Abstract
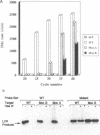
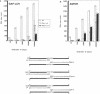
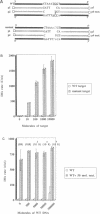
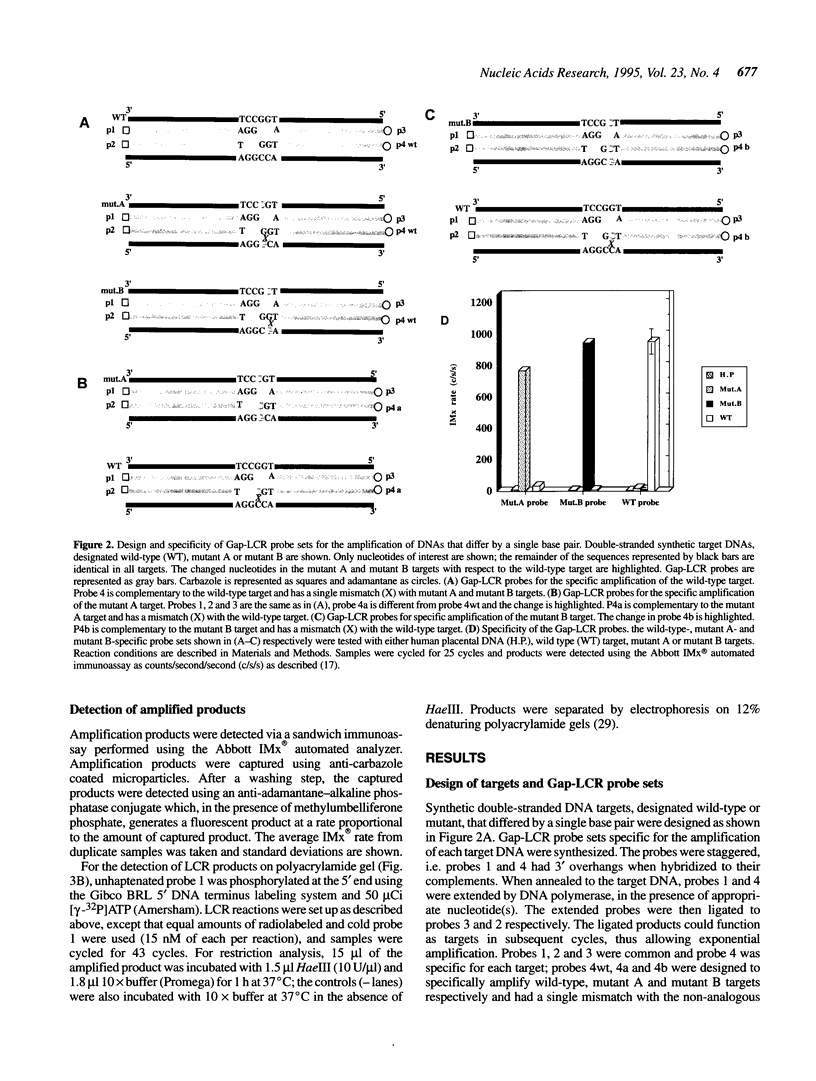
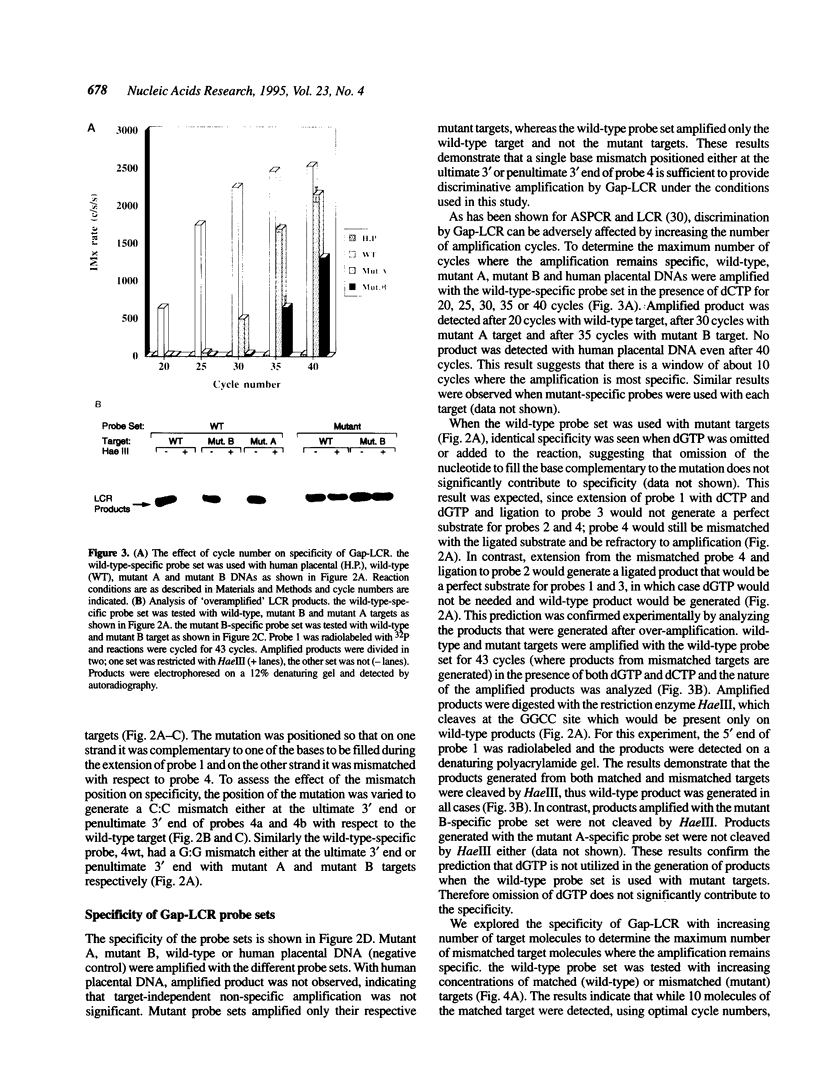
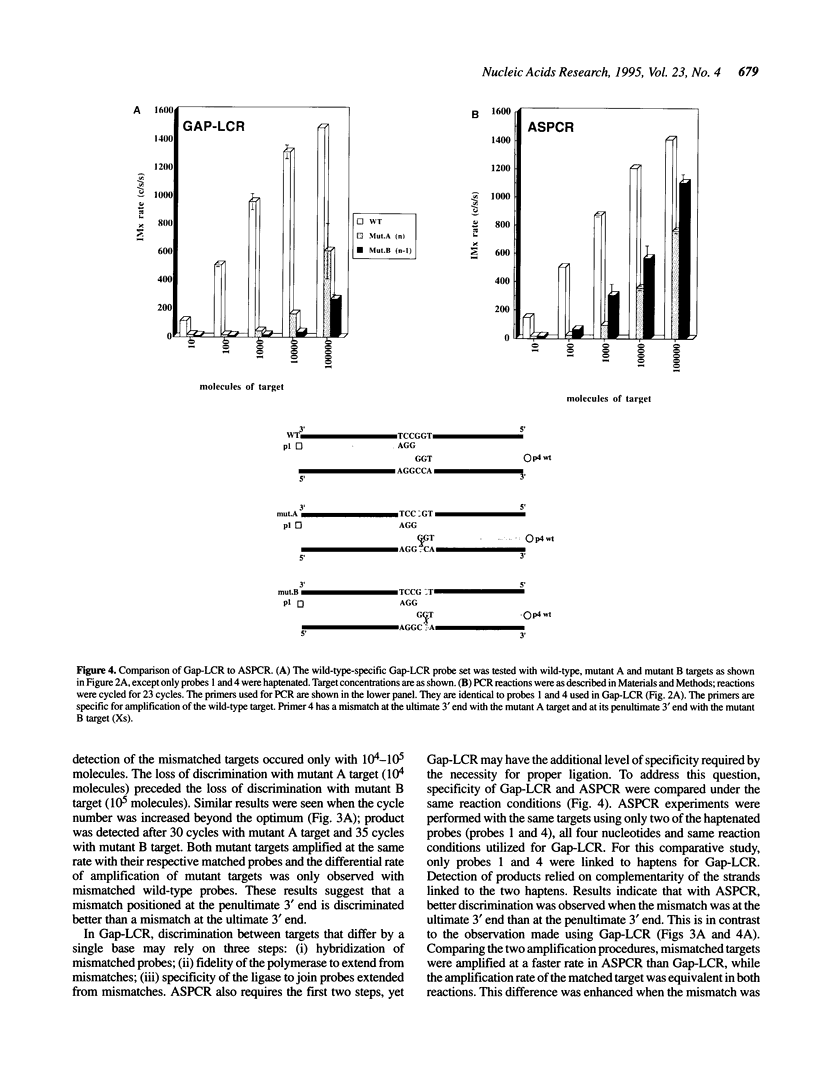
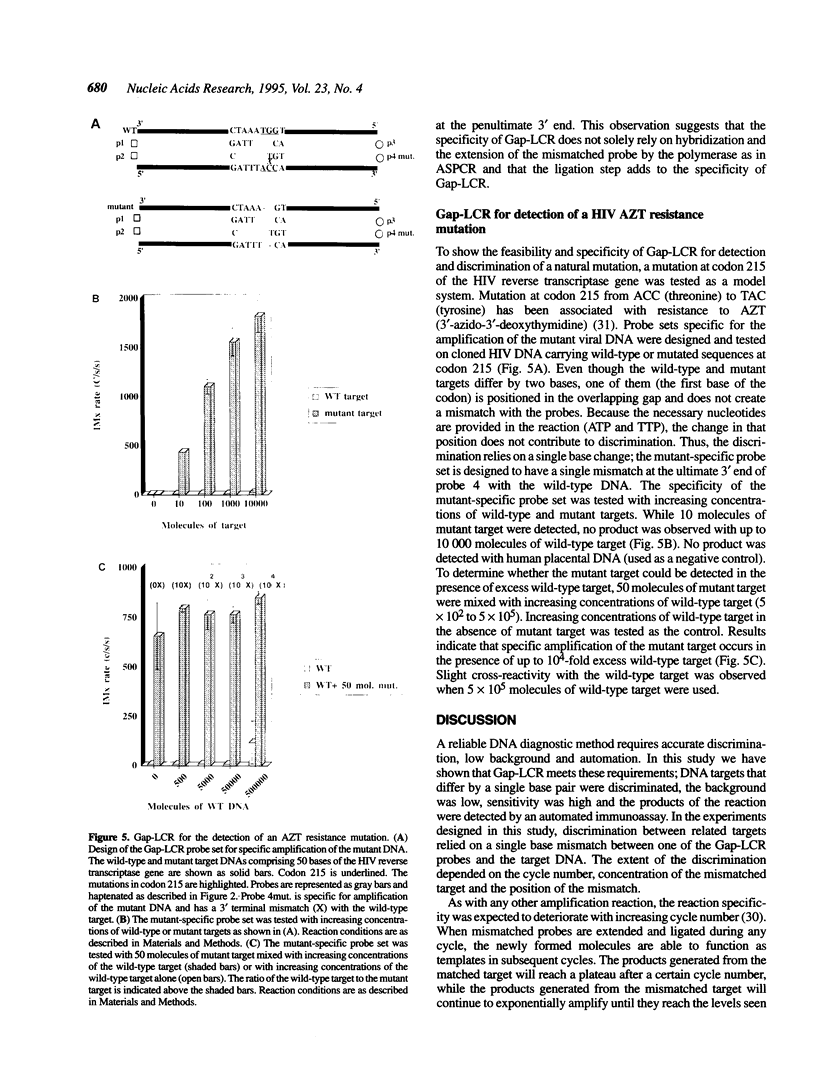
DNA amplification systems are powerful technologies with the potential to impact a wide range of diagnostic applications. In this study we explored the feasibility and limitations of a modified ligase chain reaction (Gap-LCR) in detection and discrimination of DNAs that differ by a single base. LCR is a DNA amplification technology based on the ligation of two pairs of synthetic oligonucleotides which hybridize at adjacent positions to complementary strands of a target DNA. Multiple rounds of denaturation, annealing and ligation with a thermostable ligase result in the exponential amplification of the target DNA. A modification of LCR, Gap-LCR was developed to reduce the background generated by target-independent, blunt-end ligation. In Gap-LCR, DNA polymerase fills in a gap between annealed probes which are subsequently joined by DNA ligase. We have designed synthetic DNA targets with single base pair differences and analyzed them in a system where three common probes plus an allele-specific probe were used. A single base mismatch either at the ultimate 3' end or penultimate 3' end of the allele specific probe was sufficient for discrimination, though better discrimination was obtained with a mismatch at the penultimate 3' position. Comparison of Gap-LCR to allele-specific PCR (ASPCR) suggested that Gap-LCR has the advantage of having the additive effect of polymerase and ligase on specificity. As a model system, Gap-LCR was tested on a mutation in the reverse transcriptase gene of HIV, specifically, one of the mutations that confers AZT resistance. Mutant DNA could be detected and discriminated in the presence of up to 10,000-fold excess of wild-type DNA.