Abstract
Vibrio fischeri serves as a valuable model of bacterial bioluminescence, its regulation, and its functional significance. Light output varies more than 10,000-fold in wild-type isolates from different environments, yet dim and bright strains have similar organization of the light-producing lux genes, with the activator-encoding luxR divergently transcribed from luxICDABEG. By comparing the genomes of bright strain MJ11 and the dimmer ES114, we found that the lux region has diverged more than most shared orthologs, including those flanking lux. Divergence was particularly high in the intergenic sequence between luxR and luxI. Analysis of the intergenic lux region from 18 V. fischeri strains revealed that, with one exception, sequence divergence essentially mirrored strain phylogeny but with relatively high substitution rates. The bases conserved among intergenic luxR-luxI sequences included binding sites for known regulators, such as LuxR and ArcA, and bases of unknown significance, including a striking palindromic repeat. By using this collection of diverse luxR-luxI regions, we found that expression of PluxI-lacZ but not PluxR-lacZ transcriptional reporters correlated with the luminescence output of the strains from which the promoters originated. We also found that exchange of a small stretch of the luxI-luxR intergenic region between two strains largely reversed their relative brightness. Our results show that the luxR-luxI intergenic region contributes significantly to the variable luminescence output among V. fischeri strains isolated from different environments, although other elements of strain backgrounds also contribute. Moreover, the lux system appears to have evolved relatively rapidly, suggesting unknown environment-specific selective pressures.
Bioluminescence is widespread within the Vibrionaceae; however, the selective pressures driving its evolution are not always clear (62). Further adding to the mystery of bacterial bioluminescence, light output can vary over orders of magnitude between different environmental isolates. For example, obvious visible luminescence has been frequently associated with a number of species, such as Vibrio fischeri, Vibrio harveyi, Photobacterium leiognathi, and Photobacterium phosphoreum; however, dim and even “cryptic” luminescence has also been observed upon careful examination of certain isolates, such as Vibrio salmonicida (20). Even isolates of the same species can vary in luminescence output (23, 49). Such variation is dramatically evident in V. fischeri (5, 32, 45, 62), which is a longstanding model for studying bioluminescence, its functional roles, and its regulation.
Bioluminescence in V. fischeri is directed by the lux genes. The genes responsible for bioluminescence, luxABCDE and luxG are clustered with the regulatory genes luxR and luxI and are arranged in two divergent transcripts composed of luxR and luxICDABEG (17, 18, 22, 41). The regulatory genes, luxI and luxR, constitute a pheromone-mediated regulatory mechanism referred to as quorum sensing. LuxI produces a membrane-permeable autoinducer pheromone, N-3-oxohexanoyl-l-homoserine lactone (3-oxo-C6-HSL) (15, 29). As cell density increases, 3-oxo-C6-HSL accumulates until achieving a threshold concentration, whereupon it combines with LuxR to activate transcription of luxICDABEG (17, 18, 72). V. fischeri generates two additional autoinducers, N-octanoyl-l-homoserine lactone (C8-HSL) and autoinducer 2 (AI-2), which are produced by AinS and LuxS, respectively. C8-HSL and AI-2 regulate luminescence in part through a pathway involving LuxU, LuxO, and LitR, resulting in increased LuxR at high cell density (19, 35-37, 43). We previously reviewed the interplay of these regulatory systems (68). Ultimately, all three autoinducers affect transcription from the intergenic luxR-luxI region.
For reasons that are not well understood, the brightness of different wild-type V. fischeri strains can vary greatly (5, 45). For example, a comparison of strains MJ1 (57) and ES114 demonstrated that MJ1 has ∼10,000-fold-higher luminescence (5, 8). Moreover, saturating ES114 with exogenously added autoinducer does not increase the luminescence of ES114 to the levels of MJ1, and ES114 produces little endogenous 3-oxo-C6-HSL autoinducer, even when luxI is induced fully by exogenously added autoinducer (22). These data demonstrate that different strains not only produce varied levels of luminescence but also may have different maximal capacities for luxICDABEG expression.
The evolution of such varied luminescence output and the underlying reason for the phenomenon are not well understood. There are several plausible explanations for the variable luxICDABEG expression among strains from different environments, and the mechanism(s) underpinning this variation could involve one or multiple Lux proteins, regulatory sequences at the luxI or luxR promoters, other elements of the strain background, or a combination of factors.
To explore these possibilities, and in particular to investigate the contributions of the lux locus, we systematically compared the light output, lux sequences, and other gene sequences from strains with different luminescence capacities. Our results suggest that the luxR-luxI intergenic region has been evolving rapidly and contributes significantly to the variable luminescence output among V. fischeri strains, although other elements of strain background contribute as well.
MATERIALS AND METHODS
Strains and media.
V. fischeri strains used in this study are listed in Table 1. Escherichia coli strains DH5α (24) and DH5αλpir (13) were used for cloning, with the latter being employed with plasmids bearing the R6Kγ origin of replication, including pAKD702 and its derivatives. E. coli CC118λpir (26) carrying helper plasmid pEVS104 (67) was used in triparental matings. E. coli was grown in LB medium (42), and V. fischeri was grown in LBS (66) or SWTO (7) medium. Agar at 15 mg ml−1 was added to solidify medium for plating. Chloramphenicol, kanamycin, and ampicillin were added at 20, 40, and 100 μg ml−1, respectively, for selection of E. coli on LB. Chloramphenicol was added to LBS at 2 μg ml−1 for selection of V. fischeri.
TABLE 1.
Wild-type V. fischeri strains and select corresponding plasmids used in this study
| Strain | Species or description | Source or reference | Luminescence classa | Intergenic lux cloneb | PluxR-lacZ reporterc | PluxI-lacZ reporterc |
|---|---|---|---|---|---|---|
| CG101 | Cleidopus gloriamaris | 56 | HV | pJLB156 | pJLB180 | pJLB181 |
| CG103 | Cleidopus gloriamaris | 31 | HV | pJLB26 | pJLB180 | pJLB181 |
| EM17 | Euprymna morsei | 56 | V | pJLB165 | pJLB196 | pJLB197 |
| ES114 | Euprymna scolopes | 5 | NV | pJLB29 | pJLB170 | pJLB171 |
| ES12 | Euprymna scolopes | 6 | NV | pJLB161 | pJLB188 | pJLB189 |
| ES213 | Euprymna scolopes | 6 | NV | pJLB166 | pJLB188 | pJLB189 |
| ES401 | Euprymna scolopes | 56 | NV | pJLB167 | pJLB198 | pJLB199 |
| ET101 | Euprymna tasmanica | 48 | V | pJLB163 | pJLB192 | pJLB193 |
| ET401 | Euprymna tasmanica | 47 | V | pJLB164 | pJLB194 | pJLB195 |
| H905 | Hawaii, planktonic | 33 | V | pJLB168 | pJLB200 | pJLB201 |
| MJ1Sd | Monocentris japonica | Lab collection | HV | pJLB30 | pJLB172 | pJLB173 |
| MJ1 | Monocentris japonica | E. P. Greeenberg | HV | pJLB79 | pJLB204 | pJLB205 |
| MJ11 | Monocentris japonica | 56 | HV | pJLB157 | pJLB172 | pJLB173 |
| PP3 | Hawaii, planktonic | 33 | V | pJLB25 | pJLB202 | pJLB203 |
| SA1 | Sepiola affinis | 21 | HV | pJLB159 | pJLB184 | pJLB185 |
| SR5 | Sepiola robusta | 21 | HV | pJLB162 | pJLB190 | pJLB191 |
| VLS2 | Euprymna scolopes | 31 | V | pJLB158 | pJLB182 | pJLB183 |
| WH1 | Massachusetts, planktonic | 31 | HV | pJLB160 | pJLB186 | pJLB187 |
| Mutant strains | ||||||
| JB3 | ES114; intergenic lux element from MJ1S | This study | V | NAe | pJLB174 | pJLB175 |
| JB7 | MJ1S; intergenic lux element from ES114 | This study | V | NA | pJLB176 | pJLB177 |
Strains were designated as producing nonvisible (NV), visible (V), or highly visible (HV) luminescence on SWTO plates.
lux intergenic regions were cloned in pCR-Blunt or pCR-BluntII-TOPO (see Materials and Methods).
Some strains had identical luxR-luxI intergenic sequences, and only one PluxR- and PluxI-lacZ reporter plasmid was made for each unique sequence; therefore, some reporter plasmids represent more than one strain (including CG101 and CG103, MJ1S and MJ11, and ES12 and ES213).
NA, not applicable (see Materials and Methods).
OD595 and luminescence measurements.
For measurements of growth and luminescence, overnight cultures grown in LBS were diluted 1:500 in SWTO, incubated at 24°C with shaking (200 rpm), and monitored as described previously (7, 8). Briefly, 500-μl samples were taken at regular intervals, and the optical density at 595 nm (OD595) was determined with a BioPhotometer (Brinkmann Instruments, Westbury, NY). Relative luminescence was determined using a TD-20/20 or TD-20/20n luminometer (Turner Designs, Sunnyvale, CA), immediately after vigorous shaking to oxygenate the sample. The term relative luminescence means that the units are arbitrary but that values can be used to compare samples relative to one another. The specific luminescence (relative luminescence per OD595) reported was measured for whole cells. As previously reported, even with a bright strain there is no indication that luminescence becomes oxygen limited or otherwise decays over the course of these measurements (8).
