E. Boutet, A. Buchala, and J. P. Métraux. 2003. Induced systemic resistance in Arabidopsis thaliana in response to root inoculation with Pseudomonas fluorescens CHA0. Mol. Plant Microbe Interact. 16:851-858. [DOI] [PubMed] [Google Scholar]
- 25.Isnansetyo, A., L. Cui, K. Hiramatsu, and Y. Kamei. 2003. Antibacterial activity of 2,4-diacetylphloroglucinol produced by Pseudomonas sp. AMSN isolated from a marine alga, against vancomycin-resistant Staphylococcus aureus. Int. J. Antimicrob. Agents 22:545-547. [DOI] [PubMed] [Google Scholar]
- 26.Iwaki, T., et al. 2008. Multiple functions of ergosterol in the fission yeast Schizosaccharomyces pombe. Microbiology 154:830-841. [DOI] [PubMed] [Google Scholar]
- 27.Jousset, A., E. Lara, L. G. Wall, and C. Valverde. 2006. Secondary metabolites help biocontrol strain Pseudomonas fluorescens CHA0 to escape protozoan grazing. Appl. Environ. Microbiol. 72:7083-7090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kamei, Y., and A. Isnansetyo. 2003. Lysis of methicillin-resistant Staphylococcus aureus by 2,4-diacetylphloroglucinol produced by Pseudomonas sp. AMSN isolated from a marine alga. Int. J. Antimicrob. Agents 21:71-74. [DOI] [PubMed] [Google Scholar]
- 29.Keel, C., et al. 1992. Suppression of root diseases by Pseudomonas fluorescens CHA0: importance of the bacterial secondary metabolite 2,4-diacetylphloroglucinol. Mol. Plant Microbe Interact. 5:4-13. [Google Scholar]
- 30.Kiprianova, E. A., and V. V. Smirnov. 1981. Pseudomonas fluorescens, a producer of antibiotic substance. Antibiotiki 26:135-143. [PubMed] [Google Scholar]
- 31.Kwak, Y. S., et al. 2009. Diversity, virulence, and 2,4-diacetylphloroglucinol sensitivity of Gaeumannomyces graminis var. tritici isolates from Washington State. Phytopathology 99:472-479. [DOI] [PubMed] [Google Scholar]
- 32.Leary, S. C., et al. 2002. Chronic treatment with azide in situ leads to an irreversible loss of cytochrome c oxidase activity via holoenzyme dissociation. J. Biol. Chem. 277:11321-11328. [DOI] [PubMed] [Google Scholar]
- 33.Levy, E., Z. Eyal, I. Chet, and A. Hochman. 1992. Resistance mechanisms of Septoria tritici to antifungal products of Pseudomonas. Physiol. Mol. Plant Pathol. 40:163-171. [Google Scholar]
- 34.Mahajan-Miklos, S., M. W. Tan, L. G. Rahme, and F. M. Ausubel. 1999. Molecular mechanisms of bacterial virulence elucidated using a Pseudomonas aeruginosa-Caenorhabditis elegans pathogenesis model. Cell 96:47-56. [DOI] [PubMed] [Google Scholar]
- 35.Marino, S., L. Marani, C. Nazzaro, L. Beani, and A. Siniscalchi. 2007. Mechanisms of sodium azide-induced changes in intracellular calcium concentration in rat primary cortical neurons. Neurotoxicology 28:622-629. [DOI] [PubMed] [Google Scholar]
- 36.McIntosh, T. J., and S. A. Simon. 2006. Roles of bilayer material properties in function and distribution of membrane proteins. Annu. Rev. Biophys. Biomol. Struct. 35:177-198. [DOI] [PubMed] [Google Scholar]
- 37.Morrissey, J. P., and A. E. Osbourn. 1999. Fungal resistance to plant antibiotics as a mechanism of pathogenesis. Microbiol. Mol. Biol. Rev. 63:708-724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nakaune, R., H. Hamamoto, J. Imada, K. Akutsu, and T. Hibi. 2002. A novel ABC transporter gene, PMR5, is involved in multidrug resistance in the phytopathogenic fungus Penicillium digitatum. Mol. Genet. Genomics 267:179-185. [DOI] [PubMed] [Google Scholar]
- 39.Nelson, H., S. Mandiyan, and N. Nelson. 1989. A conserved gene encoding the 57-kDa subunit of the yeast vacuolar H+-ATPase. J. Biol. Chem. 264:1775-1778. [PubMed] [Google Scholar]
- 40.Nelson, H., S. Mandiyan, and N. Nelson. 1994. The Saccharomyces cerevisiae VMA7 gene encodes a 14-kDa subunit of the vacuolar H+-ATPase catalytic sector. J. Biol. Chem. 269:24150-24155. [PubMed] [Google Scholar]
- 41.Parsons, A. B., et al. 2006. Exploring the mode-of-action of bioactive compounds by chemical-genetic profiling in yeast. Cell 126:611-625. [DOI] [PubMed] [Google Scholar]
- 42.Peier, A. M., et al. 2002. A TRP channel that senses cold stimuli and menthol. Cell 108:705-715. [DOI] [PubMed] [Google Scholar]
- 43.Phillips, D. A., T. C. Fox, M. D. King, T. V. Bhuvaneswari, and L. R. Teuber. 2004. Microbial products trigger amino acid exudation from plant roots. Plant Physiol. 136:2887-2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Raaijmakers, J. M., and D. M. Weller. 1998. Natural plant protection by 2,4-diacetylphloroglucinol-producing Pseudomonas spp. in take-all decline soils. Mol. Plant Microbe Interact. 11:144-152. [Google Scholar]
- 45.Ran, H., D. J. Hassett, and G. W. Lau. 2003. Human targets of Pseudomonas aeruginosa pyocyanin. Proc. Natl. Acad. Sci. U. S. A. 100:14315-14320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schoonbeek, H., J. M. Raaijmakers, and M. De Waard. 2002. Fungal ABC transporters and microbial interactions in natural environments. Mol. Plant Microbe Interact. 15:1165-1172. [DOI] [PubMed] [Google Scholar]
- 47.Schouten, A., et al. 2004. Defense responses of Fusarium oxysporum to 2,4-diacetylphloroglucinol, a broad-spectrum antibiotic produced by Pseudomonas fluorescens. Mol. Plant Microbe Interact. 17:1201-1211. [DOI] [PubMed] [Google Scholar]
- 48.Schouten, A., O. Maksimova, Y. Cuesta-Arenas, G. van den Berg, and J. M. Raaijmakers. 2008. Involvement of the ABC transporter BcAtrB and the laccase BcLCC2 in defense of Botrytis cinerea against the broad-spectrum antibiotic 2,4-diacetylphloroglucinol. Environ. Microbiol. 10:1145-1157. [DOI] [PubMed] [Google Scholar]
- 49.Sharma, S. C. 2006. Implications of sterol structure for membrane lipid composition, fluidity and phospholipid asymmetry in Saccharomyces cerevisiae. FEMS Yeast Res. 6:1047-1051. [DOI] [PubMed] [Google Scholar]
- 50.Thorpe, G. W., C. S. Fong, N. Alic, V. J. Higgins, and I. W. Dawes. 2004. Cells have distinct mechanisms to maintain protection against different reactive oxygen species: oxidative-stress-response genes. Proc. Natl. Acad. Sci. U. S. A. 101:6564-6569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Trombetta, D., et al. 2005. Mechanisms of antibacterial action of three monoterpenes. Antimicrob. Agents Chemother. 49:2474-2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Umemoto, N., T. Yoshihisa, R. Hirata, and Y. Anraku. 1990. Roles of the VMA3 gene product, subunit c of the vacuolar membrane H+-ATPase on vacuolar acidification and protein transport. A study with VMA3-disrupted mutants of Saccharomyces cerevisiae. J. Biol. Chem. 265:18447-18453. [PubMed] [Google Scholar]
- 53.van Etten, H., E. Temporini, and C. Wasmann. 2001. Phytoalexin (and phytoanticipin) tolerance as a virulence trait: why is it not required by all pathogens? Physiol. Mol. Plant Pathol. 59:83-93. [Google Scholar]
- 54.Weller, D. M., J. M. Raaijmakers, B. B. McSpadden Gardener, and L. S. Thomashow. 2002. Microbial populations responsible for specific soil suppressiveness to plant pathogens. Annu. Rev. Phytopathol. 40:309-348. [DOI] [PubMed] [Google Scholar]
- 55.Winzeler, E. A., et al. 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285:901-906. [DOI] [PubMed] [Google Scholar]
- 56.Xu, T., and M. Forgac. 2000. Subunit D (Vma8p) of the yeast vacuolar H+-ATPase plays a role in coupling of proton transport and ATP hydrolysis. J. Biol. Chem. 275:22075-22081. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.