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barany F. Genetic disease detection and DNA amplification using cloned thermostable ligase. Proc Natl Acad Sci U S A. 1991 Jan 1;88(1):189–193. doi: 10.1073/pnas.88.1.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkenmeyer L. G., Mushahwar I. K. DNA probe amplification methods. J Virol Methods. 1991 Nov-Dec;35(2):117–126. doi: 10.1016/0166-0934(91)90127-l. [DOI] [PubMed] [Google Scholar]
- Birkenmeyer L., Armstrong A. S. Preliminary evaluation of the ligase chain reaction for specific detection of Neisseria gonorrhoeae. J Clin Microbiol. 1992 Dec;30(12):3089–3094. doi: 10.1128/jcm.30.12.3089-3094.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha R. S., Zarbl H., Keohavong P., Thilly W. G. Mismatch amplification mutation assay (MAMA): application to the c-H-ras gene. PCR Methods Appl. 1992 Aug;2(1):14–20. doi: 10.1101/gr.2.1.14. [DOI] [PubMed] [Google Scholar]
- Cotton R. G., Rodrigues N. R., Campbell R. D. Reactivity of cytosine and thymine in single-base-pair mismatches with hydroxylamine and osmium tetroxide and its application to the study of mutations. Proc Natl Acad Sci U S A. 1988 Jun;85(12):4397–4401. doi: 10.1073/pnas.85.12.4397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dille B. J., Butzen C. C., Birkenmeyer L. G. Amplification of Chlamydia trachomatis DNA by ligase chain reaction. J Clin Microbiol. 1993 Mar;31(3):729–731. doi: 10.1128/jcm.31.3.729-731.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs R. A., Nguyen P. N., Caskey C. T. Detection of single DNA base differences by competitive oligonucleotide priming. Nucleic Acids Res. 1989 Apr 11;17(7):2437–2448. doi: 10.1093/nar/17.7.2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada K., Orgel L. E. Unexpected substrate specificity of T4 DNA ligase revealed by in vitro selection. Nucleic Acids Res. 1993 May 25;21(10):2287–2291. doi: 10.1093/nar/21.10.2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatt C., Ward M. E., Clarke I. N. Analysis of the entire nucleotide sequence of the cryptic plasmid of Chlamydia trachomatis serovar L1. Evidence for involvement in DNA replication. Nucleic Acids Res. 1988 May 11;16(9):4053–4067. doi: 10.1093/nar/16.9.4053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M. M., Arnheim N., Goodman M. F. Extension of base mispairs by Taq DNA polymerase: implications for single nucleotide discrimination in PCR. Nucleic Acids Res. 1992 Sep 11;20(17):4567–4573. doi: 10.1093/nar/20.17.4567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kan Y. W., Dozy A. M. Antenatal diagnosis of sickle-cell anaemia by D.N.A. analysis of amniotic-fluid cells. Lancet. 1978 Oct 28;2(8096):910–912. doi: 10.1016/s0140-6736(78)91629-x. [DOI] [PubMed] [Google Scholar]
- Kwok S., Kellogg D. E., McKinney N., Spasic D., Goda L., Levenson C., Sninsky J. J. Effects of primer-template mismatches on the polymerase chain reaction: human immunodeficiency virus type 1 model studies. Nucleic Acids Res. 1990 Feb 25;18(4):999–1005. doi: 10.1093/nar/18.4.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kälin I., Shephard S., Candrian U. Evaluation of the ligase chain reaction (LCR) for the detection of point mutations. Mutat Res. 1992 Oct;283(2):119–123. doi: 10.1016/0165-7992(92)90143-6. [DOI] [PubMed] [Google Scholar]
- Laffler T. G., Carrino J. J., Marshall R. L. The ligase chain reaction in DNA-based diagnosis. Ann Biol Clin (Paris) 1993;51(9):821–826. [PubMed] [Google Scholar]
- Landegren U., Kaiser R., Sanders J., Hood L. A ligase-mediated gene detection technique. Science. 1988 Aug 26;241(4869):1077–1080. doi: 10.1126/science.3413476. [DOI] [PubMed] [Google Scholar]
- Larder B. A., Kemp S. D. Multiple mutations in HIV-1 reverse transcriptase confer high-level resistance to zidovudine (AZT). Science. 1989 Dec 1;246(4934):1155–1158. doi: 10.1126/science.2479983. [DOI] [PubMed] [Google Scholar]
- Myers R. M., Larin Z., Maniatis T. Detection of single base substitutions by ribonuclease cleavage at mismatches in RNA:DNA duplexes. Science. 1985 Dec 13;230(4731):1242–1246. doi: 10.1126/science.4071043. [DOI] [PubMed] [Google Scholar]
- Myers R. M., Lumelsky N., Lerman L. S., Maniatis T. Detection of single base substitutions in total genomic DNA. Nature. 1985 Feb 7;313(6002):495–498. doi: 10.1038/313495a0. [DOI] [PubMed] [Google Scholar]
- Newton C. R., Graham A., Heptinstall L. E., Powell S. J., Summers C., Kalsheker N., Smith J. C., Markham A. F. Analysis of any point mutation in DNA. The amplification refractory mutation system (ARMS). Nucleic Acids Res. 1989 Apr 11;17(7):2503–2516. doi: 10.1093/nar/17.7.2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickerson D. A., Kaiser R., Lappin S., Stewart J., Hood L., Landegren U. Automated DNA diagnostics using an ELISA-based oligonucleotide ligation assay. Proc Natl Acad Sci U S A. 1990 Nov;87(22):8923–8927. doi: 10.1073/pnas.87.22.8923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orita M., Iwahana H., Kanazawa H., Hayashi K., Sekiya T. Detection of polymorphisms of human DNA by gel electrophoresis as single-strand conformation polymorphisms. Proc Natl Acad Sci U S A. 1989 Apr;86(8):2766–2770. doi: 10.1073/pnas.86.8.2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prosser J. Detecting single-base mutations. Trends Biotechnol. 1993 Jun;11(6):238–246. doi: 10.1016/0167-7799(93)90135-V. [DOI] [PubMed] [Google Scholar]
- Saiki R. K., Bugawan T. L., Horn G. T., Mullis K. B., Erlich H. A. Analysis of enzymatically amplified beta-globin and HLA-DQ alpha DNA with allele-specific oligonucleotide probes. Nature. 1986 Nov 13;324(6093):163–166. doi: 10.1038/324163a0. [DOI] [PubMed] [Google Scholar]
- Wiedmann M., Barany F., Batt C. A. Detection of Listeria monocytogenes with a nonisotopic polymerase chain reaction-coupled ligase chain reaction assay. Appl Environ Microbiol. 1993 Aug;59(8):2743–2745. doi: 10.1128/aem.59.8.2743-2745.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedmann M., Czajka J., Barany F., Batt C. A. Discrimination of Listeria monocytogenes from other Listeria species by ligase chain reaction. Appl Environ Microbiol. 1992 Nov;58(11):3443–3447. doi: 10.1128/aem.58.11.3443-3447.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D. Y., Ugozzoli L., Pal B. K., Wallace R. B. Allele-specific enzymatic amplification of beta-globin genomic DNA for diagnosis of sickle cell anemia. Proc Natl Acad Sci U S A. 1989 Apr;86(8):2757–2760. doi: 10.1073/pnas.86.8.2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D. Y., Wallace R. B. Specificity of the nick-closing activity of bacteriophage T4 DNA ligase. Gene. 1989;76(2):245–254. doi: 10.1016/0378-1119(89)90165-0. [DOI] [PubMed] [Google Scholar]
- Wu D. Y., Wallace R. B. The ligation amplification reaction (LAR)--amplification of specific DNA sequences using sequential rounds of template-dependent ligation. Genomics. 1989 May;4(4):560–569. doi: 10.1016/0888-7543(89)90280-2. [DOI] [PubMed] [Google Scholar]