Luminescence on solid medium was also examined qualitatively and quantitatively. Colonies were scored blind as either producing nonvisible, visible, or highly visible luminescence, based on the consensus of multiple observers (Table 1). For quantitative measurements, colonies of each strain were patched in triplicate onto SWTO plates and incubated at 24°C for 18 h. Patches were resuspended separately in Instant Ocean (Aquarium Systems, Mentor, OH), and the OD595 and luminescence were measured for each suspension as described above.
Molecular genetic techniques.
Cloning was performed using standard techniques. Plasmids were purified using Qiagen miniprep kits (Valencia, CA). PCR products were cloned into pCR-Blunt or pCR-BluntII-TOPO using the corresponding kit from Invitrogen (Carlsbad, CA). Oligonucleotides (Table 2) were synthesized by Integrated DNA Technologies (Coralville, IA). Klenow fragment, DNA ligase, and restriction enzymes were obtained from New England BioLabs (Beverly, MA). Between restriction and ligation reactions, DNA was recovered with the DNA Clean and Concentrator 5 kit (Zymo Research, Orange, CA). PCR was performed with an iCycler (Bio-Rad Laboratories, Hercules, CA) using KOD DNA polymerase (Novagen, Madison, WI). Sequences were analyzed using Sequencher (Gene Codes Corp., Ann Arbor, MI) and DNA Strider.
TABLE 2.
Oligonucleotides used in this study
| Oligonucleotide | 5′-3′ sequencea | Source or reference |
|---|---|---|
| EVS109 | CCG CCC TAG GTT ATT CAG ATA AGC ATT GAT TAA TAT C | 14 |
| EVS110 | CCG CCC TAG GGC ATG CTT AAC CTC TAT ACT CCT CCG ATG GAA | 14 |
| EVS129 | GTG TTA TCT CTT CAT CGA TAG TGA ATG | This study |
| EVS130 | GAT AAC GGA AGT TTC TAA TCG AGA AGT | This study |
| JBESMUT1 | P-TAA TAA ATT CGA TCT GGG TCA CAT TTA TGC ATC TTG | This study |
| JBESMUT2 | P-GAA TCA AAT ATC ACC AAA CCT CCA AAT TGG TG | This study |
| JBMJMUT1 | P-CAA TTA ATT GGA TTT TTG TCA CAC TAT TGT ATC GC | This study |
| JBMJMUT2 | P-GAA GCG ATG TCA ATC CAT TAC CGT TTT AAT G | This study |
| katA_2_fw | CGT GGT ATT CCT GCA ACA TAC | 39 |
| katA_3_rv | CCG ATA CCT TCA CCA TAA GC | 39 |
| katA_whole_fw | TGT CCT GTT GCA CAT AAC C | 39 |
| katA_whole_rv | CGC TTA CAT CAA TAT CAA G | 39 |
| mdh_1_fw | GGC ATT GGA CAA GCG TTA GC | 39 |
| mdh_2_rv | CGC CTC TTA GCG TAT CTA GC | 39 |
| mdh_whole_fw | AAG TAG CTG TTA TTG GTG C | 39 |
| mdh_whole_rv | CTT CGC CAA TTT TGA TAT CG | 39 |
| PconF | TTG ACA TAA AGT CTA ACC TAG GGT ATA ATT CCA TG | This study |
| PconR | GAA TTA TAC CCT AGG TTA GAC TTT ATG TCA ACA TG | This study |
| recA610Wolfwd | CTA GNH TTD AWY GSN GCN GC | 39 |
| recA610Wolrev | CTT CAC GGT TAA AAC CTT GG | 39 |
| recA_whole_fw | GAC GAT AAC AAG AAA AAA GCA CTG G | 39 |
| recA_whole_rv | CGT TTT CTT CAA TTT CWG GAG C | 39 |
P at the start of a sequence indicates 5′ phosphorylation. Underlined regions indicate AvrII sites that are absent in the lux template sequence but were incorporated in the primers to facilitate subcloning. Letters that do not correspond to standard bases refer to mixed bases at that position, following the standardized code of the International Union of Biochemistry (e.g., N is A, T, G, or C).
The lux intergenic regions from the wild-type strains listed in Table 1 were PCR amplified using primers EVS109 and EVS110 (Table 2) and inserted into pCR-Blunt(pJLB25-pJLB30) or pCR-BluntII-TOPO(pJLB79-pJLB168) for sequencing (Table 1). To generate transcriptional reporters that place PluxI or PluxR upstream of lacZ, the AvrII fragment containing the luxR-luxI intergenic region from each plasmid above was cloned into the NheI site upstream of a promoterless lacZ in pAKD702 (see the supplemental material in reference 7). The resulting reporter plasmids are listed in Table 1. In addition, the luxR-luxI intergenic regions from strains JB3 and JB7 (described below) were PCR amplified using primers EVS109 and EVS110, and the resulting products were digested with AvrII and cloned into the NheI site of pAKD702 to form reporters based on these mutant alleles in pJLB174 to pJLB177 (Table 1). To generate additional reporter controls, a consensus promoter was made by annealing oligonucleotides PconF and PconR together, and this synthetic promoter-containing insert was ligated into NheI-digested pAKD702. The resulting plasmid, in which the consensus promoter drives lacZ expression, was named pJLB207, and a plasmid with this insert in the opposite orientation was named pJLB206. Reporter plasmids were mobilized into V. fischeri from E. coli by triparental mating using pEVS104 as a helper plasmid as previously described (67).
To generate alleles with site-directed changes in the luxI-luxR intergenic region, we began with cloned copies of this region from strain ES114 or MJ1S in plasmid pEVS147 or pJLB62, respectively (8). The luxI-luxR intergenic region of ES114 was subcloned by placing a SpeI-NheI fragment of pEVS147 into the mobilizable vector pEVS118 (13), generating pEVS150. We next designed primers that would amplify around pEVS150 or pJLB62, while within the primer were sequences that would incorporate targeted sequence alterations. These primers were phosphorylated, so that PCR products could be self-ligated to generate new plasmids with the desired allelic changes and unaffected flanking regions, traits that were confirmed by sequencing. Primers JBESMUT1 and JBESMUT2 were used with the pEVS150 template, whereas primers JBMJMUT1 and JBMJMUT2 were used with the pJLB62 template; the products were each self-ligated, and the resulting plasmids were designated pJLB58 and pJLB64, respectively. We found that inclusion of a ColE1 replication origin facilitates allelic exchange in V. fischeri, and to add this origin to pJLB58, this plasmid was digested with SpeI and fused to SpeI-digested pBluescript KS(+), generating pJLB61.
Chromosomal changes in V. fischeri were engineered by allelic exchange. Alleles were mobilized into V. fischeri from E. coli by triparental mating (67), followed by selection and isolation of strains bearing single crossovers. These strains were then grown and plated nonselectively on LBS and patched onto medium containing an appropriate antibiotic to identify double-crossover mutants, which lose antibiotic resistance associated with the vector. Determination of mutant allele exchange, as opposed to reversion to wild type, was screened by PCR amplification across the intergenic luxR-luxI region and digesting the amplified fragment. Conversion of ES114 to JB3 resulted in loss of an EcoRV site, whereas the difference between MJ1S and JB7 included addition of a BsaBI site. Putative double-crossover mutants were confirmed by cloning and sequencing of such PCR products.
Several loci were used as phylogenetic markers in V. fischeri. Sequences within recA, mdh, and katA were analyzed as previously reported (39). Briefly, recA, mdh, and katA were PCR amplified using primers pairs recA_whole_fw and recA_whole_rv, mdh_whole_fw and mdh_whole_rv, or katA_whole_fw and katA_whole_rv, respectively. The subsequent products were sequenced directly with primers recA610Wolfwd and recA610Wolrev (for recA), mdh_1_fw and mdh_1_rv (for mdh), or katA_2_fw and katA_3_rv (for katA). The intergenic region between divergently transcribed genes glpA and fdh (VFA0250 and VFA0251) was amplified using primers EVS129 and EVS130, and the amplicons were cloned and sequenced.
Sequence analyses.
For the housekeeping gene concatenation, the glpA and fdh intergenic region, the luxR-luxI intergenic region, the sequences obtained above, and the corresponding sequence from V. salmonicida LFI1238 (27) were aligned using the default pairwise and multiple alignment parameters in ClustalX 2.0.11 (30). The luxI-luxR intergenic region of V. salmonicida was too divergent to provide a reasonable alignment by machine or hand alignment (data not shown) and was not included in phylogenetic reconstructions of that region. The length of the individual sequences from recA, mdh, and katA concatenated for analysis after alignment and trimming were 690, 783, and 789 bp, respectively. Table S1 of the supplemental material provides the GenBank accession numbers for sequence data from each isolate; those accession numbers that are being reported for the first time here are also provided at the end of Materials and Methods.
Phylogenetic reconstructions were completed using three methods: maximum parsimony (MP), maximum likelihood (ML), and Bayesian inference. MP reconstructions were performed by treating gaps as missing, searching heuristically using simple addition, tree bisection reconnection for swaps, and swapping on “best only” with 1,000 repetitions. For ML and Bayesian analyses, likelihood scores of 56 potential evolutionary models were tested using the Akaike information criterion (AIC) as implemented by Modeltest 3.7 (51). Based on the AIC, a general time-reversible model with a gamma shape parameter and a proportion of invariable sites (GTR+Γ+I) was chosen for recA-mdh-katA concatenation, a general time-reversible model with a gamma shape parameter (GTR+Γ) was chosen for the luxR-luxI intergenic region, and the F81 model was chosen for the glpA-fdhA intergenic sequence. Maximum likelihood reconstructions were performed by treating gaps as missing, searching heuristically using simple addition, subtree pruning and regrafting (44) for swaps, and swapping on “best only” with 1,000 repetitions as implemented by PAUP*4.0b10 (70). Bayesian inference was done with an nst of 6 and rate invgamma settings (GTR+Γ+I model) for recA-mdh-katA, an nst of 6 and rate gamma settings (GTR+Γ model) for the luxR-luxI intergenic sequences, and an nst of 1 and rates set equal (F81 model) for the glpA-fdhA intergenic sequences with heated chains set to a temperature value of 0.20 (for all data, to ensure an appropriate amount of chain swapping) using the software package MrBayes 3.1.2 (53). Statistical analysis of clade membership/presence was assessed using either 1,000 bootstrap pseudoreplicates with the above search parameters (ML and MP) or by sampling an appropriately stationary posterior probability distribution every 1,000 generations (for recA-mdh-katA), 500 generations (for the luxR-luxI intergenic region), or 200 generations (for the glpA-fdh intergenic region). For our purposes, an appropriately stationary distribution was defined, as recommended by Ronquist and colleagues (54), as an average standard deviation of split frequencies of less than 0.01 for ∼70 to 90% of samples between two independent Metropolis-coupled Markov chain Monte Carlo (MCMCMC) runs. Majority rule consensus trees drawn from the stationary sample distribution generated by MCMCMC were used for the assessment of the posterior probabilities of all clades. V. salmonicida LFI1238 was used to root all reconstructions post priori, except for those involving the luxR-luxI intergenic data, which were midpoint rooted. Graphical reconstructions were drawn and edited with Figtree 1.3.1 (http://tree.bio.ed.ac.uk/software/figtree/) and Inkscape 0.47 (http://www.inkscape.org/).
The (null) hypothesis of neutral evolution in intergenic sequences was investigated using summary statistics, specifically, Tajima's D test statistic (71) as implemented in DnaSP 5.10 (34) with segregating sites considered. The statistical significance of the test statistic was assessed using DnaSP 5.10 by comparison to an empirical distribution of test statistics generated via the coalescent process and assuming no recombination with 10,000 replicates.
To compare the DNA of lux regions from MJ11 and ES114, we used VISTA (40) with the LAGAN (9) alignment function and default settings, including a 100-bp window for calculating identity. We used BLAST (2) to align protein sequences and calculate the percent identity.
lacZ reporter expression.
To determine lacZ reporter expression, strains were grown in 25 ml SWTO in 125-ml flasks at 24°C with shaking to an OD of ∼3.0. Culture samples were taken, cells were pelleted, the supernatant was discarded, and the pellet was frozen at −20°C. The pellet was thawed and resuspended in Z-buffer for determination of β-galactosidase activity, expressed as Miller units as previously described (42). The promoterless lacZ vector pAKD702 was included as a negative control. Whether the group of PluxI-lacZ or PluxR-lacZ reporters showed significantly different activities between host strains ES114, H905, and WH1 was determined using Student's t test. Associations between promoter-reporter activity and the luminescence of the strain from which the promoter was cloned were determined using Spearman's rank correlation test.
Nucleotide sequence accession numbers.
The newly identified nucleotide sequences for recA, mdh, katA, the luxR-luxI intergenic region, and the glpA-fdhA intergenic region have been deposited with GenBank and assigned accession numbers HQ595306 to HQ595321, HQ595322 to HQ595330, HQ595331 to HQ595339, HQ436468 to HQ436485, and HQ535984 to HQ535999, respectively.
RESULTS
Comparison between bright strain MJ11 and dim strain ES114.
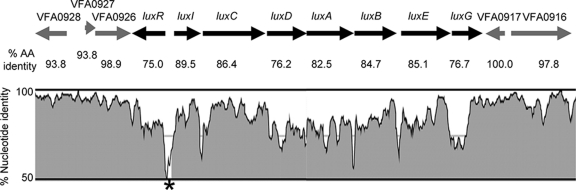
Genomic sequences of the bright V. fischeri strain MJ11 (38, 39) and the relatively dim strain ES114 (58) allowed us to compare the luxR-luxICDABEG region and several kb flanking lux, as well as global genetic interstrain differences. Thus, while previous studies had compared lux operons between strains (22), the availability of genomic data and sequence analysis tools allowed us to place such a comparison in a broader context. As we previously noted, there is a high degree of sequence conservation between ES114 and MJ11 (39). This was true in the region of chromosome II surrounding luxR-luxICDABEG; however, the lux region itself was more highly diverged, both in terms of nucleotide sequence and encoded proteins (Fig. 1). On average, orthologs in ES114 and MJ11 share greater than 95% identity, and this was true in open reading frames (ORFs) surrounding the lux region, such as VFA0916-VFA0917 and VFA0926-VFA0928 (Fig. 1). On the other hand, LuxR and the proteins encoded by the luxICDABEG operon share only 75.0 to 89.5% identity between ES114 and MJ11, while fewer than 5% of the orthologs shared by ES114 and MJ11 are so greatly diverged (Fig. 1) (see also the supplementary material in reference 39). Examination of other encoded proteins known to participate in lux regulation (Table 3) showed that most had a high degree of conservation typical of shared orthologs, with the notable exception of the AinS/AinR autoinducer system, which has also diverged more than average.
FIG. 1.
Conservation of lux and flanking regions between V. fischeri strains ES114 and MJ11. The percent identity is shown for aligned nucleotides (on the y axis of the inset graph) and for encoded proteins as the percent amino acid (AA) identity below the corresponding gene. The black and gray arrows indicate the orientation of lux genes or flanking genes, respectively. Graphic representation of the percent nucleotide identity was generated with VISTA (40) and the LAGAN alignment program (9) using default settings. An asterisk marks an unshaded region between luxI and luxR where a window in the alignment falls below the 70% identity cutoff. Conservation of regions flanking the lux genes was typical for that extending several kilobases in each direction (data not shown).
TABLE 3.
Conservation of selected luminescence regulators between ES114 and MJ11
| Regulator | ORF(s) in ES114 | Proposed regulatory rolea | % amino acid identity between ES114 and MJ11 |
|---|---|---|---|
| AinS | VF1037 | C8-HSL | 83.1 |
| AinR | VF1036 | C8-HSL | 91.7 |
| LuxS | VF0545 | AI-2 | 99.4 |
| LuxP | VF0707 | AI-2 | 98.1 |
| LuxQ | VF0708 | AI-2 | 98.2 |
| LuxU | VF0937 | AI-2/C8-HSL | 99.2 |
| LuxO | VF0938 | AI-2/C8-HSL | 99.8 |
| LitR | VF2177 | AI-2/C8-HSL | 99.5 |
| Hfq | VF2323 | AI-2/C8-HSL | 100.0 |
| qrr1b | NA | AI-2/C8-HSL | 100.0 |
| LonA | VF0798 | Degradation of LuxR | 99.7 |
| ArcB/ArcA | VF2122/VF2120 | Environmental | 98.7/100.0 |
| GacS/GacA | VF2082/VF1627 | Environmental | 99.4/100.0 |
| CRP | VF2280 | Environmental | 100.0 |
| PhoB | VF1988 | Environmental | 99.6 |
Notations indicate proposed roles in luminescence regulation through C8-HSL, AI-2, both of these autoinducers (AI-2/C8-HSL), degradation of the HSL-responsive activator LuxR, or the sensing of environmental stimuli and signal transduction.
qrr1 is a small RNA and therefore an open reading frame description is not applicable (NA), and the percent nucleotide identity (not amino acid identity) is presented.
Sequence divergence within luxR-luxICDABEG was particularly noteworthy in the intergenic luxR-luxI region (Fig. 1). Although one might expect less conservation of noncoding sequences, intergenic regions similarly situated between divergent genes were typically more highly conserved, as is illustrated by the sequences between VFA0916 and VFA0917 or between VFA0927 and VFA0928 (Fig. 1).
Analysis of the luxR-luxI intergenic region and bioluminescence among diverse strains.
To explore the evolution of intergenic luxR-luxI sequences more closely, we cloned, sequenced, and analyzed this region from a collection of strains (Table 1). This collection of diverse V. fischeri strains included isolates from fish, such as Cleidopus gloriamaris and Monocentris japonica, squid of both the Euprymna and Sepiola genera, and planktonic isolates (Table 1).
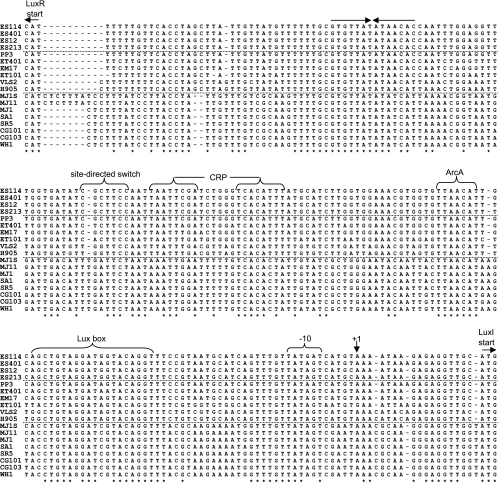
An alignment of the luxR-luxI intergenic sequences in these strains is shown in Fig. 2. Of the 222 positions in the alignment between the start codons of LuxR and LuxI, only 104 positions (47%) were conserved among all of the strains. Pairwise comparisons of this region between the strains ranged from 60 to 100% sequence identity. In many instances, conserved sequences among all strains represented established binding sites for regulators. For example, the Lux box binding site for LuxR is well conserved across the strains (Fig. 2). There are only two consistent differences in the Lux box between the nonvisible (NV) and highly visible (HV) strains, and in another study these base pair changes had relatively little influence on promoter activity, with the slight effects being roughly equal and opposite (3). Similarly, CRP was previously shown to bind within the intergenic lux region (61), where it appears to regulate luxR transcription (11, 12), and a sequence corresponding to this binding site and resembling the consensus core recognition sequence for CRP [5′-AATTGTGA(N6)TCACAATT-3′] is conserved (Fig. 2), although divergence in this sequence may also reflect varied affinities for CRP binding. Likewise, the ArcA-binding consensus core (5′-ATGTTAA-3′) is found adjacent to the Lux box on the reverse strand relative to luxI in all strains (Fig. 2), consistent with our report that ArcA binds this location and regulates luminescence (7). Another conserved feature that stood out is a palindromic repeat nearer luxR (Fig. 2), although its function is unknown. In addition, there were many differences between the strains, and perhaps the most notable was a direct repeat of 5′-CTTTATC-3′ near the luxR start codon present in MJ11 and MJ1S but lacking in other strains, including MJ1.
FIG. 2.
Alignment of lux intergenic regions from diverse V. fischeri strains. Horizontal dashed lines separate strains with luminescence on plates classified as nonvisible (ES114, ES401, ES12, and ES213), visible (PP3, ET401, EM17, ET101, VLS2, and H905), or highly visible (MJ1S, MJ11, MJ1, SA1, SR5, CG101, CG103, and WH1). Asterisks mark bases conserved across all strains. Labeled arrows mark the translational start sites for LuxI and LuxR (on the reverse strand). The transcriptional start site for luxICDABEG determined in ES114 (16) is marked +1 and with a vertical arrow, and the corresponding −10 promoter element is indicated. Core consensus sequences for CRP, ArcA, and LuxR binding are shown and are discussed further in the text. A pair of arrows above the top row of the sequences indicates the position of an inverted repeat. The region labeled the site-directed switch was engineered such that this sequence in ES114 was placed in MJ1S to generate strain JB7 and, likewise, switched from MJ1S into ES114 to generate strain JB3.
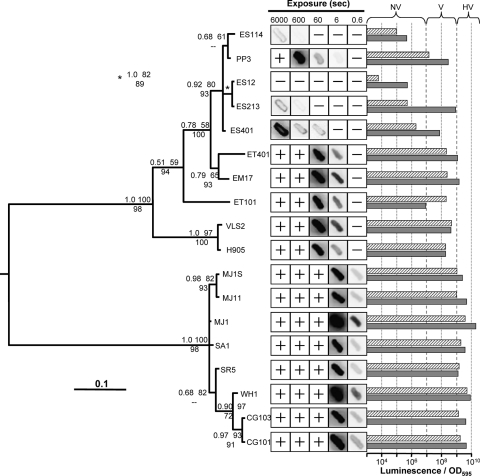
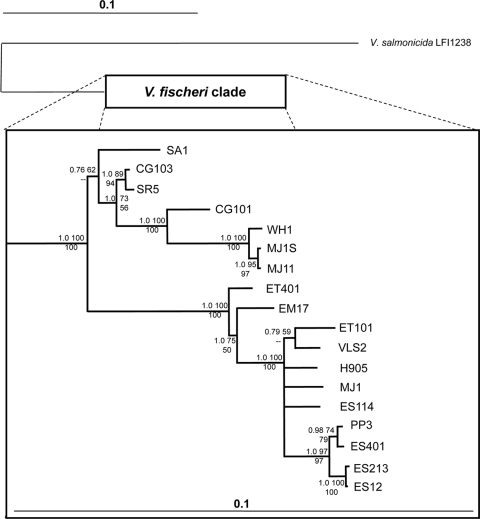
A phylogenetic reconstruction of the evolutionary relationships among intergenic luxR-luxI sequences is also shown, juxtaposed against the quantified luminescence phenotypes of the respective strains, in Fig. 3. As has been done in the past, strains were scored qualitatively as producing nonvisible (NV), visible (V), or highly visible (HV) luminescence (Fig. 3). It should be emphasized that even NV strains did exhibit detectable bioluminescence. Examination of the tree shows two distinct clades. One clade includes all the fish isolates, Sepiola squid isolates, and the planktonic strain WH1. This group encompasses all the strains classified as HV. The second clade includes all of the isolates from the Hawaiian bobtail squid, Euprymna scolopes. For the most part the isolates from E. scolopes group closely together with the Hawaiian planktonic isolate PP3, which was previously shown to resemble isolates from E. scolopes (31-33). The remainder of this clade includes isolates ET401, EM17, and ET101 from other Euprymna species found in Japan or Australia, as well as two Hawaiian isolates, VLS2 and H905, which are unusual for that locale and are not proficient colonizers of E. scolopes (31-33). The latter set of isolates includes all but one classified as visibly luminescent in Table 1, V class strain PP3.
FIG. 3.
Relatedness of the luxR-luxI intergenic sequence in V. fischeri isolates. The topology represents the 50% majority rule consensus tree generated from a Bayesian analysis. Statistical support of each node is given at the particular node based on three values: Bayesian posterior probability (upper left); ML bootstrap support values (1,000 pseudoreplicates) (upper right); MP bootstrap support values (1,000 pseudoreplicates) (lower center). Dashes indicate clade support of <50% for 1,000 MP bootstrap pseudoreplicates. The tree was mid-point rooted. Bar, 0.1 substitutions per site. Patches were grown overnight on SWTO at 24°C, and the negative images for given exposures are shown. A + or − indicates that luminescence was too high or low to see clearly, respectively. Horizontal bars display the luminescence per OD595 unit of each strain grown on plates for 18 h and resuspended (hatched bars) or grown in broth with peak luminescence reported (solid bars). Dashed vertical lines indicate approximate cutoffs that were used to distinguish strains that produced nonvisible, visible, or highly visible luminescence on SWTO plates.
Also reported in Fig. 3 is the luminescence of the V. fischeri strains on solid media and in broth. Not surprisingly, there was a strong correlation between the luminescence intensity of cells collected from solid media and luminescence classification based on observations of plates (Fig. 3). Based on these data, it appears that the threshold amount of light produced that is required for the visible classification falls somewhere between the bioluminescence produced by PP3 (V) and that of ES401 (NV). Generally, specific luminescence was higher in broth culture than during growth on solid media; however, VLS2, H905, and SR5 produced similar luminescence levels under both conditions, and ET101 had higher luminescence on solid media than in broth. Upon comparing Fig. 2 and 3, we found that ES12 and ES213 have identical luxR-luxI intergenic regions, yet ES213 produces 100-fold and 1,000-fold more light on plates and in broth, respectively. Thus, two strains having identical luxR and luxICDABEG promoters may produce very different amounts of light. Further examination of the broth cultures during growth showed that all of the strains were subject to cell density-dependent regulation of luminescence (data not shown).
Comparison of luxR-luxI intergenic divergence with that of housekeeping loci.
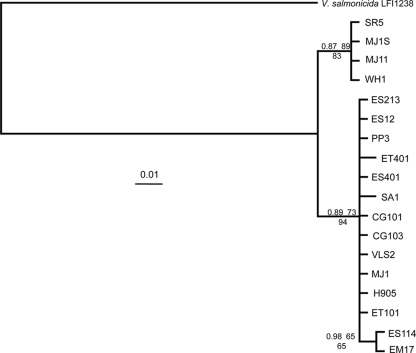
In order to better understand if there was any significance to the high nucleotide diversity among the luxI-luxR intergenic sequences, we sequenced the intergenic region between glpA and fdhA in our collection of V. fischeri strains. Like luxR and luxI, these genes are adjacent and divergently transcribed on chromosome 2. Phylogenetic reconstruction and statistical analyses of evolutionary relationships among glpA-fdhA intergenic sequences allowed an interesting comparison to results from similar analyses of the luxR-luxI intergenic region. Each of the V. fischeri strains listed in Table 1 had a 298-bp intergenic region between glpA and fdhA, and 289 bp (97%) were conserved among all the strains. Pair-wise comparisons of this region between the strains ranged from 98 to 100% sequence identity. Thus, as was the case between divergently transcribed genes near lux (VFA0916 and VFA0917 or VFA0927 and VFA0928 [Fig. 1]), this intergenic region is far more conserved than the luxR-luxI intergenic region. We generated a phylogenetic reconstruction of V. fischeri strains based on the intergenic glpA-fdhA region (Fig. 4). Given the lack of differences between the strains in this intergenic region, nodes of this tree were not supported by strong bootstrap values; however, four of the strains in one of the clades, namely, MJ1S, MJ11, WH1, and SR5, did appear to form a distinct well-supported branch (Fig. 4).
FIG. 4.
Bayesian phylogeny of the intergenic glpA-fdhA region among V. fischeri isolates. The topology was generated as described for Fig. 3, and the statistical support values given at each node were calculated and are placed in order as for Fig. 3. The tree was rooted with the corresponding sequence from V. salmonicida LFI1238. Bar, 0.01 substitutions per site. This reconstruction's low resolution is a result of low sequence diversity among isolates.
One hypothesis from the above phylogenetic results is that the luxR-luxI intergenic region is under different selective pressure than that of a “normal” housekeeping intergenic sequence on chromosome 2 of V. fischeri. To examine this hypothesis, the summary Tajima's D statistic (and its statistical significance) was calculated for each group of intergenic sequences (Table 4). The null hypothesis for this test statistic is that for a given random sample of a population, the sequence analyzed is dominated by genetic drift and has evolved under a predominantly neutral model (Tajima's D = 0). This null hypothesis was rejected for the luxR-luxI intergenic sequence, but not for the intergenic glpA-fdhA region (Table 4). This positive, statistically significant value of the D test statistic for the luxI-luxR region suggests that population demographics and/or balancing selection has caused an excess of intermediate-frequency polymorphisms in the set of sequences analyzed relative to that of an intergenic region such as the glpA-fdhA region (52).
TABLE 4.
Analysis of intergenic sequences based on Tajima's D test statistic
| Intergenic sequence | na | Sb | πc | Dd | 95% CIe | P valuef |
|---|---|---|---|---|---|---|
| luxI-luxR | 18 | 108 | 0.23558 | 2.38938 | 1.66, −1.75 | 0.002 |
| glpA-fdhA | 18 | 9 | 0.00864 | 0.01467 | 1.82, −1.79 | 0.435 |
The number of sequences.
The number of segregating sites.
π indicates nucleotide diversity.
Tajima's D test statistic.
The 95% confidence interval (CI) for distribution of 10,000 simulations of the test statistic using coalescent simulations.
The P value was assessed using the coalescent generated test statistic distribution.
To better compare the phylogeny of our strain collection to the evolutionary history of lux intergenic sequences, we sequenced portions of recA, katA, and mdh. The concatenated sequences of these housekeeping genes were used to reconstruct the evolutionary relationships among V. fischeri strains in a manner independent from intergenic sequences (Fig. 5). The topology of the phylogenetic reconstruction inferred from these housekeeping genes was generally similar to the topology of the luxI-luxR intergenic sequence reconstruction (Fig. 3). One notable exception was strain MJ1, which groups with other HVL strains according to lux sequence (Fig. 3) but not in phylogeny (Fig. 5). This is consistent with the observation noted above that the intergenic glpA-fdhA region of MJ1 is distinct from that of MJ1S and MJ11. Moreover, this general pattern seen with the glpA-fdhA region and the concatenated sequences was also observed for phylogenetic reconstructions made from each of the recA, katA, and mdh loci individually (data not shown). Overall, the phylogenetic reconstruction of this strain collection appears to show two distinct clades of V. fischeri, which resembles the clear bifurcation reported in other recent studies (4, 39).
FIG. 5.
Bayesian phylogeny of concatenated recA-katA-mdh sequences among V. fischeri isolates. The topology was generated as described for Fig. 3, and the statistical support values given at each node were calculated and placed in order as for Fig. 3. The tree was rooted with the corresponding sequences from V. salmonicida LFI1238. Bars, 0.1 substitutions per site; the V. fischeri clade was expanded in order to more easily view fine-scale topological resolution among all isolates.
Differences in intergenic luxR-luxI sequences affect luxI promoter strength and bioluminescence.
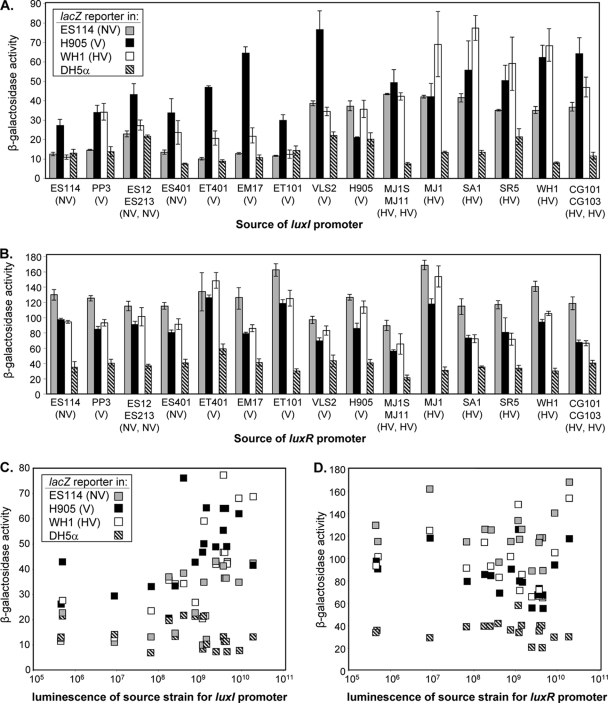
Given the high substitution rate in the luxR-luxI intergenic region (Fig. 1, 2, and 3) and the correlation between these sequences and luminescence output (Fig. 3), we hypothesized that the strength of the luxR and/or luxI promoters might underlie some of the differences in luminescence. To begin testing this hypothesis, we constructed PluxI-lacZ and PluxR-lacZ reporter plasmids (Table 1). These reporters were derived from V. fischeri plasmid pES213 (13), which is very stable in strain ES114 (14); however, we found that they were not stable in all V. fischeri (data not shown). For example, our pES213 derivatives were not stable in MJ11, which was recently discovered to harbor a very large and unique plasmid absent in ES114 (38). Accordingly, we only considered data from strains in which the reporters are stably maintained in the absence of selection. We also considered the possibility that differences in plasmid copy number or endogenous β-galactosidase activity (1) might invalidate interstrain comparisons using these reporters. Therefore, as controls we included a promoterless reporter as well as plasmids with lacZ strongly expressed from a consensus promoter or weakly expressed from a construct where the consensus promoter was oriented away from lacZ. We found that background β-galactosidase activity was undetectable and that activities from our synthetic promoter constructs were similar in different V. fischeri backgrounds (data not shown), indicating that it would be reasonable to make comparisons using PluxI-lacZ and PluxR-lacZ reporter plasmids in distinct V. fischeri strains. On the other hand, β-galactosidase activities from the synthetic promoter-lacZ constructs were somewhat lower in E. coli (data not shown), and so the lower activity of the PluxI-lacZ and PluxR-lacZ reporters in E. coli versus V. fischeri is not a meaningful comparison.
The PluxI-lacZ and PluxR-lacZ reporters were examined in E. coli DH5α as well as in V. fischeri strains ES114, H905, and WH1, which represent NV, V, and HV luminescence classes, respectively. The β-galactosidase activities from the 15 distinct sets of PluxI-lacZ and PluxR-lacZ reporters in the four host strains are shown along with the source of the promoter sequence in Fig. 6A and B. The activities of these reporters are also displayed (Fig. 6C and D) as a function of the luminescence of the strain(s) serving as the source of the PluxI or PluxR promoter in the reporter. Expression from the PluxI-lacZ reporters showed a significant (P < 0.01) trend toward higher activity in strains H905 and WH1 than in ES114 (Fig. 6A), which perhaps not surprisingly indicates that in general luxI promoters from any source are more highly expressed in relatively bright strains. More interestingly, in each of the V. fischeri host strains, expression from the luxI reporters tended to correlate (P < 0.02 in each V. fischeri host) with the luminescence of the strain from which the promoter was derived (Fig. 6C), suggesting that the intrinsic strength of the luxI promoter tends to be higher when it is from a brighter strain, regardless of the V. fischeri host strain for the reporter. This trend was not evident (P = 0.99) when the reporters were placed in E. coli (Fig. 6C). Moreover, neither the luminescence of the host strain nor that of the source strain(s) for luxR promoters was positively correlated with reporter activity, and if anything, luxR reporter expression was slightly higher in the relatively dim strain ES114 (Fig. 6B and D).
FIG. 6.
Activity of lux promoter reporters. The β-galactosidase activities (in Miller units) from specific PluxI-lacZ and PluxR-lacZ constructs are shown for reporters carried in V. fischeri strains ES114 (gray), H905 (black), and WHI (open) as well as in E. coli DH5α (hatched lines). The luminescence class of the V. fischeri strains (Table 1) is indicated in parentheses as NV, V, or HV. (A and B) The strain(s) corresponding to the promoter sequence in PluxI-lacZ and PluxR-lacZ constructs, respectively, and these are arranged (left to right) in the same order as the strains shown in Fig. 3 (top to bottom). (C and D) Reporter activity is plotted as a function of the luminescence of the source strain(s) for PluxI-lacZ and PluxR-lacZ promoters, respectively. Data from different experiments with treatments in common and with similar results were pooled, and averages are reported (panels A and B also show standard errors [n = 3]).
The results above led us to hypothesize that differences in the luxR-luxI intergenic region might underlie a considerable amount of the strain-specific variation in luminescence output. To test this directly, we exchanged a short (7- to 8-bp) sequence on the chromosome in the luxR-luxI intergenic region between the relatively bright (HV) strain MJ1S and the relatively dim (NV) strain ES114. The sequence exchanged between these strains is shown in Fig. 2 (labeled site-directed switch). This sequence was targeted because it was suspected of involvement in FNR-mediated regulation that might be strain specific; however, more recently we (and others) found no evidence of a direct regulation of lux promoters by FNR (60). This short intergenic sequence from ES114 was placed in MJ1S to generate strain JB7, and likewise, the corresponding sequence from MJ1S was placed in ES114 to generate strain JB3. The striking results were that JB3 is more than 100-fold brighter than its parent ES114, and JB7 is more than a 100-fold dimmer than its parent MJ1S (Fig. 7). Indeed, JB3 is more than 10-fold brighter than JB7 (Fig. 7), despite the fact that JB3 is derived from the much dimmer ES114. These shifts in luminescence were observed regardless of the fnr genotype under both well-aerated and anaerobic conditions (data not shown). This result indicates that changes in the luxR-luxI intergenic region can have large effects on luminescence output in an otherwise-isogenic background.
FIG. 7.
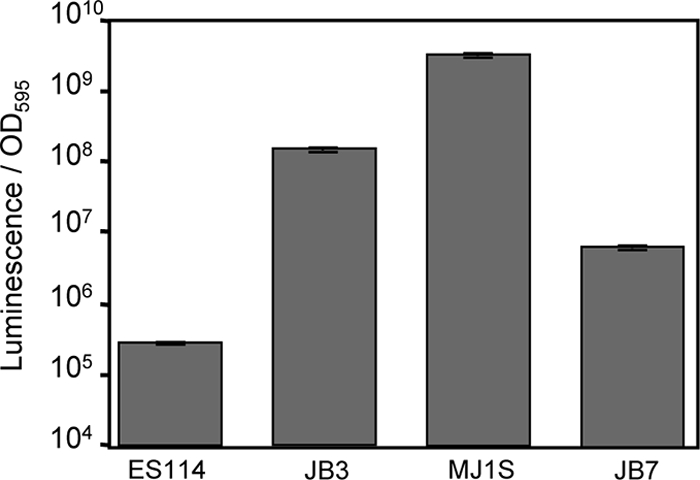
Exchanging a short stretch of the luxI-luxR intergenic region between ES114 and MJ1S largely reverses their relative brightness. Luminescence per OD595 unit is shown for cultures of ES114, JB3, MJ1S, and JB7 grown in SWTO broth at 24°C with shaking (200 rpm). Data are the average peak luminescence levels per OD595 unit, with standard errors (n = 2).
DISCUSSION
The functional significance of bioluminescence for light-producing bacteria remains a controversial topic (62), and it is made all the more mysterious by the wide variation in light output by different environmental isolates, ranging from brightly visible to “cryptic.” Some bioluminescent members of the Vibrionaceae are not known to have any specific light organ hosts to make use of the light they produce. These strains can be free living but are also isolated frequently from fish gut tracts, where their luminescence may be valuable in making fecal matter visible to potential new hosts (46). Such strains can be bright or dim, and it is not clear how or why any of them would have evolved and retained attenuated dim luminescence. V. fischeri does have mutualistic symbiotic hosts, and strains of this species also vary considerably in luminescence (Fig. 3). At least for V. fischeri, there appears to be a plausible explanation for differences in luminescence output. Specifically, some V. fischeri strains have evolved symbioses with fishes for which light is thought to be used by the host in bright signaling behaviors, whereas in other strains luminescence may provide a dim counterilluminating camouflage (28, 46, 65). However, this does not explain the mechanistic underpinnings of luminescence variability.
Despite the wide variation in luminescence output by different environmental isolates of V. fischeri, the general makeup and arrangement of the genes responsible for luminescence appear to be conserved in dim and bright strains (22), raising the question of how this phenotypic divergence evolved. We have found that the lux and ainSR loci of bright and dim V. fischeri strains have diverged in sequence far more than most orthologs. This observation is consistent with selective pressure(s) on luminescence output and suggests relatively rapid evolution of the loci underlying bioluminescence and pheromone production. The divergence of the luxI-luxR intergenic region is particularly striking (Fig. 1 and 2), and we have shown that variation in this region has resulted in a correlation between luxI promoter strength and strain brightness (Fig. 6). Moreover, exchanging a small (<10-bp) part of the luxI-luxR intergenic region between two strains largely reversed their relative luminescence levels. Although it is clear that other factors influence luminescence output, as discussed below, our data clearly demonstrate that much of the difference in strain luminescence can be attributed to this regulatory control region.
Conservation in the luxR-luxI intergenic region highlights key regulatory regions.
Comparison of the luxR-luxI intergenic region further revealed conserved regulatory sites. While some, such as the Lux box and ArcA-binding sites, were previously known, at least one potential new regulatory site was identified. Specifically, there is a region near luxR that includes an absolutely conserved 5′-TTTGC-3′ sequence adjacent to a highly conserved palindromic repeat of 5′-GTGTTAT/ATAACAC-3′ (Fig. 2). This palindrome is perfectly conserved in all NV and some V strains, has one mismatch in other V strains, and two mismatches in VL strains. The function of this conserved sequence, if any, is unknown; however, its location is consistent with it serving as the binding site for LitR, which previously was mapped to a 427-bp fragment on the luxR side of the Lux box (19). Little is known about LitR, but its homolog in Vibrio harveyi (named LuxR in that organism, and referred to here as LuxRVh) has been analyzed more extensively. Swarzman and Meighan proposed a 5′-AGTGTTAC-3′ consensus binding sequence for LuxRVh (69), which is remarkably similar to half of the palindrome described above. On the other hand, a more recent and thorough analysis of LuxRVh binding revealed a quite different recognition sequence (50). The approach used in the earlier study leaves open the possibility that the consensus sequence those authors reported reflects another regulatory sequence that does not directly interact with LuxRVh but is overlapped by its footprint in the same promoters. Similarly, the conserved palindrome in V. fischeri may reflect a regulatory mechanism that works in concert with LitR rather than by binding LitR itself. Further experimentation will be required to distinguish between these possibilities.
Importance of sequences outside the luxR-luxI intergenic region.
Our results also indicate that regions outside the luxI and luxR promoters can influence luminescence. This was illustrated in a comparison of strains ES12 and ES213, which have identical luxR-luxI intergenic regions but differ in luminescence output by orders of magnitude. The disparate luminescence in these strains, and others, may be due in part to functional differences in the Lux proteins, different levels of other lux-regulating proteins, posttranscriptional regulation, or differences in cell physiology. In this regard, a comparison of coding sequences between ES114 and MJ11 showed notable divergence in LuxI, LuxR, the light-generating Lux proteins, and the AinS/AinR system, and it seems likely that these differences may underlie some of the disparity seen in luminescence output.
Convergence of directed and real-world evolution in LuxR.
Another noteworthy example of interstrain differences outside the luxR-luxI intergenic region involves LuxR. Directed evolution of luxR from MJ1 in the laboratory has revealed LuxR variants with altered responsiveness to various homoserine lactones (10, 25), and two LuxR alleles (T33A and I96V) that were isolated based on increased responsiveness to the V. fischeri autoinducer C8-HSL (10) are also found in the native ES114 LuxR. In other words, one of the differences we found between the LuxRs encoded by ES114 and MJ1 was already discovered as conferring responsiveness to C8-HSL on MJ1's LuxR. ES114 produces more C8-HSL than does MJ1 (68), so perhaps C8-HSL and 3-oxo-C6-HSL have evolved different roles in distinct V. fischeri clades, and this may be reflected in the LuxR proteins.
V. fischeri as a model for future evolutionary studies of luminescence.
Bioluminescence is a trait scattered throughout the family Vibrionaceae; however, the selective pressures driving the evolution of bioluminescence are not always clear (62). As noted here, the mystery deepens in that light output can vary by over several orders of magnitude between different environmental isolates of V. fischeri, and this is true for other Vibrio species as well (23, 49). The work here provides evidence that lux has evolved relatively rapidly in V. fischeri and helps provide a foundation for understanding the mechanistic differences leading to dimmer or brighter strains. Moreover, bioluminescence contributes to fitness of V. fischeri in its symbiotic environment (8, 73), and this symbiosis can be reconstituted in the laboratory (55, 63), affording the opportunity to assess the consequences of genetic changes to the lux system under ecologically relevant growth conditions. Interestingly, when MJ11 colonized squid, the animals were much brighter than those colonized by native symbionts (39, 59), which would theoretically disrupt the advantages of dim counterillumination. Preliminary evidence was recently presented that suggested that when MJ11 was grown for multiple generations in association with its nonnative host E. scolopes, most of the squid-adapted strains became dimmer, resembling natural isolates from E. scolopes (59). This could indicate a selection against the energetic costs of bright bioluminescence when it confers no advantage; however, the relatively rapid loss of bright bioluminescence in the squid as opposed to findings in culture suggests a positive selection for dimness in this host. Further work with such evolved strains, along with more mechanistic studies, will help shed light on the evolution and importance of bioluminescence in the model bacterium V. fischeri.
Supplementary Material
Acknowledgments
We thank Edward G. Ruby for helpful discussions, E. Peter Greenberg for providing MJ1, Danielle Britt for technical assistance, and several volunteers who ranked strain bioluminescence levels in a darkroom.
This work was supported by the National Science Foundation (NSF) under grant CAREER MCB-0347317 to E.V.S. M.S.W. was supported by a grant (RR12294) to E.G. Ruby from the National Institutes of Health (NIH) and by a training grant from the NIH (5 T32 GM007215-35). A.N.S. was supported by a National Defense Science and Engineering Graduate Fellowship.
Genomic information was provided through the support of the W. M. Keck Foundation (strain ES114) and the Gordon and Betty Moore Foundation (strain MJ11).
Footnotes
Published ahead of print on 11 February 2011.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Adin, D. M., K. L. Visick, and E. V. Stabb. 2008. Identification of a cellobiose utilization gene cluster with cryptic beta-galactosidase activity in Vibrio fischeri. Appl. Environ. Microbiol. 74:4059-4069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Antunes, L. C., R. B. Ferreira, C. P. Lostroh, and E. P. Greenberg. 2008. A mutational analysis defines Vibrio fischeri LuxR binding sites. J. Bacteriol. 190:4392-4397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ast, J. C., H. Urbanczyk, and P. V. Dunlap. 2009. Multi-gene analysis reveals previously unrecognized phylogenetic diversity in Aliivibrio. Syst. Appl. Microbiol. 32:379-386. [DOI] [PubMed] [Google Scholar]
- 5.Boettcher, K. J., and E. G. Ruby. 1990. Depressed light emission by symbiotic Vibrio fischeri of the sepiolid squid Euprymna scolopes. J. Bacteriol. 172:3701-3706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boettcher, K. J., and E. G. Ruby. 1994. Occurrence of plasmid DNA in the sepiolid squid symbiont Vibrio fischeri. Curr. Microbiol. 29:279-286. [Google Scholar]
- 7.Bose, J. L., et al. 2007. Bioluminescence in Vibrio fischeri is controlled by the redox-responsive regulator ArcA. Mol. Microbiol. 65:538-553. [DOI] [PubMed] [Google Scholar]
- 8.Bose, J. L., C. S. Rosenberg, and E. V. Stabb. 2008. Effects of luxCDABEG induction in Vibrio fischeri: enhancement of symbiotic colonization and conditional attenuation of growth in culture. Arch. Microbiol. 190:169-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brudno, M., et al. 2003. LAGAN and Multi-LAGAN: efficient tools for large-scale multiple alignment of genomic DNA. Genome Res. 13:721-731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Collins, C. H., F. H. Arnold, and J. R. Leadbetter. 2005. Directed evolution of Vibrio fischeri LuxR for increased sensitivity to a broad spectrum of acyl-homoserine lactones. Mol. Microbiol. 55:712-723. [DOI] [PubMed] [Google Scholar]
- 11.Dunlap, P. V., and E. P. Greenberg. 1985. Control of Vibrio fischeri luminescence gene expression in Escherichia coli by cyclic AMP and cyclic AMP receptor protein. J. Bacteriol. 164:45-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dunlap, P. V., and E. P. Greenberg. 1988. Control of Vibrio fischeri lux gene transcription by a cyclic AMP receptor protein-LuxR protein regulatory circuit. J. Bacteriol. 170:4040-4046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dunn, A. K., M. O. Martin, and E. V. Stabb. 2005. Characterization of pES213, a small mobilizable plasmid from Vibrio fischeri. Plasmid 54:114-134. [DOI] [PubMed] [Google Scholar]
- 14.Dunn, A. K., D. S. Millikan, D. M. Adin, J. L. Bose, and E. V. Stabb. 2006. New rfp- and pES213-derived tools for analyzing symbiotic Vibrio fischeri reveal patterns of infection and lux expression in situ. Appl. Environ. Microbiol. 72:802-810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eberhard, A., et al. 1981. Structural identification of autoinducer of Photobacterium fischeri luciferase. Biochemistry 20:2444-2449. [DOI] [PubMed] [Google Scholar]
- 16.Egland, K. A., and E. P. Greenberg. 1999. Quorum sensing in Vibrio fischeri: elements of the luxl promoter. Mol. Microbiol. 31:1197-1204. [DOI] [PubMed] [Google Scholar]
- 17.Engebrecht, J., K. Nealson, and M. Silverman. 1983. Bacterial bioluminescence: isolation and genetic analysis of functions from Vibrio fischeri. Cell 32:773-781. [DOI] [PubMed] [Google Scholar]
- 18.Engebrecht, J., and M. Silverman. 1984. Identification of genes and gene products necessary for bacterial bioluminescence. Proc. Natl. Acad. Sci. U. S. A. 81:4154-4158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fidopiastis, P. M., C. M. Miyamoto, M. G. Jobling, E. A. Meighen, and E. G. Ruby. 2002. LitR, a new transcriptional activator in Vibrio fischeri, regulates luminescence and symbiotic light organ colonization. Mol. Microbiol. 45:131-143. [DOI] [PubMed] [Google Scholar]
- 20.Fidopiastis, P. M., H. Sorum, and E. G. Ruby. 1999. Cryptic luminescence in the cold-water fish pathogen Vibrio salmonicida. Arch. Microbiol. 171:205-209. [DOI] [PubMed] [Google Scholar]
- 21.Fidopiastis, P. M., S. von Boletzky, and E. G. Ruby. 1998. A new niche for Vibrio logei, the predominant light organ symbiont of squids in the genus Sepiola. J. Bacteriol. 180:59-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gray, K. M., and E. P. Greenberg. 1992. Physical and functional maps of the luminescence gene cluster in an autoinducer-deficient Vibrio fischeri strain isolated from a squid light organ. J. Bacteriol. 174:4384-4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grim, C. J., et al. 2008. Occurrence and expression of luminescence in Vibrio cholerae. Appl. Environ. Microbiol. 74:708-715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 25.Hawkins, A. C., F. H. Arnold, R. Stuermer, B. Hauer, and J. R. Leadbetter. 2007. Directed evolution of Vibrio fischeri LuxR for improved response to butanoyl-homoserine lactone. Appl. Environ. Microbiol. 73:5775-5781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Herrero, M., V. De Lorenzo, and K. N. Timmis. 1990. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J. Bacteriol. 172:6557-6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hjerde, E., et al. 2008. The genome sequence of the fish pathogen Aliivibrio salmonicida strain LFI1238 shows extensive evidence of gene decay. BMC Genomics 9:616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jones, B. W., and M. K. Nishiguchi. 2004. Counterillumination in the Hawaiian bobtail squid, Euprymna scolopes Berry (Mollusca: Cephalopoda). Mar. Biol. 144:1151-1155. [Google Scholar]
- 29.Kaplan, H. B., and E. P. Greenberg. 1985. Diffusion of autoinducer is involved in regulation of the Vibrio fischeri luminescence system. J. Bacteriol. 163:1210-1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Larkin, M. A., et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947-2948. [DOI] [PubMed] [Google Scholar]
- 31.Lee, K.-H. 1994. Ecology of Vibrio fischeri, the light organ symbiont of the Hawaiian sepiolid squid Euprymna scolopes. Ph.D. thesis. University of Southern California, Los Angeles, CA.
- 32.Lee, K.-H., and E. G. Ruby. 1994. Competition between Vibrio fischeri strains during initiation and maintenance of a light organ symbiosis. J. Bacteriol. 176:1985-1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee, K.-H., and E. G. Ruby. 1992. Detection of the light organ symbiont, Vibrio fischeri, in Hawaiian seawater using lux gene probes. Appl. Environ. Microbiol. 58:942-947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Librado, P., and J. Rozas. 2009. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25:1451-1452. [DOI] [PubMed] [Google Scholar]
- 35.Lupp, C., and E. G. Ruby. 2004. Vibrio fischeri LuxS and AinS: comparative study of two signal synthases. J. Bacteriol. 186:3873-3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lupp, C., and E. G. Ruby. 2005. Vibrio fischeri uses two quorum-sensing systems for the regulation of early and late colonization factors. J. Bacteriol. 187:3620-3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lupp, C., M. Urbanowski, E. P. Greenberg, and E. G. Ruby. 2003. The Vibrio fischeri quorum-sensing systems ain and lux sequentially induce luminescence gene expression and are important for persistence in the squid host. Mol. Microbiol. 50:319-331. [DOI] [PubMed] [Google Scholar]
- 38.Mandel, M. J., E. V. Stabb, and E. G. Ruby. 2008. Comparative genomics-based investigation of resequencing targets in Vibrio fischeri: focus on point miscalls and artefactual expansions. BMC Genomics 9:138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mandel, M. J., M. S. Wollenberg, E. V. Stabb, K. L. Visick, and E. G. Ruby. 2009. A single regulatory gene is sufficient to alter bacterial host range. Nature 458:215-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mayor, C., et al. 2000. VISTA: visualizing global DNA sequence alignments of arbitrary length. Bioinformatics 16:1046-1047. [DOI] [PubMed] [Google Scholar]
- 41.Meighen, E. A. 1994. Genetics of bacterial bioluminescence. Annu. Rev. Genet. 28:117-139. [DOI] [PubMed] [Google Scholar]
- 42.Miller, J. H. 1992. A short course in bacterial genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 43.Miyashiro, T., M. S. Wollenberg, X. Cao, D. Oehlert, and E. G. Ruby. 2010. A single qrr gene is necessary and sufficient for LuxO-mediated regulation in Vibrio fischeri. Mol. Microbiol. 77:1556-1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Morrison, D. A. 2007. Increasing the efficiency of searches for the maximum likelihood tree in a phylogenetic analysis of up to 150 nucleotide sequences. Syst. Biol. 56:988-1010. [DOI] [PubMed] [Google Scholar]
- 45.Nealson, K. H. 1977. Autoinduction of bacterial luciferase. Occurrence, mechanism and significance. Arch. Microbiol. 112:73-79. [DOI] [PubMed] [Google Scholar]
- 46.Nealson, K. H., and J. W. Hastings. 1979. Bacterial bioluminescence: its control and ecological significance. Microbiol. Rev. 43:496-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nishiguchi, M. K. 2002. Host-symbiont recognition in the environmentally transmitted sepiolid squid-Vibrio mutualism. Microb. Ecol. 44:10-18. [DOI] [PubMed] [Google Scholar]
- 48.Nishiguchi, M. K., E. G. Ruby, and M. J. McFall-Ngai. 1998. Competetive dominance among strains of luminous bacteria provides an unusual form of evidence for parallel evolution in Sepiolid squid-Vibrio symbioses. Appl. Environ. Microbiol. 64:3209-3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.O'Grady, E. A., and C. F. Wimpee. 2008. Mutations in the lux operon of natural dark mutants in the genus Vibrio. Appl. Environ. Microbiol. 74:61-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pompeani, A. J., et al. 2008. The Vibrio harveyi master quorum-sensing regulator, LuxR, a TetR-type protein is both an activator and a repressor: DNA recognition and binding specificity at target promoters. Mol. Microbiol. 70:76-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Posada, D., and T. R. Buckley. 2004. Model selection and model averaging in phylogenetics: advantages of Akaike information criterion and Bayesian approaches over likelihood ratio tests. Syst. Biol. 53:793-808. [DOI] [PubMed] [Google Scholar]
- 52.Pybus, O. G., and B. Shapiro. 2009. Natural selection and adaptation of molecular sequences, p. 413-415. In P. Lemey, M. Salemi, and A.-M. Vandamme (ed.), A phylogenetic handbook: a practical approach to phylogenetic analysis and hypothesis testing, 2nd ed. Cambridge University Press, Cambridge, United Kingdom.
- 53.Ronquist, F., and J. P. Huelsenbeck. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572-1574. [DOI] [PubMed] [Google Scholar]
- 54.Ronquist, F., P. van der Mark, and J. P. Huelsenbeck. 2009. Bayesian phylogenetic analysis using MrBayes, p. 210-266. In P. Lemey, M. Salemi, and A.-M. Vandamme (ed.), The phylogenetic handbook: a practical approach to phylogenetic analysis and hypothesis testing, 2nd ed. Cambridge University Press, Cambridge, United Kingdom.
- 55.Ruby, E. G. 1996. Lessons from a cooperative bacterial-animal association: the Vibrio fischeri-Euprymna scolopes light organ symbiosis. Annu. Rev. Microbiol. 50:591-624. [DOI] [PubMed] [Google Scholar]
- 56.Ruby, E. G., and K.-H. Lee. 1998. The Vibrio fischeri-Eupyrmna scolopes light organ association: current ecological paradigms. Appl. Environ. Microbiol. 64:805-812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ruby, E. G., and K. H. Nealson. 1976. Symbiotic association of Photobacterium fischeri with the marine luminous fish Monocentris japonica: a model of symbiosis based on bacterial studies. Biol. Bull. 151:574-586. [DOI] [PubMed] [Google Scholar]
- 58.Ruby, E. G., et al. 2005. Complete genome sequence of Vibrio fischeri: a symbiotic bacterium with pathogenic congeners. Proc. Natl. Acad. Sci. U. S. A. 102:3004-3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schuster, B. M., L. A. Perry, V. S. Cooper, and C. A. Whistler. 2010. Breaking the language barrier: experimental evolution of non-native Vibrio fischeri in squid tailors luminescence to the host. Symbiosis 51:85-96. [Google Scholar]
- 60.Septer, A. N., J. L. Bose, A. K. Dunn, and E. V. Stabb. 2010. FNR-mediated regulation of bioluminescence and anaerobic respiration in the light-organ symbiont Vibrio fischeri. FEMS Microbiol. Lett. 306:72-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shadel, G. S., J. H. Devine, and T. O. Baldwin. 1990. Control of the lux regulon of Vibrio fischeri. J. Biolumin. Chemilumin. 5:99-106. [DOI] [PubMed] [Google Scholar]
- 62.Stabb, E. V. 2005. Shedding light on the bioluminescence “paradox.” ASM News 71:223-229. [Google Scholar]
- 63.Stabb, E. V. 2006. The Vibrio fischeri-Euprymna scolopes light organ symbiosis, p. 204-218. In F. L. Thompson, B. Austin, and J. Swings (ed.), The biology of vibrios. ASM Press, Washington, DC.
- 64.Stabb, E. V., M. S. Butler, and D. M. Adin. 2004. Correlation between osmolarity and luminescence of symbiotic Vibrio fischeri strain ES114. J. Bacteriol. 186:2906-2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stabb, E. V., and D. S. Millikan. 2009. Is the Vibrio fischeri-Euprymna scolopes symbiosis a defensive mutualism?, p. 85-98. In J. F. White, Jr., and M. S. Torres (ed.), Defensive mutualism in microbial symbiosis. Taylor and Francis, Boca Raton, FL.
- 66.Stabb, E. V., K. A. Reich, and E. G. Ruby. 2001. Vibrio fischeri genes hvnA and hvnB encode secreted NAD+-glycohydrolases. J. Bacteriol. 183:309-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stabb, E. V., and E. G. Ruby. 2002. RP4-based plasmids for conjugation between Escherichia coli and members of the Vibrionaceae. Methods Enzymol. 358:413-426. [DOI] [PubMed] [Google Scholar]
- 68.Stabb, E. V., A. Schaefer, J. L. Bose, and E. G. Ruby. 2008. Quorum signaling and symbiosis in the marine luminous bacterium Vibrio fischeri, p. 233-250. In S. C. Winans and B. L. Bassler (ed.), Chemical communication among microbes. ASM Press, Washington, DC.
- 69.Swartzman, E., and E. A. Meighen. 1993. Purification and characterization of a poly(dA-dT) lux-specific DNA-binding protein from Vibrio harveyi and identification as LuxR. J. Biol. Chem. 268:16706-16716. [PubMed] [Google Scholar]
- 70.Swofford, D. L. 2003. PAUP*: phylogenetic analysis using parsimony (*and other methods). Sinauer Associates, Sunderland, MA.
- 71.Tajima, F. 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123:585-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Urbanowski, M. L., C. P. Lostroh, and E. P. Greenberg. 2004. Reversible acyl-homoserine lactone binding to purified Vibrio fischeri LuxR protein. J. Bacteriol. 186:631-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Visick, K. L., J. Foster, J. Doino, M. McFall-Ngai, and E. G. Ruby. 2000. Vibrio fischeri lux genes play an important role in colonization and development of the host light organ. J. Bacteriol. 182:4578-4586. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.