Abstract
In Dictyostelium discoideum, extracellular K+ or Ca2+ at a concentration of 40 or 20 mM, respectively, facilitates motility in the absence or presence of a spatial gradient of chemoattractant. Facilitation results in maximum velocity, cellular elongation, persistent translocation, suppression of lateral pseudopod formation, and myosin II localization in the posterior cortex. A lower threshold concentration of 15 mM K+ or Na or 5 mM Ca2+ is required for chemotactic orientation. Although the common buffer solutions used by D. discoideum researchers to study chemotaxis contain sufficient concentrations of cations for chemotactic orientation, the majority contain insufficient levels to facilitate motility. Here it has been demonstrated that Nhe1, a plasma membrane protein, is required for K+ but not Ca2+ facilitation of cell motility and for the lower K+ but not Ca2+ requirement for chemotactic orientation.
INTRODUCTION
Extracellular Ca2+ and K+, ubiquitous in the soluble environment of cells, both free-living and in multicellular organisms (5, 15, 21, 36, 56, 79, 80, 84, 91), have long been known to affect the motility of cells ranging in complexity from bacterial to human (7, 8, 19, 25, 31, 60, 62, 65, 98). In Dictyostelium discoideum, which serves as a powerful model for studying animal cell motility (4, 6, 20, 34, 41, 42, 43, 45, 50, 82, 87, 86, 92, 98), either extracellular K+ or Ca2+ at an optimum concentration of 40 mM or 20 mM, respectively, enhances most aspects of basic cell motility in the absence of chemoattractant, including cell elongation, uropod formation, the suppression of lateral pseudopod formation, velocity, directional persistence, and localization of myosin II in the posterior cell cortex (51, 78). These facilitating concentrations of K+ or Ca2+ cannot be replaced with other monovalent or divalent cations (51) and are within the range of the soluble concentrations of the two cations found in soil (1, 5, 32, 44, 49, 56) and manure (9, 66, 83), two common niches in which soil amoebae thrive (12, 27, 35, 67, 71). These concentrations of K+ and Ca2+ are close to those found to be optimum for enhancing the motility of a variety of other animal cells. For instance, 9 mM Ca2+ has been found to be optimum for flagellated sea urchin sperm motility (99), and Ca2+ ranging in concentration from 1 to 10 mM has been found optimum for vertebrate cell motility (28, 58, 63). Concentrations of K+ greater than 100 mM have been found to be optimum for the polarization and motility of human polymorphonuclear leukocytes (52, 69).
The surface molecules and mechanisms regulating K+ and Ca2+ facilitation in D. discoideum have not been elucidated. However, Patel and Barber (64) reported a defective behavioral phenotype for cells of the nhe1 null mutant in a facilitating concentration of K+ that was strikingly similar to the behavior of wild-type cells in a nonfacilitating concentration of K+ (51). Nhe1 is a plasma membrane protein related to cation/H+ exchangers (64). We therefore tested the possibility that Nhe1 mediated K+ facilitation. First, to define and illustrate the process of cationic facilitation, we compared cell motility and chemotaxis among buffer solutions containing a facilitating concentration of K+ or Ca2+ and buffer solutions commonly used by D. discoideum researchers to study motility and chemotaxis. The majority of the latter solutions contain nonfacilitating cation concentrations. The comparative study revealed that the majority of common buffer solutions support chemotactic orientation but are not optimum for motility, thus illustrating cationic facilitation. This comparison also revealed for the first time that a threshold concentration of 15 mM K+ or Na2+ or a threshold concentration of 5 mM Ca2+ was required for chemotactic orientation. We then tested the role of Nhe1 by analyzing the nhe1− null mutant. Our results revealed that Nhe1 mediates both K+ facilitation and the K+/Na+ requirement for chemotactic orientation, but plays no role in Ca2+ facilitation or the Ca2+ requirement for chemotactic orientation. The possible mechanisms through which Nhe1 might mediate these monovalent cation effects, including the possibility that Nhe1 acts as a monovalent cation sensor, are discussed.
MATERIALS AND METHODS
Strain maintenance and development.
The parent strain Ax2, the nhe1− deletion strain, and the nhe1−/nhe1+ (nhe1−/[act15]:nhe1:HA) complemented strain were obtained from the Dictyostelium stock center (http://dictybase.org/StockCenter/StockCenter.html) and subcloned. The mutant strains were originally generated by Patel and Barber (64). To select for mutant strains, the nhe1− cells were grown in the presence of 10 μg/ml of blasticidin S (Sigma-Aldrich, St. Louis, MO), and nhe1−/nhe1+ cells were grown in blasticidin S plus 10 μg/ml G418 (Sigma-Aldrich, St. Louis, MO). To obtain aggregation-competent amoebae, cells were developed on filters according to methods described previously (76, 94, 96). Developmental filters were saturated with a buffer containing 20 mM KCl, 2.5 mM MgCl2, 20 mM KH2PO4, and 5 mM Na2HPO4, pH 6.4 (81). For experimental purposes, cells were washed four times in the above buffer solution or in Tricine buffer (TB) (pH 7.0) (51).
Analysis of basic motile behavior and chemotaxis.
Cells were perfused with test solution in a Sykes-Moore chamber according to the methods previously described (88). For chemotaxis, cells were distributed on the bridge of a Plexiglas gradient chamber according to methods previously described (51, 89). Gradients were generated by filling one trough bordering the bridge with buffer solution lacking cAMP and in the other trough with buffer solution containing 10−6 M cAMP. Cell were incubated for 5 min, and then images were digitally acquired every 4 s for a 10-min period.
DIAS analysis.
Cell images were digitally acquired and analyzed with two-dimensional (2D)-DIAS software (DIAS) as previously described (77, 96). Movies were exported to a QuickTime format and imported into DIAS. Instantaneous velocity, percent cells moving at ≥9 μm per min, directional persistence, chemotactic index (CI), and percent positive chemotaxis were computed from centroid positions (75, 77). “Instantaneous velocity” was computed as the average velocity (distance between two consecutive centroids divided by interval time) for two consecutive intervals (54). It is computed for the two intervals a to b and b to c and then b to c and c to d. It then prescribes the average of each pair to frame b and c, respectively. It is a method for smoothing the velocities computed for consecutive intervals (77). The parameter “% cells ≥ 9 μm/min” was computed as the proportion of cells in the analyzed population moving with an instantaneous velocity equal to or greater the 9 μm per minute. “Directional persistence” was computed as the net distance between the first and last centroids of a centroid track divided by the summed distances between all intervals. The intervals between centroids for calculating motility and chemotaxis parameters were 4 s and are based on the stability of persistence measurements, which will be dealt with in a subsequent methods paper. (D. R. Soll, D. Wessels, and S. Kuhl, unpublished data). The “chemotactic index” was computed as the net distance moved up a cAMP gradient divided by the total distance (the sum of the distances at 4-s intervals). A CI of −1.00 represented direct movement down the gradient, +1.00 represented direct movement up the gradient, 0.00 represented random movement, and +0.20 to +0.99 represented positive chemotaxis. The “% positive chemotaxis” was computed as the proportion of cells in a population with a chemotactic index greater than 0.00. “Positive flow” was computed by calculating the region in the second of two consecutive cell parameter images not overlapping the first and converting that area to the percentage of the area of the first image. The interval between centroids for calculating motility and chemotaxis parameters was 4 s and is based on the stability of persistence measurements, which will be dealt with in a subsequent methods paper. (Soll et al., unpublished). The percent suppression of lateral pseudopods was determined from the expansion zones present in difference pictures generated by DIAS according to the methods of Wessels et al. (96).
Strains expressing MhcA-GFP.
To generate a nhe1− derivative expressing MhcA-green fluorescent protein (GFP), the plasmid containing mhcA-GFP (57) was isolated from the mhcA−/mhcA-GFP strain, a generous gift from Thomas Egelhoff of Case Western University, Cleveland, OH, using the Rapid-Plasmid kit (Marligen Biosciences, Ijamsville, MD) according to the manufacturer's instructions. The plasmid, containing a neomycin resistance marker (61), was transformed into DH5α cells (Invitrogen, Carlsbad, CA) and subcloned, and the isolated plasmid was sequenced. The plasmid was then transformed into both the parental strain, Ax2, and the nhe1− deletion mutant according to the methods of Eichinger and Rivero (30). The concentration of G418, a substitute for neomycin, was subsequently increased over a period of 2 to 3 weeks from 2 to 20 μg/ml to select for transformants.
Fluorescence imaging of GFP-labeled cells.
Live imaging of GFP-labeled cells was performed as previously described (94) using a Bio-Rad Radiance 2100 MP laser scanning confocal microscopy and a 40× Nikon S Fluor 1.30 oil objective. Simultaneous fluorescent and differential interference contrast (DIC) images were generated using the 488-nm and 476-nm argon lasers, respectively. The LaserSharp 2000 (release 5.2) software program was used for image acquisition and for measuring the pixel intensities of line profiles across labeled cells as described by Lusche et al. (51).
Nucleotide sequence accession numbers.
The GenBank accession numbers for Nhe2 to -4 cDNAs from D. discoideum strain Ax2 are as follows: for Nhe2 (NheB), HQ730876; for Nhe3 (NheC), HQ730877; and for Nhe4 (NheD), HQ730878.
RESULTS
Cationic facilitation of motility.
To obtain optimum motility, either the Ca2+ concentration must be 10 to 20 mM or the K+ concentration 40 mM (51). Combining 20 mM Ca2+ and 40 mM K+ does not increase motility parameters above that achieved by either alone (see Table S1 in the supplemental material). We have referred to the optimization of motility by 20 mM Ca2+ or 40 mM K+ as “the cationic facilitation of cell motility.” Since the objective here was to test whether Nhe1 specifically mediated K+ facilitation, it was paramount that cationic facilitation was clearly defined and illustrated. One way to do this was to compare cell behavior in solutions containing a facilitating concentration of K+ or Ca2+ with cell behavior in buffers commonly used by researchers of D. discoideum, the majority of which do not contain facilitating concentrations of K+ or Ca2+. This comparison could not be performed retrospectively from the published literature since the conditions and strains used in different studies varied dramatically. Moreover, a variety of methods have been used to calculate motility parameters, which can compromise comparisons. Differences in the interval time between data points (see Fig. S1A and B) and the velocity thresholds for subtracting nonmotile cells (see Fig. S1C and D) can have a significant impact on computed motility parameter.
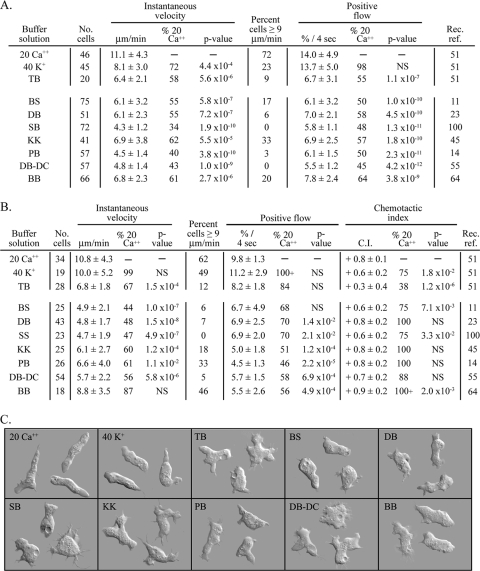
Therefore, to compare cell behavior in different buffer solutions, we used the single laboratory strain Ax2, uniform culture conditions, the same perfusion chamber for assessing behavior in the absence of a chemoattractant (88), the same gradient chamber for assessing chemotaxis (89), the same interval time (4 s) between data points, and the same instantaneous velocity threshold (≥3 μm per min) for identifying motile cells in a population (77). The three motility parameters (average instantaneous velocity, percent cells with speeds of ≥9 μm per min, and average percent positive flow) are defined in Materials and Methods. In the absence of cAMP, these parameters were maximal in 20 mM Ca2+ solution, as they were in 20 mM Ca2+ in a cAMP gradient (Fig. 1A and B, respectively). We therefore used the values in 20 mM Ca2+ solution as the references for comparison with cells translocating in the other solutions. In the absence of a chemoattractant, the three motility parameters in 40 mM K+ solution and in the buffer solution (BB) used by Patel and Barber (61), were below that in 20 mM Ca2+ solution but still on average higher than that in the six other buffer solutions (Fig. 1A). Of the six other buffer solutions, the mean instantaneous velocity was highest in KK buffer (Fig. 1A) (45), which contained the highest concentration of K+ (24.3 mM) (Table 1). The motility parameters in the five test buffers BS, DB, SS, PB, and DB−DC were no better on average than that in Tricine buffer (TB) alone (Fig. 1A), which contains a nonfacilitating K+ concentration of 5 mM (Table 1). Cell shape also reflected facilitation. Cells translocating in 20 mM Ca2+ solution or 40 mM K+ solution were more elongate on average than cells in the other test solutions (Fig. 1C).
Fig. 1.
Motile behavior of Dictyostelium discoideum, analyzed in buffers commonly used by researchers studying motility and chemotaxis. The composition of the solutions is provided in Table 1. Cell motility and cell polarization were maximal in solutions containing facilitating concentrations of Ca2+ and K+ but not in the majority of commonly used buffers, which do not contain facilitating concentrations of either cation. This was true in both the absence and the presence of a cAMP gradient. Chemotactic orientation in a cAMP gradient, however, was equally efficient in all tested buffers. (A) Motility parameters in test solutions in the absence of cAMP. (B) Motility and chemotaxis parameters in a cAMP spatial gradient. (C) Differential interference contrast microscopy images of cells crawling in the buffer solutions indicated in the upper left corner of each panel in the absence of cAMP. Parameters in panels A and B are defined in Materials and Methods. Values in panels A and B are the means ± standard deviations for the population. No. cells, number of cells analyzed; Rec. ref., Recent reference for buffer; see Table 1 for abbreviations. The P value for parameters in each test solution versus 20 mM Ca2+ were determined by the Student t test; NS, not significant for P values of >5.0 × 10−2.
Table 1.
Compositions of tested buffer solutions
| Buffer solution (abbra) | Composition | [Na+] (mM) | [K+] (mM) | [Total monovalent cations] (mM) | [Ca2+] (mM) | [Mg2+] (mM) | Reference |
|---|---|---|---|---|---|---|---|
| Tricine buffer (TB) | 5 mM Tricine, 5 mM KCl | 5 | 5 | 51 | |||
| 20 mM Ca2+ solution (20 Ca2+) | 5 mM Tricine, 5 mM KCl, 20 mM CaCl2 | 5 | 5 | 20 | 51 | ||
| 40 mM K+ solution (40 K+) | 2.5 mM MgCl2, 20 mM KH2PO4, 20 mM KCl, 5 mM Na2HPO4 | 5 | 40 | 45 | 2.5 | 51 | |
| Bonner solution (BS) | 2.7 mM CaCl2, 10 mM KCl, 10 mM NaCl | 10 | 10 | 20 | 2.7 | 11 | |
| Developmental buffer (DB) | 5 mM Na2HPO4, 5 mM NaH2PO4, 2 mM MgSO4, 200 μM CaCl2 | 15 | 15 | 0.2 | 2.5 | 23 | |
| Sorenson buffer (SB) | 2.7 mM Na2HPO4, 14 mM KH2PO4 | 5.4 | 14 | 19.4 | 100 | ||
| KK2 (KK) | 3.9 mM K2HPO4, 16.5 mM KH2PO4, 2 mM MgSO4, 0.1 mM CaCl2 | 24.3 | 24.3 | 0.1 | 2.5 | 46 | |
| Phosphate buffer (PB) | 4 mM Na2HPO4, 7.4 mM KH2PO4 | 8 | 7.4 | 15.4 | 14 | ||
| DB minus divalent cations (DB-DC) | 5 mM Na2HPO4, 5 mM NaH2PO4 | 15 | 15 | 55 | |||
| Barber buffer (BB) | 20 mM KH2PO4, 20 mM K2HPO4 | 60 | 60 | 64 |
abbr, abbreviation.
When a spatial gradient of cAMP was generated in the tested buffer solutions, the three measured motility parameters were maximal in 40 mM K+ solution and 20 mM Ca2+ solution (Fig. 1B). Two of the three parameters were close to maximal in solution BB (64) (Fig. 1B). All three parameters were reduced in the remaining six buffer solutions and TB (Fig. 1B). In marked contrast, the chemotactic index (CI) was on average as high in the seven test solutions as it was in 20 mM Ca2+ or 40 mM K+ solution (Fig. 1B). These results indicate that although the majority of commonly used buffer solutions do not contain high enough cation concentrations to facilitate motility, they all support chemotactic orientation maximally and similarly.
The low CI in TB compared to that in the other test buffers (Fig. 1B) also suggested that a monovalent cation concentration threshold also existed for maximum chemotactic orientation and that this threshold was lower than that for the facilitation of motility. To identify the threshold for orientation, we compared the CIs of cells in TB containing 5, 10, and 15 mM Na+. The computed CIs were +0.48 ± 0.38 (number of cells, 43), +0.43 ± 0.36 (number of cells, 50), and +0.67 ± 0.32 (number of cells, 92), respectively. The CI in 15 mM Na+ was significantly different from the lower CIs in 5 and 10 mM Na+ (P = 0.05 and 0.002, respectively). Together, our results indicate that a monovalent cation (K+ and/or Na+) concentration threshold of 15 mM is necessary for maximum chemotactic orientation. Interestingly, all of the buffer solutions empirically formulated by various researchers to study D. discoideum chemotaxis contained concentrations of K+ and/or Na2+ that were 15 mM or greater (Table 1).
We previously demonstrated that in a low concentration of monovalent cations, a Ca2+ threshold also existed for optimum chemotactic orientation (51). This threshold was 5 mM, again below the threshold concentration for Ca2+ facilitation of motility. These results, therefore, illustrate the cationic facilitation of cell motility and reveal that most researchers use buffers containing concentrations of cations that are suboptimal for cell motility. These results also reveal these buffers do contain concentrations of Na+ and/or K+ equal to or above the threshold for maximum chemotactic orientation.
Nhe1, a candidate for mediating K+ facilitation.
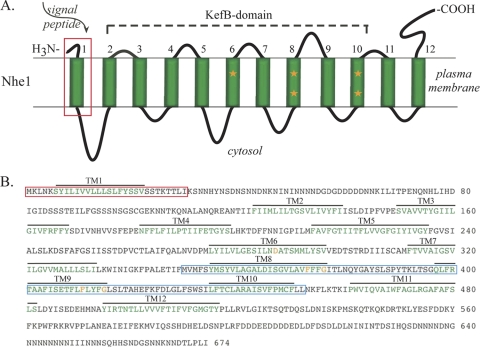
We considered the possibility that the putative monovalent cation/H+ exchanger Nhe1 was the surface molecule mediating the facilitation of motility and the monovalent cation requirement for chemotactic orientation, because the behavioral phenotype of the nhe1− null mutant in a facilitating concentration of K+ (64) was similar to that of wild-type cells in a nonfacilitating concentration of K+ (51). A BLAST search (http://blast.ncbi.nm.nih.gov/Blast.cgi) of the D. discoideum genome revealed that Nhe1 was a member of a family of four related proteins, Nhe1 (NheA), Nhe2 (NheB), Nhe3 (NheC), and Nhe4 (NheD). Patel and Barber (64) previously noted that Nhe1 of D. discoideum was distinct from Nhe1s in other species but was similar to Nhe8 in a variety of other species, including humans. The 12 transmembrane domains of Nhe1 were identified with the software program TMHMM (http://www.cbs.dtu.dk/services/TMHMM/), and the signal peptide was identified with the software program PSORT II (http://psort.ihgc.jp) (Fig. 2A and B). Nhe1 and Nhe2 belong to the CPA1 family of exchangers (Transport DB database; http://www.membranetransport.org), members of which can exchange Na+ or K+ for H+ without a change in membrane potential (17, 22, 47). This characteristic has been considered indicative of coupled receptors, including divalent cation-sensing receptors (16, 37). Nhe1 also contains a region homologous to KefB domains (Fig. 2A), which have been shown in bacteria to interact with K+ in transport systems (13, 26, 33, 47, 68). Five amino acids that are highly conserved in KefB domains according to the Conserved Domain Database (http://www.ncbi.nlm.nih.gov/structure/cdd/cdd.shtml) (53) were detected in D. discoideum Nhe1 (Fig. 2A and B). The first 30 amino acids of the N-terminal domain of Nhe1 has a signal sequence that is common to the first transmembrane domain of a class of trimeric G protein-coupled receptors found in higher eukaryotes (2, 97) (Fig. 2A and B). Finally, Nhe1 contains a 100-amino-acid region that is homologous to a similarly positioned region in a family of chemosensory receptors in Caenorhabditis elegans, according to a motif scan at http://myhits.isb-sib.ch (40) and confirmed with the Worm Database (http://www.wormbase.org) (70, 85) (Fig. 2B, blue box). We have used the same Nhe1, rather than NheA, the correct name according to D. discoideum nomenclature (http://dictybase.org), because Nhe1 has been used by Barber and coworkers in previous papers (24, 64).
Fig. 2.
Deduced structure of Dictyostelium discoideum Nhe1. (A) The model for Nhe1 in the plasma membrane. The signal peptide is boxed in red, the 12 transmembrane domains are numbered and colored green, and orange stars indicate amino acids in transmembrane domains 6, 8, and 9 that are conserved in the KefB-domain of the superfamily. (B) Deduced amino acid sequence of Dictyostelium discoideum Nhe1 (accession number XP_643611). The amino acids in the signal peptide are boxed in red, the amino acids in the 12 transmembrane (TM) domains are colored green, the region homologous to the C. elegans chemoreceptor domain is boxed in blue, and the five conserved amino acids in the KefB domain are colored orange.
K+ facilitation is lost in nhe1− cells in the absence of cAMP.
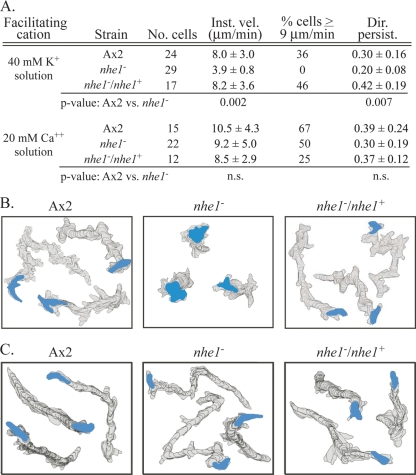
If Nhe1 is the plasma membrane protein that mediates K+ facilitation, then deleting it should result in the loss of K+ facilitation. Cells of the parental strain Ax2 and a complemented nhe1 strain, the nhe1−/nhe1+ strain, translocated with mean instantaneous velocities of 8.0 ± 3.0 and 8.2 ± 3.6 μm per minute, respectively, in 40 mM K+ solution (Fig. 3A). In marked contrast, nhe1− cells translocated with a mean instantaneous velocity of 3.9 ± 0.8 μm per minute, less than half that of control cells and only 0.9 μm per minute above the threshold of 3 μm per minute used to discriminate motile cells (Fig. 3A). The motility defect in the velocity of nhe1− cells was most evident in comparisons of high-end velocities (i.e., the proportion of cells with velocities of ≥9 μm per minute). While the proportions of cells of the parental and complemented control strains were 36 and 46%, respectively, the proportion of nhe1− cells was 0% (Fig. 3A). Finally, the directional persistence of nhe1− cells, defined in Materials and Methods, was 33% lower than that of Ax2 cells and 52% lower than that of nhe1−/nhe1+ cells (Fig. 3A). These behavioral defects were evident in perimeter tracks of nhe1− cells in 40 mM K+ solution in the absence of chemoattractant (Fig. 3B). The motile phenotype of nhe1− cells translocating on a substrate in 40 mM K+ solution in the absence of a chemoattractant (Fig. 3A and B) was therefore highly similar to that of parental control cells in TB alone containing K+ at the nonfacilitating concentration of 5 mM (Fig. 1A and C). These results demonstrate that in the absence of cAMP, the motile behavior of nhe1− cells was not facilitated by 40 mM K+ (51).
Fig. 3.
Cells of the nhe1− mutant have lost K+ facilitation but have retained Ca2+ facilitation. (A) Motility parameters for parental control strain Ax2, the nhe1− mutant, and the nhe1−/nhe1+ complemented control in the absence of chemoattractant either in 40 mM K+ solution or 20 mM Ca2+ solution. P values were obtained by Student's t test. Parameters are defined in Materials and Methods. (B) 2D-DIAS-generated stacked perimeter tracks of three representative Ax2, nhe1−, and nhe1−/nhe1+ cells in 40 mM K+ solution. Perimeters were drawn at 12-s intervals over a 10-min period. The final outline of the cell is color coded blue. (C) 2D-DIAS-generated stacked perimeter tracks of three representative Ax2, nhe1−, and nhe1−/nhe1+ cells in 20 mM Ca2+ solution with perimeters drawn as described for panel B. No. cells, the number of cells analyzed; Inst. vel., instantaneous velocity; % cells > 9 μm/min, the proportion of cells moving with average velocities > 9 μm/min; Dir. persist., directional persistence; ns, not significant.
Ca2+ facilitation is intact in nhe1− cells in the absence of chemoattractant.
If Nhe1 specifically mediates K+ facilitation, then deletion of the gene nhe1 should not affect Ca2+ facilitation. We tested this prediction in 20 mM Ca2+ solution in the absence of a chemoattractant. The mean velocities of parental and complemented control cells were 10.5 ± 4.3 and 8.5 ± 2.9 μm per min, respectively (Fig. 3A). The mean velocity of nhe1− cells was 9.2 ± 5.0 μm per min (Fig. 3A), which was statistically indistinguishable from that of the two control strains. The velocity in 20 mM Ca2+ solution was more than twice that in 40 mM K+ solution (Fig. 3A). The percentage of nhe1− cells with velocities of ≥9 μm per minute in 20 mM Ca2+ solution was high (50%), like that of control strains (67 and 25%, respectively) (Fig. 3A). It differed markedly from that of nhe1− cells in 40 mM K+ solution, which was 0% (Fig. 3A). The directional persistence parameter of nhe1− cells in 20 mM Ca2+ solution was also similar to that of control cells (Fig. 3A). The similarity between the behavior of nhe1− cells and that of control cells in 20 mM Ca2+ solution was evident in a comparison of perimeter tracks (Fig. 3C). A repeat of this experiment in 10 mM Ca2+ provided similar results (data not shown). Together, these results demonstrate that in the absence of a chemoattractant, deletion of nhe1 results in the selective loss of K+, but not Ca2+, facilitation.
K+ facilitation and chemotactic orientation are defective in nhe1− cells in a cAMP gradient.
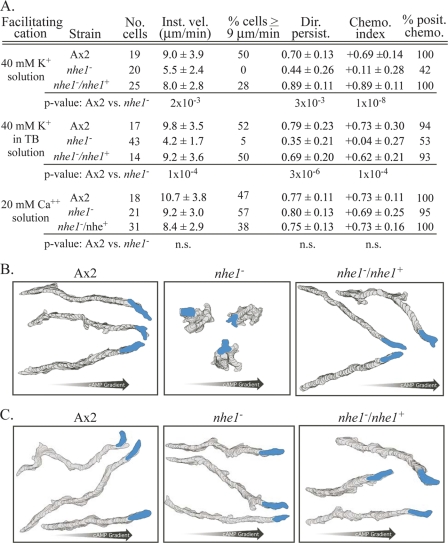
In a spatial gradient of the chemoattractant cAMP generated in 40 mM K+ solution in a chamber designed based on that of Zigmond (89, 101), the velocities of nhe1− cells were 39% and 31% lower than those of parental and complemented control cells, respectively (Fig. 4A). The proportion of cells with velocities of ≥9 μm per min was 0%, compared to 50 and 28% for the two respective control strains (Fig. 4A). Directional persistence was also far lower than that of either of the control strains (Fig. 4A). The same motility defects were observed in a cAMP gradient generated in 40 mM K+ in TB (Fig. 4A).
Fig. 4.
Cells of the nhe1− mutant have lost the capacity to undergo K+ facilitation and K+-dependent chemotactic orientation in a spatial gradient of cAMP but have retained Ca2+ facilitation and Ca2+-dependent chemotactic orientation. (A) Motility parameters for parental control strain Ax2 and the nhe1− and nhe1−/nhe1+ strains in 40 mM K+ solution and 20 mM Ca2+ solution. Chemo. index, chemotactic index; % posit. chemo. proportion of cells with positive chemotactic index. See the legend to Fig. 3 for remaining definitions of abbreviations. Parameters are defined in Materials and Methods. (B) 2-D DIAS-generated stacked perimeter tracks of three representative Ax2, nhe1−, and nhe1−/nhe1+ cells in a spatial gradient of cAMP generated in 40 mM K+ solution. (C) 2D-DIAS-generated stacked perimeter tracks of three representative Ax2, nhe1−, and nhe1−/nhe1+ cells in a spatial gradient of cAMP generated in 20 mM Ca2+ solution. See the Fig. 3 legend for additional explanations.
To assess chemotactic orientation, the chemotactic index and the percentage of cells exhibiting a positive chemotactic index (defined in Materials and Methods) were compared between the mutant and the two control strains. Cells of the nhe1− mutant strain exhibited weak to negligible chemotactic orientation in a cAMP gradient generated in 40 mM K+ solution, formulated in phosphate buffer (Table 1) or TB. The mean chemotactic index of nhe1− cells in the two buffers was +0.11 ± 0.28 and +0.04 ± 0.27, respectively, compared to +0.69 ± 0.14 and +0.73 ± 0.30, respectively, for parental control cells and +0.89 ± 0.11 and +0.62 ± 0.21 for complemented control cells (Fig. 4A). The percent positive chemotaxis of nhe1− cells in 40 mM K+ in the two buffers was 42 and 53%, respectively, compared to 100 and 94%, respectively, for parental control cells and 100 and 93%, respectively, for complemented control cells (Fig. 4A). Perimeter tracks reflected these results (Fig. 4B). These results demonstrate first that 40 mM K+ does not facilitate motility in nhe1− cells in a spatial gradient of cAMP and second that nhe1− cells have lost not only K+ facilitation but also K+-dependent chemotactic orientation in a cAMP gradient. These quantitative results obtained in gradients generated in a chemotaxis chamber are in agreement with the decrease in chemotactic efficiency observed earlier by Patel and Barber (64).
We also tested whether nhe1− cells also lost chemotactic orientation in a simple phosphate buffer solution, DB−DC, used by McCann et al. (55), which contained 15 mM Na+ (Table 1). The chemotactic index of nhe1− cells in this solution was negligible (+0.04 ± 0.29; n = 63), and the percent positive chemotaxis was 61%. Hence, neither 40 mM K+ solution nor a phosphate buffer solution containing 15 mM Na+ supported chemotactic orientation in nhe1− cells. These results demonstrate that Nhe1 is essential not only for K+ facilitation but also for the K+ or Na+ requirement for chemotactic orientation in a cAMP gradient.
Ca2+ facilitation is intact in nhe1− cells in spatial gradient of cAMP.
In contrast to the aberrant behavior observed in a spatial gradient of cAMP generated in 40 mM K+ solution or in the Na+-based buffer solution DB−DC (Table 1), both the motility and chemotactic orientation of nhe1− cells were indistinguishable from those of control cells in a cAMP gradient generated in 20 mM Ca2+ solution (Fig. 4A and C). The instantaneous velocity, percent cells with velocities of ≥9 μm per minute, directional persistence, chemotactic index, and percent cells with a positive chemotactic index were statistically similar to those of control strains (Fig. 4A). Cell perimeter tracks reinforced these findings (Fig. 4C). Nhe1, therefore, is essential for K+- but not Ca2+-dependent chemotactic orientation.
Cell elongation and the suppression of lateral pseudopods.
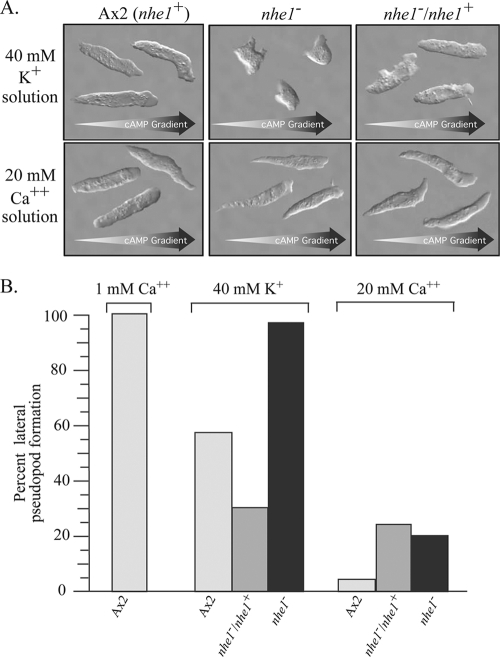
Both 40 mM K+ solution and 20 mM Ca2+ solution have been shown to enhance cell elongation and suppress lateral pseudopod formation during wild-type cell migration (51). These facilitated changes are important in attaining maximum velocity (51, 78). The effects of 40 mM K+ and 20 mM Ca2+ on cell elongation and lateral pseudopod formation were therefore compared between nhe1− and control cells in a cAMP gradient. Differential interference contrast microscopy revealed that either 40 mM K+ or 20 mM Ca2+ induced parental and complemented control cells to elongate, move in the direction of the single anterior pseudopod, suppress lateral pseudopod formation, extend new pseudopods primarily at the anterior end, and form a tapered uropod (Fig. 5A). However, while 20 mM Ca2+ induced these same changes in nhe1− cells, 40 mM K+ did not (Fig. 5A).
Fig. 5.
Cells of the mutant nhe1− do not respond to 40 mM K+ by elongating and suppressing lateral pseudopod formation, but they do respond to 20 mM Ca2+. (A) Differential interference microscopy images of three representative Ax2 (nhe1+), nhe1−, and nhe1−/nhe1+ cells in spatial gradients of cAMP generated in solutions containing either 40 mM K+ (top panels) or 20 mM Ca2+ (bottom panels). Arrows at the bottom of each panel indicate the direction of the cAMP gradient. (B) Percent lateral pseudopod formation was determined by dividing the number obtained in 40 mM K+ or 20 mM Ca2+ solution for Ax2, nhe1−/nhe1+, and nhe1− cells by the number obtained for Ax2 cells in a nonfacilitating 1 mM Ca solution and multiplying the fraction by 100. Light-gray bars, Ax2 control (nhe1+); dark-gray bars, nhe1−/nhe1+ complemented control; black bars, nhe1− strain.
Lateral pseudopods have previously been defined as new projections emanating from the flank of the cell body (78, 93, 94) or new anterior pseudopods resulting from anterior pseudopod bifurcation, a process also referred to as “splitting” (3). For a measure of the unsuppressed frequency, pseudopod formation was assessed in a cAMP gradient generated in 1 mM Ca2+ (51). The “frequency of lateral pseudopod formation” was computed as the frequency in a test solution divided by the frequency in the reference solution, multiplied by 100. In 40 mM K+ solution, the frequencies of lateral pseudopod formation of parental control (Ax2) and complemented control (nhe1−/nhe1+) cells were 55 and 30%, respectively, compared to those in 1 mM Ca2+ solution (Fig. 5B). The frequency of lateral pseudopod formation of nhe1− cells in 40 mM K+, however, was almost the same as that of control cells in 1 mM Ca2+ solution (Fig. 5B). These results indicate that 40 mM K+ does not suppress lateral pseudopod formation in nhe1− cells as it does in control cells. In 20 mM Ca2+ solution, however, lateral pseudopod formation in Ax2, nhe1−/nhe1+, and nhe1− cells was suppressed by 97%, 80%, and 85%, respectively (Fig. 5B) These results support the conclusion that the motile behavior of nhe1− cells is facilitated by 20 mM Ca2+ but not 40 mM K+.
Induced cortical localization of myosin II.
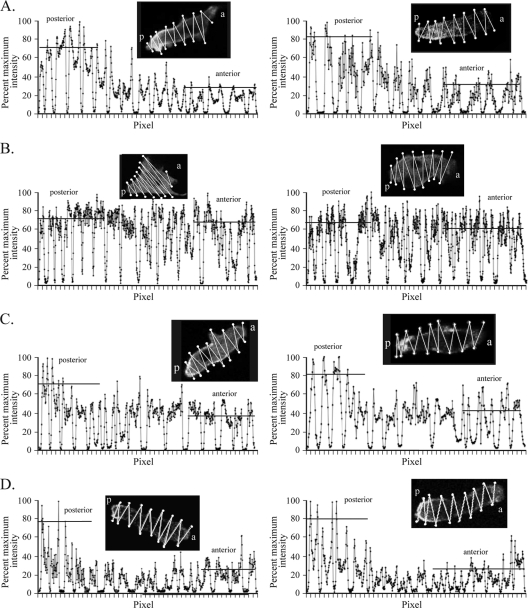
In the process of cationic facilitation, myosin II localizes to the posterior cell cortex (51). Posterior localization is believed to be important for cell elongation, the suppression of lateral pseudopod formation, uropod formation, efficient translocation, and efficient chemotaxis (29, 39, 48, 73, 78, 95). One would therefore expect 20 mM Ca2+ solution but not 40 mM K+ solution to induce posterior localization of myosin II in nhe1− cells. To test this prediction, parental control and nhe1− cells were transformed with a plasmid containing GFP-tagged myosin II (57) and the pattern of GFP fluorescence was analyzed by confocal microscopy. Projections of the center five images of a Z-series of 16 scans of each cell were stacked and analyzed for combined pixel intensity through a zigzag track protocol diagrammed in the upper portion of each panel in Fig. 6 (51). Intensity plots of representative control cells in a spatial gradient of cAMP generated in 40 mM K+ solution (Fig. 6A) or 20 mM Ca2+ solution (Fig. 6C) revealed broad peaks and troughs, the former representing the scans through the cell body and the latter the extracellular regions. In both cases, the intensity was highest in the cortex of the posterior region of the cell body (Fig. 6A and C). Intensity plots of representative nhe1− cells in a spatial gradient of cAMP generated in 40 mM K+ solution (Fig. 6B) or 20 mM Ca2+ solution (Fig. 6D) also revealed broad peaks and troughs. In 20 mM Ca2+, the intensity was higher in the posterior cortex of the cell body of nhe1− cells (Fig. 6D), just as it was in control cells (Fig. 6C). However, in 40 mM K+, the intensities along a nhe1− cell were similar in the anterior and posterior cell cortex (Fig. 6B). In these cells, the posterior end of apolar nhe1− cells was identified by the position of tail fibers (38), not shown here. These results demonstrate that 20 mM Ca2+ but not 40 mM K+ induces the localization of myosin II to the posterior cortex of nhe1− cells.
Fig. 6.
Myosin II does not localize normally in the posterior cortex of nhe1− cells in chemotaxis when K+ is the facilitating cation, but it does when Ca2+ is the facilitating cation. The maximum intensity for a scan was normalized to 100%. (A) Ax2 control cells in 40 mM K+ solution. (B) nhe1− cells in 40 mM K+ solution. (C) Ax2 control cells in 20 mM Ca2+ solution. (D) nhe1− cells in 20 mM Ca2+ solution. a, anterior; p, posterior. Horizontal bars in each graph indicate the average peak intensities for the posterior and anterior regions of the representative cell images. Insets show the zigzag path of intensity measurements along the cell that is plotted in the corresponding graph below.
K+ concentration.
The preceding results demonstrating that nhe1− cells have selectively lost K+ facilitation were based on tests performed with 40 mM K+, the optimum concentration for the facilitation of wild-type cell motility (51). To test the possibility that K+ might facilitate motility and polarity in nhe1− cells but at a different concentration, we analyzed the effects of higher and lower K+ concentrations. We found that in the range of 5 to 80 mM, K+ did not facilitate cell motility or mediate chemotactic orientation in nhe1− cells (Table 2).
Table 2.
The nhe1− mutant is insensitive to K+ at concentrations ranging from 5 to 80 mM
| [K+] (mM)a | Strain name or description | No. of cells | Instantaneous velocity (mm/min) | Directional persistence | Chemotactic index |
|---|---|---|---|---|---|
| 5 | Ax2 | 28 | 6.8 ± 1.8 | 0.47 ± 0.22 | +0.34 ± 0.35 |
| nhe1− | 19 | 3.7 ± 1.3 | 0.15 ± 0.11 | +0.03 ± 0.12 | |
| 20 | Ax2 | 27 | 7.4 ± 2.5 | 0.63 ± 0.17 | +0.59 ± 0.21 |
| nhe1− | 36 | 4.7 ± 1.3 | 0.42 ± 0.21 | +0.03 ± 0.34 | |
| 40 | Ax2 | 17 | 9.8 ± 3.5 | 0.79 ± 0.23 | +0.77 ± 0.30 |
| nhe1− | 43 | 4.2 ± 1.7 | 0.35 ± 0.21 | +0.04 ± 0.27 | |
| 60 | Ax2 | 21 | 6.7 ± 2.6 | 0.68 ± 0.21 | +0.63 ± 0.27 |
| nhe1− | 45 | 3.2 ± 0.2 | 0.37 ± 0.14 | +0.01 ± 0.28 | |
| 80 | Ax2 | 54 | 6.4 ± 3.0 | 0.66 ± 0.24 | +0.56 ± 0.24 |
| nhe1− | 40 | 3.5 ± 0.7 | 0.37 ± 0.19 | +0.02 ± 0.25 |
The K+ concentration was varied in Tricine buffer, and each solution was used as the buffer for generating a cAMP gradient.
DISCUSSION
Previously we demonstrated that 40 mM K+ or 20 mM Ca2+ facilitated cell motility (51). To illustrate the process of facilitation, we compared motility in solutions containing facilitating concentrations of K+ and Ca2+ with that in buffer solutions commonly employed by researchers studying motility and chemotaxis in D. discoideum. Six of the seven contained nonfacilitating concentrations of Na+ and/or K+, ranging from 15 to 24 mM. None contained a facilitating concentration of Ca2+. Our results first demonstrated that the six buffer solutions were suboptimal for cell motility, but all performed as well as 40 mM K+ or 20 mM Ca2+ solution in supporting chemotactic orientation. However, chemotactic orientation in TB alone, which contains 5 mM K+, was reduced dramatically. This suggested that there was a monvalent cationic requirement for chemotactic orientation that was above 5 mM but below 40 mM. By varying the Na+ concentration, we found this cationic requirement to be 15 mM, the minimum concentration in the seven tested buffer solutions, suggesting that in the independent development of the seven buffer solutions used to study D. discoideum motility and chemotaxis, an unrecognized consensus had emerged that a minimum monovalent cation concentration of 15 mM was necessary for efficient chemotactic orientation. While 15 mM Na+ or K+ will fulfill the requirement for orientation, only 40 mM K+ will facilitate motility. Na+ at 40 mM is inhibitory (51). Furthermore, 5 mM Ca2+ also fulfills the requirement for orientation (51).
There are three possible ways in which Nhe1 may mediate the effects of extracellular monovalent cations: through changes in cytosolic pH, through changes in the concentration of cytosolic monvalent cations, or as a coupled sensor. Whatever the mechanism, Nhe1 must function by assessing differences in the steady-state concentration of extracellular monovalent cations. Nhe1 has been classified as a Na+/H+ exchanger (64, 90). However, the steady-state intracellular pH (pHi) of wild-type, complemented mutant, and mutant cells in the absence of cAMP was measured by Patel and Barber (64), using the fluorescent pH indicator 2′,7′-bis(2-carboxyethyl)-5(6)-carboxyfluorescein (BCECF). They reported them to be 6.58 ± 0.05, 6.63 ± 0.07, and 6.48 ± 0.06, respectively. Since the pHi of complemented mutant cells differed from that of wild-type cells by half the difference between those of wild-type and nhe1− cells, it was not clear if the latter difference was functionally meaningful. Moreover, the cytosolic specificity of BCECF fluorescence is in question, because it has been demonstrated that BCECF stains organelles and vesicles, as well as the cytosol (10, 18, 59, 72). Therefore, it is not clear if Nhe1 regulates intracellular pH. Alternately, Nhe1 may function by increasing intracellular monovalent cations. It has been demonstrated that incremental increases in the concentration of an extracellular monovalent cation result in increases in the cytosolic concentration of that cation (74). Therefore, Nhe1 might function by regulating the cytosolic concentration of monovalent cations. Finally, and perhaps more interestingly, Nhe1 may function as a coupled monovalent cation sensor. Nhe1 belongs to the group CPA1, members of which do not cause changes in membrane potential, a characteristic of a group of G protein-coupled receptors that includes Ca2+-sensing receptors (16, 37). Nhe1 has a KefB domain that has been demonstrated in other molecules to bind to K+ (13, 68). Nhe1 also contains a signal sequence in the first 30 amino acids of the N terminus that is common to a class of trimeric G protein-coupled receptors (2, 97). Finally, a 100-amino-acid region of Nhe1 is homologous to similarly positioned regions of chemosensory receptors in C. elegans (70, 85). Therefore, Nhe1 may function by sensing the extracellular concentration of monovalent cations. In that case, the signal would be transduced through molecules interacting with Nhe1. However, there are no data that unequivocally distinguish between the three alternative mechanisms.
Our results therefore demonstrate that Nhe1 mediates K+ facilitation of efficient cell motility and the monovalent cation requirement for chemotactic orientation. Nhe1 mediates these effects at constant extracellular concentrations of monovalent cations. In the absence of these concentrations of K+ or K+/Na+, respectively, 20 mM Ca2+ will facilitate efficient cell motility and at 5 mM Ca2+ will support chemotactic orientation (51). Again, these last effects are achieved at constant extracellular Ca2+ concentrations. The surface molecule that mediates the Ca2+ effects has not been identified. However, the pathways transducing the alternative K+/Na+ and Ca2+ signals must ultimately converge, since the induced responses are similar.
Supplementary Material
ACKNOWLEDGMENTS
We thank the dictyBase stock center (http://dictybase.org/StockCenter/StockCenter.html) for supplying strains.
This research was supported by the Developmental Studies Hybridoma Bank at the University of Iowa, a National Resource initiated by the NIH.
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
Published ahead of print on 14 January 2011.
REFERENCES
- 1. Adams F. 1971. Ionic concentrations and activities in soil solutions. Soil Sci. Soc. Am. J. 35:420–426 [Google Scholar]
- 2. Alken M., et al. 2009. The sequence after the signal peptide of the G protein-coupled endothelin B receptor is required for efficient translocon gating at the endoplasmic reticulum membrane. Mol. Pharmacol. 75:801–811 [DOI] [PubMed] [Google Scholar]
- 3. Andrew N., Insall R. H. 2007. Chemotaxis in shallow gradients is mediated independently of PtdIns 3-kinase by biased choices between random protrusions. Nat. Cell Biol. 9:193–200 [DOI] [PubMed] [Google Scholar]
- 4. Annesley S. J., