Abstract
Legionella pneumophila, a bacterium that replicates within aquatic amoebae and persists in the environment as a free-living microbe, is the causative agent of Legionnaires' disease. Among the many Legionella species described, L. pneumophila is associated with 90% of human disease, and within the 15 serogroups (Sg), L. pneumophila Sg1 causes more than 84% of Legionnaires' disease worldwide. Thus, rapid and specific identification of L. pneumophila Sg1 is of the utmost importance for evaluation of the contamination of collective water systems and the risk posed. Previously we had shown that about 20 kb of the 33-kb locus carrying the genes coding for the proteins involved in lipopolysaccharide biosynthesis (LPS gene cluster) by L. pneumophila was highly specific for Sg1 strains and that three genes (lpp0831, wzm, and wzt) may serve as genetic markers. Here we report the sequencing and comparative analyses of this specific region of the LPS gene cluster in L. pneumophila Sg6, -10, -12, -13, and -14. Indeed, the wzm and wzt genes were present only in the Sg1 LPS gene cluster, which showed a very specific gene content with respect to the other five serogroups investigated. Based on this observation, we designed primers and developed a classical and a real-time PCR method for the detection and simultaneous identification of L. pneumophila Sg1 in clinical and environmental isolates. Evaluation of the selected primers with 454 Legionella and 38 non-Legionella strains demonstrated 100% specificity. Sensitivity, specificity, and predictive values were further evaluated with 209 DNA extracts from water samples of hospital water supply systems and with 96 respiratory specimens. The results showed that the newly developed quantitative Sg1-specific PCR method is a highly specific and efficient tool for the surveillance and rapid detection of high-risk L. pneumophila Sg1 in water and clinical samples.
Legionella pneumophila is ubiquitous in natural, aqueous environments, where it parasitizes aquatic protozoa (1). From these environments Legionella can contaminate artificial water circuits and then grow and proliferate to high numbers, in particular in hot water systems, including aerosol-producing devices such as air conditioning systems or cooling towers (18, 19, 37). Upon aerosol formation via man-made water systems, Legionella can enter the human lung and cause a severe form of pneumonia. Since transmission of Legionella from person to person has never been observed, prevention needs to concentrate on the elimination of this pathogen from water and aerosol-producing systems. Thus, rapid and precise detection of Legionella in water systems is of the utmost importance for risk prediction and the elimination of Legionella from possible infection sources.
Water contamination by Legionella is currently monitored by culture-based methods approved by the International Organization for Standardization (29) and the French organization for standardization (2). The alert and action thresholds defined by the national authorities that oblige installation operators and policy authorities to intervene are based on culture methods. But, like all culture-based methods, identification of Legionella by culture is time-consuming, requiring as long as 8 days to obtain reliable and definitive results. Furthermore, the sensitivity of Legionella detection based on culture methods depends largely on the physiological state of the cells. Another drawback is that viable but nonculturable bacteria, or legionellae present within their protozoan hosts, that might be in the tested water sample cannot be detected by culture, leading to exposure to an undetected Legionella source (12, 24). Therefore, it is important to improve current testing for Legionella and to develop rapid and accurate tests.
Whereas 53 Legionella species have been identified to date and more than 70 serogroups have been described (5, 19, 32), many studies have reported an unequal distribution of the clinical cases among the species and subgroups of Legionella. L. pneumophila is responsible for more than 90% of clinical cases, and among the 15 serogroups (Sg) characterized within the species, L. pneumophila Sg1 is responsible for about 85% of all cases worldwide (16, 26, 43). Given this epidemiological background, it is necessary to identify rapidly not only the presence of Legionella in water samples but simultaneously also the species and serogroups, in order to estimate the risk of legionellosis.
Thus, research into the development of molecular tools to improve the detection of Legionella in water, especially quantitative real-time PCR (qPCR) methods, is very active (3, 17, 33, 38, 39, 40). Indeed, real-time PCR allows rapid detection and quantification of DNA with high sensitivity and specificity. Moreover, it allows the detection of viable but nonculturable cells (17). qPCR is presently an important complement to the standard culture-based method but does not substitute for it, since discussions as to whether the detection of nonviable bacteria by PCR may lead to an overestimation of the risk of legionellosis are still ongoing (33). However, to rely only on culture-based methods may, in contrast, lead to an underestimation of the risk. Furthermore, several of these PCR-based assays have also been tested on clinical pulmonary specimens and have shown good results (13, 14, 25, 31).
Until now, all qPCR methods developed for Legionella have been available only for genus-specific (based on the16S rRNA gene of Legionella spp.) or species-specific (based on the mip gene, encoding the macrophage infectivity potentiator of L. pneumophila) detection (39). Other genome targets, such as the 5S rRNA gene (25), the 23S rRNA gene (34), or the rpoB gene (41), have also been used, but no qPCR method specific for the detection of high-risk isolates of L. pneumophila Sg1 is available yet, since no specific targets for Sg1 have been known.
We showed previously that about 20 kb of the 33-kb locus carrying the genes coding for the proteins involved in lipopolysaccharide biosynthesis (LPS gene cluster) in L. pneumophila is highly specific for Sg1 strains (8). Furthermore, multigenome analysis by comparative hybridization identified three genes (lpp0831, lpp0837/wzm, and lpp0838/wzt) present in a single copy each in all Sg1 strains and only Sg1 strains (8). Here we sequenced this region of the LPS gene cluster in L. pneumophila Sg6, -10, -12, -13, and -14 strains as well, and we compared these regions to the LPS gene cluster of Sg1. Indeed, two (wzm and wzt) of the genes identified previously were present only in the Sg1 LPS gene cluster. Sequence comparisons allowed the definition of primers for the amplification of an Sg1-specific region and the development of a real-time PCR assay to detect and simultaneously identify L. pneumophila Sg1 in environmental and clinical samples. The sensitivity and specificity values obtained during this study demonstrate the potential of our rapid molecular assay for the surveillance and detection of L. pneumophila Sg1 isolates in community water supply systems and in clinical specimens.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
A total of 454 Legionella strains were selected from the Culture Collection of the National Reference Centre (Lyon, France) and the lab collections of the Biology of Intracellular Bacteria Unit at the Institut Pasteur and the Hospital Raymond Poincaré, Garches, France. Four hundred four L. pneumophila strains, 19 L. longbeachae strains, and 31 strains of other Legionella species were selected. Furthermore, 38 bacterial strains that do not belong to the genus Legionella but are frequently present in aquatic environments were included in the study as recommended by French standards (Normes AFNOR) (Table 1; see also Table S1 in the supplemental material). The Legionella strains were grown at 37°C on buffered charcoal-yeast extract (BCYE) agar or in BYE broth adjusted to pH 6.9 [10 g of N-(2-acetamido)-2-aminoethanesulfonic acid (ACES) buffer/liter, 10 g of yeast extract/liter, 0.4 g of l-cysteine/liter, 0.25 g of ferric pyrophosphate/liter] overnight under agitation before DNA extraction. Non-Legionella strains were grown on brain heart infusion (BHI) broth at 25°C or 37°C depending on the species growth requirements.
TABLE 1.
Strains used in this study
| Species | Serogroup | No. of strains tested | Total |
|---|---|---|---|
| Legionella species | |||
| Legionella pneumophila | 1 | 257 | 404 |
| 6 | 52 | ||
| 3 | 19 | ||
| Other | 76 | ||
| Legionella longbeachae | 19 | 50 | |
| Other | 31 | ||
| Non-Legionella species | 38 | ||
| Serratia sp. | 22 | ||
| Other | 16 | ||
| Total | 492 |
Sequencing and analysis of the LPS gene cluster.
Genomic DNA was isolated from L. pneumophila Sg6, -10, -12, -13, and -14 strains (Table 2) using the DNeasy Blood & Tissue kit (Qiagen, Hilden, Germany). The LPS gene cluster was amplified using a Long Range PCR kit (Qiagen, Hilden, Germany) in a final volume of 50 μl according to the manufacturer's instructions. The PCR fragment obtained was gel purified and used as a template for establishing a shotgun library (inserts, 1 to 3 kb) according to the protocol described by Cazalet and colleagues in 2010 (7). Two hundred clones were sequenced from both ends for each LPS gene cluster with an ABI Prism BigDye Terminator cycle sequencing ready reaction kit and a 3730xl genetic analyzer (Applied Biosystems, Foster City, CA). Sequences were assembled and finished as described previously (23). Coding sequences were defined, and annotation was done, using CAAT-Box software (21). For sequence comparison the Artemis Comparison Tool (ACT), freely available at the Sanger Centre, was used (6).
TABLE 2.
Strains and primers used for amplification of the LPS gene cluster region between lpp0826 (rmlA) and lpp0845 (csrA homologue) in Sg6, -10, -12, -13, and -14
| L. pneumophila strain | Sg | Primer designation | Primer sequence | Size of amplicon (kb) |
|---|---|---|---|---|
| ATCC 33215 | 6 | Csra-LpsLR1 | CTTCACGATGAACAGAAACATCTTTTGGAGCA | 28.5 |
| RmlA-LPS-LR1 | GGGAAGAGGTTTTGTCTGGCTGGATATGGG | |||
| ATCC 43283 | 10 | Csra-LpsLR1 | CTTCACGATGAACAGAAACATCTTTTGGAGCA | 30.8 |
| RmlA-LPS-LR1 | GGGAAGAGGTTTTGTCTGGCTGGATATGGG | |||
| ATCC 43290 | 12 | Csra-LpsLR1 | CTTCACGATGAACAGAAACATCTTTTGGAGCA | 28.3 |
| RmlA-LPS-LR1 | GGGAAGAGGTTTTGTCTGGCTGGATATGGG | |||
| ATCC 43736 | 13 | Csra-LpsLR3 | CTTCGCGATGAACAGAAACATCTTTTGGTGC | 28.7 |
| RmlA-LPS-LR1 | GGGAAGAGGTTTTGTCTGGCTGGATATGGG | |||
| ATCC 43073 | 14 | Csra-LpsLR1 | CTTCACGATGAACAGAAACATCTTTTGGAGCA | 30.7 |
| RmlA-LPS-LR1 | GGGAAGAGGTTTTGTCTGGCTGGATATGGG |
Collection of samples from hot water supply systems.
A representative sampling of hospital buildings (21 buildings in 14 hospitals [Assistance Publique-Hôpitaux de Paris {AP-HP}] in the Paris area) was conducted between June and November 2009. The samples were taken by each hospital hygiene team responsible for the management of the hospital hot water supply system (15). For each sample, 2 liters of water was collected into sterile bottles containing sodium thiosulfate (20 mg/liter) to neutralize any residual chlorine. The water samples were sent to the laboratory in less than 24 h. One liter was filtered for examination by the culture method, and 1 liter was filtered for DNA extraction for qPCR. DNA extracts were kept at −20°C until use.
Collection of samples from clinical cases.
A total of 96 respiratory samples, including 27 samples from patients with confirmed legionellosis cases and 69 samples from patients with nonlegionellosis pneumonia, were collected from hospital laboratories throughout France between July 2007 and April 2010. These samples were tested both by our Sg1-qPCR and by the commercialized Legionella species & Legionella pneumophila multiplex real-time PCR kit (Diagenode, Liège, Belgum). Of the 27 Legionnaires' disease cases, 10 were diagnosed by culture, 12 by urinary antigen testing (Binax Now immunochromatographic test), and 5 (negative by the other tests) by the multiplex real-time PCR kit.
Analysis of water samples by culture.
Viable legionellae were quantified by culture according to the French standard method T90-431 (2). A 200-μl volume of each water sample was directly plated onto glycine-vancomycin-polymyxin B-cycloheximide (GVPC) medium (Oxoid, Dardilly, France). Water samples were filtered through a polycarbonate membrane (pore size, 0.45 μm; Millipore). The membrane was sonicated for 10 min in 5 ml of sterile water. A 100-μl volume of the concentrate was plated onto GVPC medium, directly or after a 10-fold dilution, an acid treatment (5 min at pH 2), or a heat treatment (30 min at 50°C). The plates were incubated at 37°C, and colonies were counted after 3 days and 8 days. For each sample in which Legionella growth was detected, five colonies were further analyzed by propagation on BCYE agar at 37°C, compared to propagation on Columbia agar with 5% sheep blood (COS) (Oxoid, Dardilly, France). The Legionella strains isolated on BCYE were typed using a latex agglutination test (Oxoid, Dardilly, France) specific for Legionella spp., L. pneumophila Sg2 to Sg14, or L. pneumophila Sg1. Subsequently, isolates defined as belonging to Sg2 to Sg14 were further typed by immunofluorescence using homemade polyclonal rabbit sera (National Reference Center for Legionella, Lyon, France) and monovalent reagents for each serogroup (Sg2 to Sg15) (bioMérieux, Craponne, France) (36). Legionella species other than L. pneumophila were identified by PCR amplification and sequencing of the mip gene using the European Working Group for Legionella Infections (EWGLI) protocol adapted from the work of Ratcliff and colleagues (35).
Primer and probe design.
Primers for endpoint PCR were designed with Primer3 software (http://frodo.wi.mit.edu/) for the lpp0831, wzm, and wzt genes on regions identified as specific for L. pneumophila Sg1 with respect to the available Legionella sequences. Primers and probes for qPCR were designed using Primer Express software (Applied Biosystems, Foster City, CA). The primers were synthesized by Sigma-Proligo (Sigma-Aldrich, Lyon, France), and the 3′ Black Hole Quencher (BHQ) Double-Dye probe was supplied by Eurogentec (Angers, France) (Table 3).
TABLE 3.
Primers and probe specific for L. pneumophila Sg1 used in this study
| Primer or probe | Function | Sequence (5′-3′)a | Amplicon length (bp) |
|---|---|---|---|
| P1 | PCR forward primer | TTACCGCTTGCTTTTATGGA | 294 |
| P2 | PCR reverse primer | CCTATCAACGCTCTTGGAAA | |
| P65 | qPCR forward primer | CAAAGGGCGTTACAGTCAAACC | 75 |
| P66 | qPCR reverse primer | CAAACACCCCAACCGTAATCA | |
| sg1-pb | qPCR probe | FAM-TCTTGGGATTGGGTTGGGTTATTTTAACTCCT-BHQ1 |
FAM, 6-carboxyfluorescein.
DNA extraction from pure culture.
DNA was extracted from pure cultures with the DNeasy Blood & Tissue kit (Qiagen, Hilden, Germany). Two milliliters of liquid culture was centrifuged. The pellet was resuspended in 180 μl of buffer ATL, and extraction was performed according to the manufacturer's instructions. DNA concentrations were determined by spectrophotometry at 260 nm (Nanodrop, Wilmington, DE). The DNA was diluted at 1 ng/μl and was distributed into 96-well plates. Two microliters was used for endpoint PCR and qPCR to test the specificity of the primers.
DNA extraction from water samples for qPCR.
For each sample, 1 liter of water was filtered through a 0.40-μm-pore-size polycarbonate membrane (Millipore, Molsheim, France). After 10 filtrations, 1 liter of sterile water (water for injections; CDM Lavoisier, Paris, France) was filtered as a negative control for the filtration step. DNA extraction was performed directly on the membrane by using the Aquadien kit (Bio-Rad, Marnes-la-Coquette, France) according to the manufacturer's instructions. DNA was eluted in 100 μl of elution buffer. For each serial extraction, a negative control for the extraction step was performed by following the same protocol without the polycarbonate membrane. DNA extracts were kept at −20°C until they were used for qPCR assays.
DNA extraction from clinical samples for qPCR.
DNA was extracted with the automated MagNA Pure Compact system (Roche Diagnostics, Meylan, France) and Nucleic Acid Isolation Kit I. Prior to automated extraction, samples were liquefied as follows: 22 μl of dithiothreitol (DTT) was added to 200 μl of the sample, and the mixture was incubated for 10 min at 37°C; then 180 μl of Bacteria Lysis Buffer and 20 μl of proteinase K (20 mg/ml) were added to 200 μl of the liquefied sample, and the mixture was incubated for 10 min at 65°C. We used the manufacturer's DNA extraction protocol with an elution volume of 100 μl. DNA extracts were kept at −20°C until they were used for qPCR assays.
Sg1-endpoint PCR conditions.
The amplification mixtures contained 5 μl of GeneAmp 10× PCR buffer II, 4 μl of MgCl2 at 25 mM, 1 μl of deoxynucleoside triphosphate (dNTP) mixture at 10 mM for each dNTP, 0.3 μl of AmpliTaq DNA polymerase at 5 U/μl (Applied Biosystems, Foster City, CA), 3 μl of each primer (P1 and P2 [Table 3]) at 10 μM, 32 μl of purified PCR-grade water (Applied Biosystems, Foster City, CA), and 2 μl of DNA at 1 ng/μl. The thermal profile used was 1 min at 94°C; 35 cycles of 30 s at 94°C, 30 s at 55°C, and 30 s at 72°C; and a final step of 4 min at 72°C for final elongation. A positive control (L. pneumophila Sg1 strain ATCC 33152) and a negative control (purified PCR-grade water) were included in all PCR assays. The PCR products were run in 1% agarose gels for interpretation.
qPCR conditions. (i) Sg1-qPCR.
Reaction mixtures contained 2 μl of DNA, 800 nM each primer (P65 and P66 [Table 3]), 200 nM Sg1-specific probe, 10 μl of 2× TaqMan universal PCR master mix (Applied Biosystems, Foster City, CA), 2 μl of 10× Exogenous Internal Positive Control mix (Applied Biosystems, Foster City, CA), 0.4 μl of 50× Exogenous Internal Positive Control DNA (Applied Biosystems, Foster City, CA), and purified PCR-grade water to a final volume of 20 μl. Amplification was performed on a Chromo4 system (Bio-Rad, Marnes-la-Coquette, France) with the following parameters: a first step at 50°C for 2 min to activate the DNA polymerase; one step at 95°C for 10 min (initial denaturation); and 50 cycles of 15 s at 95°C and 1 min at 60°C. Each test was performed in duplicate. For clinical samples, the same PCR protocol was used, and amplification products were detected with the LightCycler DNA Master Hybridization Probe kit, used according to the manufacturer's instructions (Roche). If PCR inhibition occurred (no amplification of the internal positive control or a threshold cycle [CT] of >40), the DNA samples were diluted 2- to 32-fold in PCR-grade water, and Sg1-qPCR was performed with 5 μl of these dilutions (in a final volume of 50 μl). If false-negative results were suspected (compared to culture results), DNA samples obtained by the method described previously (Aquadien kit; Bio-Rad) were purified again and concentrated with a cleanup method (QIAamp DNA Micro; Qiagen). Then Sg1-qPCR was performed with 5 μl of these new DNA samples (in a final volume of 50 μl), and after 2- to 32-fold dilution in PCR-grade water if necessary.
(ii) Commercial qPCR.
L. pneumophila qPCR was performed as recommended by the kit supplier (iQcheck Quanti Legionella; Bio-Rad, Marnes- la-Coquette, France). For qPCR, the DNA was quantified in genome units (GU) by using the external standard solutions supplied with the commercial kit (iQCheck Quanti Legionella; Bio-Rad).
Estimation of the detection and quantification limits of the Sg1-qPCR.
The detection limit (DL) of the Sg1-qPCR assay (DLPCR) was defined as the smallest number of GU per qPCR that gave a positive result in at least 90% of cases. The quantification limit (QL) of the Sg1-qPCR assay (QLPCR) was defined as the smallest number of GU per qPCR that produced a coefficient of variation below 25% (30). The DL and QL of the Sg1-qPCR method applied to water samples (DLmeth and QLmeth, expressed in GU per liter) were calculated as follows: DLmeth = DLPCR × (f × F)/v; QLmeth = QLPCR × (f × F)/v. f refers to the proportion of concentrate used for DNA extraction (e.g., 1/1.6 for 1 ml of 1.6 ml), and F refers to the proportion of DNA extract used in the qPCR assay (e.g., 1/20, for 5 μl of 100 μl). v is the volume of the water sample filtered, expressed in liters. For DLmeth, the result was divided by 2, because the qPCR was performed in duplicate for each DNA extracted from a water sample. The results obtained by our Sg1-qPCR were quantified using the “iQCheck Analysis” software supplied by Bio-Rad (Marnes-la-Coquette, France).
Statistical analysis.
Agreement between qPCR and culture was estimated by Cohen's kappa coefficient. Because a gold standard was lacking, we used the Hui-Walter paradigm (28), which is the commonly accepted standard method for the evaluation of diagnostic tests. It introduces a latent class approach, which requires two (or more) tests evaluated in two (or more) subpopulations. This model assumes that (i) the prevalence of the disease is different within each population; (ii) the tests have the same properties across populations; and (iii) the tests are conditionally independent given the disease status. In this study, the two tests (culture and qPCR) had to be compared under various conditions (size of the building, age of the water supply system, hot water production method, localization, temperature, and chlorination), which define subpopulations of water samples and which may not be independent of each other. Therefore, we performed a multiple correspondence analysis of multiway tables formed by the combination of all conditions, followed by hierarchical clustering on principal components. The resulting binary classification was used for the main comparison of culture and qPCR. Furthermore, the chi-square test and a stratified McNemar test were used to analyze building and water sample characteristics.
Nucleotide sequence accession numbers.
LPS gene cluster sequences were submitted to GenBank and were assigned accession numbers FR749891 (ATCC 33215), FR749890 (ATCC 43283), FR747826 (ATCC 43290), FR747827 (ATCC 43736), and FR733637 (ATCC 43073).
RESULTS
L. pneumophila Sg1 encodes a specific gene cluster present within the genes encoding lipopolysaccharide biosynthesis proteins.
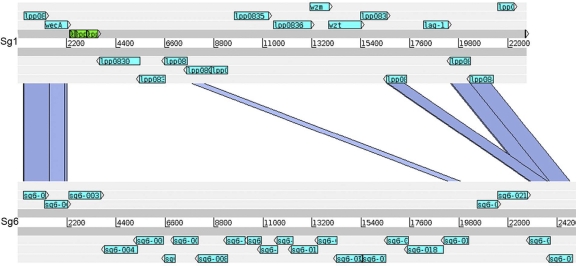
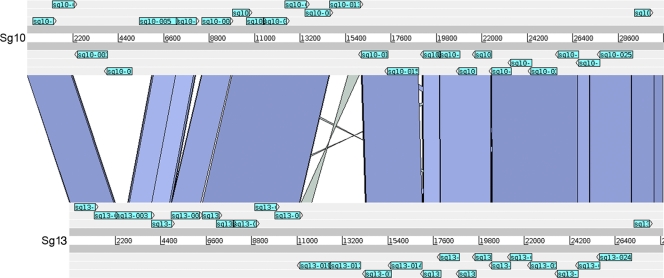
A comparative genomic hybridization analysis of 217 L. pneumophila strains belonging to the 15 serogroups present in the species found a highly specific correlation between Sg1 and the gene content of a 33-kb segment encoding genes involved in LPS biosynthesis. The LPS gene cluster of Sg1 can be subdivided into a 13-kb and a 20-kb region. While the 13-kb region was well conserved, the 20-kb region spanning lpp0827 to lpp0843 was highly specific to Sg1 strains, and three genes, wzm, wzt, and lpp0831, were identified as present in all Sg1 strains and only Sg1 strains (8). To confirm the hybridization results and to study the LPS gene clusters of different L. pneumophila serogroups in more detail, we undertook sequence analysis of this 20-kb region in six additional L. pneumophila serogroups. Using long-range PCR and subsequent shotgun sequencing, we were able to amplify and sequence this region in strains of Sg6, -10, -12, -13, and -14. As shown in Table 2, the size of this region ranged from 28.5 to 30.8 kb. Indeed, except for the flanking regions and a few genes within the region analyzed, the sequence of the Sg1 LPS gene cluster was highly divergent from those of the newly sequenced serogroups (Table 4; Fig. 1), further confirming the specificity of the LPS of Sg1 relative to those of other L. pneumophila serogroups. In contrast, comparison of the sequenced regions of non-Sg1 strains revealed high levels of similarity among some. In particular, Sg6 and Sg12 shared high similarity on the DNA as well as the protein level (Table 5), as did Sg10, Sg13, and Sg14 (Fig. 2). When our previously defined genetic markers for Sg1 (wzm, wzt, and lpp0831) were analyzed, homologous lpp0831 sequences were identified in the LPS clusters of Sg10, -13, and -14, encoding proteins with 40% amino acid similarity to Lpp0831. This gene was not detected by our array-based study, because the nucleotide identity is below the detection limit of 70%. In conclusion, our new results further substantiate the finding that L. pneumophila Sg1 encodes a conserved, specific region within its LPS gene cluster comprising at least two genetic markers of Sg1 strains, wzm and wzt.
TABLE 4.
The L. pneumophila Sg1-specific region of the LPS gene cluster (lpp0826 [rmlA] to lpp0844) is highly specific relative to those of Sg6, -10, -12, -13, and -14
| Gene name in L. pneumophila strain Paris | % Amino acid identitya for: |
||||
|---|---|---|---|---|---|
| Sg6 | Sg10 | Sg12 | Sg13 | Sg14 | |
| lpp0827 | 61 | 60 | 60 | 61 | 60 |
| lpp0828/wecA | 55 | 56 | 56 | 58 | 56 |
| lpp0830 | − | − | − | − | − |
| lpp0831 | − | 40 | − | 40 | 40 |
| lpp0832 | − | 35 | − | 35 | 35 |
| lpp0833 | 25 | 59 | 25 | 59 | 59 |
| lpp0834 | − | 60 | − | 60 | 60 |
| lpp0835 | − | − | − | − | − |
| lpp0836 | − | − | − | − | − |
| lpp0837/wzm | − | − | − | − | − |
| lpp0838/wzt | − | − | − | − | − |
| lpp0839 | 29 | 27 | 30 | 27 | 27 |
| lpp0840 | 64 | 65 | 65 | 65 | 65 |
| lpp0841/lag-1 | − | − | − | − | − |
| lpp0842 | 65 | 66 | 66 | 67 | 67 |
| lpp0843 | 69 | 69 | 69 | 69 | 70 |
Determined by BlastP comparison. −, the gene is absent from the LPS cluster of the indicated serogroup.
FIG. 1.
Comparison of the LPS genes lpp0827 to lpp0843 of L. pneumophila Sg1 strain Paris to those of L. pneumophila Sg6 strain ATCC 33215. The comparison of the two genomic regions is based on TBlastX. Blue bars indicate homologous regions in the two gene clusters based on a level of protein similarity greater than 25%. Light blue, genes predicted in this region; green, pseudogenes.
TABLE 5.
The L. pneumophila Sg6-specific region of the LPS gene cluster (lpp0826 [rmlA] to lpp0844) is specific relative to those of Sg10, -13, and -14 but highly similar to that of Sg12
| Gene name | % Amino acid identitya for: |
|||
|---|---|---|---|---|
| Sg10 | Sg12 | Sg13 | Sg14 | |
| sg6-001 | 96 | 96 | 98 | 96 |
| sg6-002 | 97 | 97 | 87 | 97 |
| sg6-003 | − | − | − | − |
| sg6-004 | 98 | 99 | 33 | 98 |
| sg6-005 | 79 | 99 | − | 79 |
| sg6-006 | − | 97 | − | − |
| sg6-007 | − | 97 | − | − |
| sg6-008 | − | 99 | − | − |
| sg6-009 | − | 99 | − | − |
| sg6-010 | − | 99 | − | − |
| sg6-011 | − | 100 | − | − |
| sg6-012 | − | 100 | − | − |
| sg6-013 | 27 | 99 | 28 | 27 |
| sg6-014 | − | 97 | − | − |
| sg6-015 | − | 98 | − | − |
| sg6-016 | − | 96 | − | − |
| sg6-017 | − | 99 | − | − |
| sg6-018 | − | 100 | − | − |
| sg6-019 | 25 | 100 | 25 | 25 |
| sg6-020 | − | 100 | − | − |
| sg6-021 | − | 99 | − | − |
| sg6-022 | 93 | 99 | 91 | 91 |
| sg6-023 | 97 | 97 | 97 | 96 |
| sg6-024 | 74 | 99 | 74 | 74 |
| sg6-025 | 26 | 99 | 26 | 26 |
| sg6-026 | 72 | 100 | 72 | 72 |
Determined by BlastP comparison. −, the gene is absent from the LPS cluster of the indicated serogroup.
FIG. 2.
Comparison of the LPS gene region of L. pneumophila Sg10 strain ATCC 43283 with that of L. pneumophila Sg13 strain ATCC 43736. The comparison of the two genomic regions is based on TBlastX. Blue bars indicate homologous regions in the two gene clusters based on a level of protein similarity greater than 25%. Light blue, genes predicted in this region.
Primers targeting wzm, encoding the transmembrane component of the ABC transporter of the O-antigenic polysaccharide of LPS, are genetic markers for Sg1 strains.
In order to investigate whether the three specific genes can be exploited as targets for Sg1-specific PCR assays, we designed primers best suited for differentiating L. pneumophila Sg1 from other L. pneumophila serogroups and other Legionella spp. In total, 11 primer pairs were tested for their specificity on 492 strains, of which 404 belonged to L. pneumophila Sg1 to Sg14, 50 to Legionella spp., and 38 to non-Legionella sp. strains (Table 1; see also Table S1 in the supplemental material). The best amplification was obtained with the P1-P2 primer pair (for endpoint PCR) (Table 3), targeting wzm; with this primer pair, all L. pneumophila Sg1 strains were indeed amplified (n = 257). None of the non-Sg1 strains showed a positive amplification signal. Thus, the specificity of primer set P1-P2 is 100%.
Development of an Sg1-qPCR method.
Our PCR screening of 492 strains suggested that the 294-bp region of the wzm gene, spanning nucleotides 99 to 392, is a highly specific genetic marker for Sg1 strains. We thus designed a primer pair and a probe for a qPCR assay targeting this region. This primer pair (P65-P66) (Table 3) showed the same specificity as P1-P2 on the 492 strains tested (100%). It was thus chosen for further development of the Sg1-qPCR. The detection limit (DLPCR) and the quantification limit (QLPCR) were revealed to be equal to those of the commercial qPCR (80 and 480 GU/liter, respectively).
The Sg1-qPCR is highly sensitive for the surveillance of water supply systems for contamination with L. pneumophila Sg1.
No method exists that allows rapid, specific, and simultaneous identification and typing of L. pneumophila contaminating water systems such as hospital water supplies, cooling towers, or water supplies of retirement homes. We thus tested our Sg1-PCR for its performance on water samples from hospitals. In total, 209 samples were collected from the water supply systems of 14 hospitals located in the Paris, France, area. Among the 209 samples tested, 60 were positive by our Sg1-qPCR; of these, 33 were confirmed by culture. This corresponds to a sensitivity of 84.6% and a positive predictive value (PPV) of 55% relative to culture as the gold standard (Table 6; see also Table S2 in the supplemental material). We then further investigated the 27 samples that were positive with the Sg1-qPCR but negative for L. pneumophila Sg1 by culture. Seven samples out of these 27 were culture positive for L. pneumophila Sg2-14. For the remaining 20 of the 27 Sg1-qPCR-positive samples, no colony was growing on the agar plate. However, when tested by the commercial qPCR identifying L. pneumophila (Bio-Rad), 26 out of the 27 Sg1-qPCR-positive but culture-negative samples tested positive (data not shown), suggesting that our Sg1-qPCR result was correct. Probably nonculturable L. pneumophila and/or DNA of nonviable Legionella was present in these samples. For 7 out of the 27 samples, culture confirmed the presence of L. pneumophila Sg2 to -14 but not Sg1. This suggests that these samples were contaminated with L. pneumophila Sg2 to -14 and that the Sg1 colonies had been overlooked when the culture was tested, since only 5 colonies are selected. Taken together, if we consider the 20 samples confirmed by the two different PCR methods as positive, only 7 out of 60 Sg1-positive samples contained non-Sg1 Legionella strains according to culture. However, this result might also be due to the fact that these samples had been contaminated by both Sg1 and Sg2 to -14 isolates, and L. pneumophila Sg1 may indeed have been present.
TABLE 6.
Qualitative comparison of Sg1-qPCR to culture for L. pneumophila Sg1
| Result by Sg1-qPCR | No. of samples with the following result by culture for L. pneumophila Sg1: |
||
|---|---|---|---|
| Positive | Negative | Total | |
| Positive | 33 | 27 | 60 |
| Negative | 6 | 143 | 149 |
| Total | 39 | 170 | 209 |
One hundred forty-nine of the 209 samples tested were negative according to our Sg1-qPCR (Table 6). This corresponds to a specificity of 84.1% and a negative predictive value of 95.5% relative to culture as the gold standard. qPCR inhibition was observed in 11.9% of samples (25/209) for Sg1-qPCR. Inhibitory activity was easily resolved by dilution of the DNA samples (see Materials and Methods). Surprisingly, 6 of the 149 Sg1-qPCR-negative samples were culture positive (Table 6). As shown in Table 7, one of these six samples was highly contaminated, as deduced from results obtained by culture, and was nonquantifiable by the Bio-Rad Legionella sp. qPCR (data not shown). The remaining five Sg1-qPCR-negative but culture-positive samples were nonquantifiable by culture and also negative or nonquantifiable by the Bio-Rad L. pneumophila qPCR (data not shown). This suggests problems with the DNA extraction for these samples.
TABLE 7.
Characterization of Sg1-qPCR-negative but culture-positive samples
| Culture quantification (CFU/liter)a | Culture identification (latex agglutination) | qPCR result for: |
No. of samples | |
|---|---|---|---|---|
| L. pneumophila | L. pneumophila Sg1 | |||
| NQ | L. pneumophila Sg1 | Negative | Negative | 2 |
| NQ | L. pneumophila Sg1 | NQ | Negative | 3 |
| 5,300 | L. pneumophila Sg1 | NQ | Negative | 1 |
| Total | 6 | |||
NQ, nonquantifiable.
In conclusion, Sg1-qPCR and culture results were concordant for 182 out of 209 samples tested. The kappa coefficient was 0.57, which might be considered good agreement (20). According to the Hui and Walter method, the prevalence of Sg1 was estimated as 0.29, with sensitivities and specificities of 1 and 0.99 for qPCR and 0.55 and 0.96 for culture, respectively.
Contamination of water supply systems depends on sample and building characteristics.
The buildings from which we collected the 209 water samples investigated were chosen according to several criteria known as risk factors for the propagation of Legionella: continuous chlorination, age (less or more than 30 years), the size of the water supply system (less or more than 25% of hospital beds), and the hot water production method (Table 8). For each building, 10 sanitary water samples were collected: (i) from the hot water supply outset (n = 1), (ii) from the hot water supply return loop (n = 1), (iii) from representative taps (n = 3), (iv) from disadvantaged taps (n = 4), and (v) a cold sanitary water sample from a representative tap without hot water (n = 1).
TABLE 8.
Distribution of positive L. pneumophila Sg1 water samples (by culture or qPCR) with respect to building and sample characteristics
| Characteristic | Total no. of samples | Positive samples |
P (culture vs qPCR)b | No. (%) of samples positive by Sg1-qPCR but negative by Sg1 culture | |||
|---|---|---|---|---|---|---|---|
| By Sg1 culture |
By Sg1-qPCR |
||||||
| No. (%) | Pa | No. (%) | Pa | ||||
| Size of the building | |||||||
| Principal | 109 | 20 (18) | NS | 31 (28) | NS | 0.02 | 14 (13) |
| Secondary | 100 | 19 (19) | 29 (29) | 13 (13) | |||
| Age of the water supply system | |||||||
| >30 yr | 109 | 20 (18) | NS | 31 (28) | NS | 0.02 | 15 (14) |
| <30 yr | 100 | 19 (19) | 29 (29) | 12 (12) | |||
| Production of hot water | |||||||
| Instantaneous | 149 | 30 (20) | NS | 47 (32) | NS | 0.02 | 21 (14) |
| Storage | 60 | 9 (15) | 13 (22) | 6 (10) | |||
| Type of sample | |||||||
| Hot water | |||||||
| Representative | 65 | 9 (14) | NS | 21 (32) | NS | 0.01 | 13 (20) |
| Disadvantaged | 81 | 19 (23) | 25 (31) | 9 (11) | |||
| Outset | 21 | 1 (5) | 4 (19) | 3 (14) | |||
| Return loop | 21 | 5 (24) | 6 (29) | 2 (10) | |||
| Cold water | 21 | 5 (24) | 4 (19) | 0 (0) | |||
| Temp | |||||||
| >23°C and <55°C | 129 | 28 (22) | NS | 37 (29) | NS | <0.01 | 13 (10) |
| <23°C or >55°C | 80 | 11 (14) | 23 (29) | 14 (18) | |||
| Chlorination | |||||||
| Yes | 108 | 27 (25) | <0.01 | 43 (40) | <0.01 | <0.01 | 20 (19) |
| No | 101 | 12 (12) | 17 (17) | 7 (7) | |||
Determined by the chi-square test. NS, not significant.
Determined by a stratified McNemar test.
Despite French rules to control the risk of Legionella contamination, 13 of the 21 (62%) buildings tested yielded samples positive by Sg1-qPCR and L. pneumophila Sg1 culture. All types of samples (hot water supply outset, return loop, representative taps, disadvantaged taps, and cold sanitary water) were found positive for L. pneumophila Sg1 at least once by both methods.
Compared to L. pneumophila Sg1 culture, Sg1-qPCR was significantly more frequently positive for samples collected from chlorinated buildings (40% versus 25% [P < 0.01]), for samples with temperatures between 23°C and 55°C (29% versus 22% [P < 0.01]), and for samples from disadvantaged taps (31% versus 23% [P < 0.01]). When samples collected from buildings with chlorinated and nonchlorinated water were compared, samples from buildings with chlorinated water were positive significantly more often than samples from buildings with nonchlorinated water by L. pneumophila Sg1 culture (25% versus 12% [P < 0.01]) and Sg1-qPCR (40% versus 17% [P < 0.01]). Finally, neither the size and age of the building nor the water production mode had a significant impact on positive results for L. pneumophila Sg1 either by the culture method or by Sg1-qPCR. Taken together, chlorination, water temperature, and the frequency of use of the system seem to be the most important factors for contamination with L. pneumophila Sg1.
Detection of L. pneumophila Sg1 by Sg1-qPCR is a rapid and reliable method for testing clinical samples.
For evaluation of the performance of the Sg1-qPCR developed here for the detection of L. pneumophila Sg1 in clinical cases, 96 respiratory samples from patients presenting with pneumonia were tested. A total of 22 out of the 96 samples tested were positive for L. pneumophila Sg1 by conventional methods (culture and/or urinary antigen testing); 3 samples were L. pneumophila PCR positive; and 2 samples were Legionella sp. PCR positive (Table 9). When our Sg1-qPCR was used, 22 out of the 96 samples tested indicated pneumonia due to L. pneumophila Sg 1. These comprised the 21 culture-proven samples and 1 of the 3 L. pneumophila PCR-positive samples. The only Sg1-qPCR-negative sample among the 22 diagnosed legionellosis cases was the sample identified by the urinary L. pneumophila Sg 1 antigen test. However, the Sg1-PCR might well be correct, since this case was not confirmed by any other method. This respiratory sample was negative by culture, negative by L. pneumophila PCR, and negative by nested sequence-based typing (SBT) (no amplification for 7 alleles as described by Ginevra and colleagues [22] [data not shown]). Thus, considering that the two Sg1-qPCR-negative but L. pneumophila PCR-positive samples were indeed L. pneumophila Sg2 to Sg15 strains (which cannot be confirmed, because no culture is available), and that the one Sg1-qPCR-positive and L. pneumophila PCR-positive sample was indeed an L. pneumophila Sg1 strain, our Sg1-qPCR shows a very high level of performance, with a sensitivity of 95.6% (22/23) and a specificity of 100% (73/73), for use in the diagnosis of legionellosis in clinical samples.
TABLE 9.
Analysis of 96 pulmonary samples by Sg1-qPCR and the urinary antigen test
| Type of case | No. of samples with the following Sg1-qPCR result: |
Total no. of samples | |
|---|---|---|---|
| Positive | Negative | ||
| Legionellosis, confirmed by: | |||
| Culture for L. pneumophila Sg1 or urinary antigen test for L. pneumophila Sg1 | 21 | 1a | 22 |
| L. pneumophila PCR | 1 | 2b | 3 |
| Legionella sp. PCR | 0 | 2 | 2 |
| Total legionellosis cases | 22 | 5 | 27 |
| Nonlegionellosis pneumonia | 0 | 69 | 69 |
| Total cases | 96 | ||
Urinary antigen positive, culture negative, L. pneumophila PCR negative, nested-SBT negative (no amplification for 7 alleles).
Culture and urinary antigen data were negative or not available for these samples, but positive results by L. pneumophila PCR or nested SBT show that these cases were caused by strains of L. pneumophila Sg1 or L. pneumophila Sg2 to Sg15.
DISCUSSION
Many studies have shown that qPCR for the detection of Legionella species in the environment is a promising method that could be used instead of culture (3, 4, 17, 33, 39, 40). However, in contrast to culture, available qPCR methods do not allow the identification of L. pneumophila Sg1, the L. pneumophila serogroup responsible for more than 85% of human infections. Furthermore, although the urinary antigen test for L. pneumophila Sg1 performs very well for community-acquired cases, the Sg1-qPCR method described here should also be useful for diagnosing clinical samples, in particular in the context of nosocomial legionellosis, for which the sensitivity of the urinary antigen test was reported to be less than 50% (27), as well as in epidemiological investigations implicating L. pneumophila strains of unknown serogroups.
Sequencing and comparative genomics by hybridization identified three genes coding for proteins implicated in lipopolysaccharide (LPS) biosynthesis as specific to Sg1 strains (8, 9). Our work thus focused on the in-depth sequence characterization of the specific genomic region of the LPS cluster in different L. pneumophila serogroups and subsequently on the development of a qPCR-based method allowing rapid and accurate detection and identification of L. pneumophila Sg1 in water samples, a tool needed for comprehensive surveillance. Analysis of the LPS gene cluster in five additional L. pneumophila strains belonging to Sg6, -10, -12, -13, and -14 revealed that the previously defined 20-kb region of the LPS gene cluster spanning lpp0827 to lpp0843 in L. pneumophila strain Paris (8) is indeed highly specific for Sg1 strains. For example, when this region is compared in Sg1 and Sg6, the second common serogroup in human disease (16), only three genes (lpp0828, lpp0840, and lpp0842) show an amino acid identity of >50%. The remaining genes are highly divergent or absent (Table 4). Sequence comparison of the different regions thus confirmed that the genes identified previously, in particular wzm, are possibly specific targets for a new Sg1-qPCR method.
Based on these results, we developed an Sg1-PCR and an Sg1-qPCR method that were tested on 492 Legionella strains and 209 water samples collected in hospital settings. The results showed that this assay was highly specific and sensitive for the detection and identification of L. pneumophila Sg1 in DNA samples extracted from culture (100%). Furthermore, the qPCR method is much faster (4 h versus 8 days) and easier than culture and allows, in addition, the direct quantification of the amount of L. pneumophila Sg1 present. In contrast, culture allows only an approximation of the quantity of L. pneumophila Sg1, by counting colonies by immunofluorescence or by the agglutination of some colonies (at least five colonies according to the standard method, AFNOR T90-431). Obviously, the presence of L. pneumophila Sg1 can be missed with the culture method, since the results depend on the colonies selected for further analysis. In our study, we identified seven Sg1-qPCR-positive but culture-negative samples, containing L. pneumophila strains of other serogroups that probably constituted the majority in culture.
The probable misidentification of L. pneumophila serogroups in some samples, combined with the problem of culture-negative but PCR-positive samples, represents the current challenge for PCR usage in the surveillance of community water supply systems. Twenty out of 60 Sg1-qPCR-positive specimens were culture negative in our study (12.9% of the water samples tested). However, the confirmation of the positivity of all but one sample by the Bio-Rad L. pneumophila qPCR (data not shown) indicates that these results may correspond to the detection of the DNA of misidentified colonies in the culture, viable but nonculturable cells, cells present within amoebae, or dead cells. Although it might be difficult to associate such results directly with a risk for legionellosis, they provide important information, because they can reveal the presence of a niche in the water supply system that may one day represent a risk. This hidden risk would not have been identified by culture until favorable conditions of growth were restored (such as a dip in the temperature or the failure of the chlorination system) and L. pneumophila could proliferate to high numbers. The combination of qPCR with extraction methods that allow the exclusion of dead bacteria could improve the significance of a positive signal (10-12, 17).
The Sg1-qPCR developed here showed a high level of performance; however, six water samples positive by culture gave negative results by Sg1-qPCR. This might have been due to the presence of PCR inhibitors in these water samples, as previously described (4), or to an insufficient amount of DNA obtained. The nature of the possible inhibitors was not investigated, but an additional purification step and/or dilutions of the DNA samples often allowed the removal of the inhibition and the restoration of the sensitivity of the Sg1-qPCR. Furthermore, the six Sg1-qPCR false-negative results were obtained for very weakly contaminated water samples (5/6) or for water samples for which commercial L. pneumophila qPCR (Bio-Rad) was only weakly positive, suggesting an extraction problem (1 out of the 6 samples was nonquantifiable by the commercial qPCR, but 5,300 CFU/liter were counted after culture). Consequently, the detection thresholds of our Sg1-qPCR and the culture method were similar; however, the Sg1-qPCR identified additional samples not detected by the culture method. In the future, the concentration and extraction phases probably need to be improved so as to discard PCR inhibitors in one step and to obtain a larger amount of DNA from the water samples.
In a second step, we undertook a random multicenter study to evaluate prospectively the contamination of hospital buildings, which are very often implicated in nosocomial legionellosis. Despite French recommendations and regulations to prevent Legionella growth in hospital water supply systems (15), an important proportion of the samples taken from hospitals was positive for L. pneumophila and L. pneumophila Sg1 by culture and Sg1-qPCR. Surprisingly, hospital water supply systems with continuous chlorination were more often found positive for L. pneumophila Sg1 by culture as well as by Sg1-qPCR than nonchlorinated systems. Continuous chlorination (1 to 2 mg/liter) of hospital hot water systems has been implemented in France over the past 10 years to combat Legionella contamination. However, our results indicate that this procedure is not efficient at totally eliminating L. pneumophila Sg1 and that the many false-negative culture results (25% positive by Sg1 culture but 40% positive by Sg1-qPCR) may miss many contaminated water systems and thus represent a danger to the patients. A similar result was obtained for water samples with a higher temperature (<23°C or >55°C), suggesting that slight heating ensured that Legionella could not be recovered by culture but could be identified by Sg1-qPCR. Thus, the implementation of molecular approaches might allow earlier and more-efficient action for Legionella eradication in contaminated water pipes. Combining the Sg1-qPCR newly described here, the first quantitative test for L. pneumophila Sg1 available, with the already commercialized qPCR specific for L. pneumophila should allow the correct evaluation of the risk of a hospital water installation. For example, if a hot water supply or cooling tower is positive for L. pneumophila Sg1, even at a low level, more-drastic decisions could be taken to control further proliferation of Legionella. Regulatory alerts or action levels could be associated with qualitative information about the amount of L. pneumophila Sg1 present. This test could also be very useful for quickly tracking an environmental source in case of a legionellosis outbreak, mainly due to L. pneumophila Sg1 (43).
That the LPS gene cluster contains serogroup-specific regions was also recently shown by Thürmer and colleagues, who developed an endpoint PCR-based method specific for L. pneumophila Sg1 that can be used instead of the agglutination test for the serotyping of L. pneumophila strains (38). The genes that were targeted by their study were lpp0833 and lpp0831, two genes that did not appear to be Sg1 specific in our combined analyses. However, since the nucleotide similarity between the orthologous genes is low, these genes might indeed be good markers and could be used to develop a qPCR method further. Thürmer and colleagues also proposed specific sequences that could distinguish between monoclonal subgroup Knoxville and the closely related subgroups Benidorm and Bellingham, which, again, are located in the LPS gene cluster (38). Very recently, Yong and colleagues reported on the identification of a serogroup-specific gene, lpg0744 (lpp0833), again part of the LPS gene cluster, by subtractive hybridization (42). Combined with the identification of an L. pneumophila species-specific gene encoding an apyrase, lpg1905 (lpp1880), these newly designed tests could significantly enhance our ability to detect and identify species and subgroups of Legionella rapidly and accurately (42). Furthermore, in the future, additional specific primers can be defined for the sequences of the LPS gene cluster regions of other L. pneumophila non-Sg1 serogroups, and in particular also for L. longbeachae, which also carries very specific LPS-encoding genes (7). In the long term, this study opens new perspectives for the diagnosis of legionellosis in that it allows simultaneous Sg1-qPCR and L. pneumophila qPCR on clinical specimens. This approach can be used to complement bacteriological culture and antigen detection, allowing rapid and specific diagnosis.
Supplementary Material
Acknowledgments
This work received financial support from the Institut Pasteur-APHP program CA-01234 (PTR284) and the Centre National de la Recherche (CNRS). L. Gomez-Valero is the holder of an Institut Carnot postdoctoral research fellowship. The development of ACT and Artemis is funded by the Wellcome Trust, through its support of the Pathogen Genomics Group at the Sanger Centre, United Kingdom.
Footnotes
Published ahead of print on 30 December 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Albert-Weissenberger, C., C. Cazalet, and C. Buchrieser. 2007. Legionella pneumophila—a human pathogen that co-evolved with fresh water protozoa. Cell. Mol. Life Sci. 64:432-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Association Française de Normalisation. 2003. Water quality. Detection and enumeration of Legionella spp. and Legionella pneumophila. Method by direct inoculation and after concentration by membrane filtration of centrifugation. AFNOR NF T90-431. Association Française de Normalisation, La Plaine Saint-Denis, France.
- 3.Association Française de Normalisation. 2006. Water quality. Detection and quantification of Legionella and/or Legionella pneumophila by concentration and genic amplification by polymerase chain reaction (PCR). AFNOR XP T90-471. Association Française de Normalisation, La Plaine Saint-Denis, France.
- 4.Behets, J., P. Declerck, Y. Delaedt, B. Creemers, and F. Ollevier. 2007. Development and evaluation of a Taqman duplex real-time PCR quantification method for reliable enumeration of Legionella pneumophila in water samples. J. Microbiol. Methods 68:137-144. [DOI] [PubMed] [Google Scholar]
- 5.Benson, R. F., and B. S. Fields. 1998. Classification of the genus Legionella. Semin. Respir. Infect. 13:90-99. [PubMed] [Google Scholar]
- 6.Carver, T. J., et al. 2005. ACT: the Artemis Comparison Tool. Bioinformatics (Oxford, England) 21:3422-3423. [DOI] [PubMed] [Google Scholar]
- 7.Cazalet, C., et al. 2010. The Legionella longbeachae genome and transcriptome uncovers unique strategies to cause Legionnaires' disease. PLoS Genet. 6:e1000851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cazalet, C., et al. 2008. Multigenome analysis identifies a worldwide distributed epidemic Legionella pneumophila clone that emerged within a highly diverse species. Genome Res. 18:431-441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cazalet, C., et al. 2004. Evidence in the Legionella pneumophila genome for exploitation of host cell functions and high genome plasticity. Nat. Genet. 36:1165-1173. [DOI] [PubMed] [Google Scholar]
- 10.Chang, B., et al. 2009. Specific detection of viable Legionella cells by combined use of photoactivated ethidium monoazide and PCR/real-time PCR. Appl. Environ. Microbiol. 75:147-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen, N. T., and C. W. Chang. 2010. Rapid quantification of viable legionellae in water and biofilm using ethidium monoazide coupled with real-time quantitative PCR. J. Appl. Microbiol. 109:623-634. [DOI] [PubMed] [Google Scholar]
- 12.Delgado-Viscogliosi, P., L. Solignac, and J. M. Delattre. 2009. Viability PCR, a culture-independent method for rapid and selective quantification of viable Legionella pneumophila cells in environmental water samples. Appl. Environ. Microbiol. 75:3502-3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Diederen, B. M., et al. 2007. Evaluation of real-time PCR for the early detection of Legionella pneumophila DNA in serum samples. J. Med. Microbiol. 56:94-101. [DOI] [PubMed] [Google Scholar]
- 14.Diederen, B. M., J. A. Kluytmans, C. M. Vandenbroucke-Grauls, and M. F. Peeters. 2008. Utility of real-time PCR for diagnosis of Legionnaires' disease in routine clinical practice. J. Clin. Microbiol. 46:671-677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Direction Générale de la Santé. 2002. Circulaire DGS/SD7A/SD5C/DHOS/E4 no. 2002/243 du 22 avril 2002 relative à la prévention du risque lié aux légionelles dans les établissements de santé. Bulletin officiel. Direction Générale de la Santé, Paris, France.
- 16.Doleans, A., et al. 2004. Clinical and environmental distributions of Legionella strains in France are different. J. Clin. Microbiol. 42:458-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dusserre, E., et al. 2008. A PCR-based method for monitoring Legionella pneumophila in water samples detects viable but noncultivable legionellae that can recover their cultivability. Appl. Environ. Microbiol. 74:4817-4824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fields, B. S. 2008. Legionella in the environment, p. 85-91. In P. Hoffmann, H. Friedman, and M. Bendinelli (ed.), Legionella pneumophila: pathogenesis and immunity. Springer, New York, NY.
- 19.Fields, B. S., R. F. Benson, and R. E. Besser. 2002. Legionella and Legionnaires' disease: 25 years of investigation. Clin. Microbiol. Rev. 15:506-526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fleiss, J. L. 1981. Statistical methods for rates and proportions, 2nd ed., p. 38-46. John Wiley & Sons, Inc., New York, NY.
- 21.Frangeul, L., et al. 2004. CAAT-Box, Contigs-Assembly and Annotation Tool-Box for genome sequencing projects. Bioinformatics 20:790-797. [DOI] [PubMed] [Google Scholar]
- 22.Ginevra, C., et al. 2009. Evaluation of a nested-PCR-derived sequence-based typing method applied directly to respiratory samples from patients with Legionnaires' disease. J. Clin. Microbiol. 47:981-987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Glaser, P., et al. 2001. Comparative genomics of Listeria species. Science 294:849-852. [DOI] [PubMed] [Google Scholar]
- 24.Hay, J., D. V. Seal, B. Billcliffe, and J. H. Freer. 1995. Non-culturable Legionella pneumophila associated with Acanthamoeba castellanii: detection of the bacterium using DNA amplification and hybridization. J. Appl. Bacteriol. 78:61-65. [DOI] [PubMed] [Google Scholar]
- 25.Hayden, R. T., et al. 2001. Direct detection of Legionella species from bronchoalveolar lavage and open lung biopsy specimens: comparison of LightCycler PCR, in situ hybridization, direct fluorescence antigen detection, and culture. J. Clin. Microbiol. 39:2618-2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Helbig, J. H., et al. 2002. Pan-European study on culture-proven Legionnaires' disease: distribution of Legionella pneumophila serogroups and monoclonal subgroups. Eur. J. Clin. Microbiol. Infect. Dis. 21:710-716. [DOI] [PubMed] [Google Scholar]
- 27.Helbig, J. H., et al. 2003. Clinical utility of urinary antigen detection for diagnosis of community-acquired, travel-associated, and nosocomial Legionnaires' disease. J. Clin. Microbiol. 41:838-840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hui, S. L., and S. D. Walter. 1980. Estimating the error rates of diagnostic tests. Biometrics 36:167-171. [PubMed] [Google Scholar]
- 29.International Organization for Standardization. 1998. Water quality—detection and enumeration of Legionella. ISO11731:1998. International Organization for Standardization, Geneva, Switzerland.
- 30.Joly, P., et al. 2006. Quantitative real-time Legionella PCR for environmental water samples: data interpretation. Appl. Environ. Microbiol. 72:2801-2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koide, M., et al. 2006. Detection of Legionella species in clinical samples: comparison of polymerase chain reaction and urinary antigen detection kits. Infection 34:264-268. [DOI] [PubMed] [Google Scholar]
- 32.Lück, P. C., et al. 2010. Legionella dresdenensis sp. nov., isolated from river water. Int. J. Syst. Evol. Microbiol. 60:2557-2562. [DOI] [PubMed] [Google Scholar]
- 33.Morio, F., et al. 2008. Real-time PCR assay for the detection and quantification of Legionella pneumophila in environmental water samples: utility for daily practice. Int. J. Hyg. Environ. Health 211:403-411. [DOI] [PubMed] [Google Scholar]
- 34.Nazarian, E. J., D. J. Bopp, A. Saylors, R. J. Limberger, and K. A. Musser. 2008. Design and implementation of a protocol for the detection of Legionella in clinical and environmental samples. Diagn. Microbiol. Infect. Dis. 62:125-132. [DOI] [PubMed] [Google Scholar]
- 35.Ratcliff, R. M., J. A. Lanser, P. A. Manning, and M. W. Heuzenroeder. 1998. Sequence-based classification scheme for the genus Legionella targeting the mip gene. J. Clin. Microbiol. 36:1560-1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reyrolle, M., et al. 2004. Rapid identification of Legionella pneumophila serogroups by latex agglutination. Eur. J. Clin. Microbiol. Infect. Dis. 23:864-866. [DOI] [PubMed] [Google Scholar]
- 37.Steinert, M., U. Hentschel, and J. Hacker. 2002. Legionella pneumophila: an aquatic microbe goes astray. FEMS Microbiol. Rev. 26:149-162. [DOI] [PubMed] [Google Scholar]
- 38.Thürmer, A., J. H. Helbig, E. Jacobs, and P. C. Lück. 2009. PCR-based ‘serotyping’ of Legionella pneumophila. J. Med. Microbiol. 58:588-595. [DOI] [PubMed] [Google Scholar]
- 39.Wellinghausen, N., C. Frost, and R. Marre. 2001. Detection of legionellae in hospital water samples by quantitative real-time LightCycler PCR. Appl. Environ. Microbiol. 67:3985-3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yaradou, D. F., et al. 2007. Integrated real-time PCR for detection and monitoring of Legionella pneumophila in water systems. Appl. Environ. Microbiol. 73:1452-1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yong, S. F., F. N. Goh, and Y. F. Ngeow. 2010. Legionella species and serogroups in Malaysian water cooling towers: identification by latex agglutination and PCR-DNA sequencing of isolates. J. Water Health 8:92-100. [DOI] [PubMed] [Google Scholar]
- 42.Yong, S. F. Y., et al. 2010. Molecular detection of Legionella: moving on from mip. Front. Cell. Infect. Microbiol. 1:123. doi: 10.3389/fmicb.2010.00123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu, V. L., et al. 2002. Distribution of Legionella species and serogroups isolated by culture in patients with sporadic community-acquired legionellosis: an international collaborative survey. J. Infect. Dis. 186:127-128. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.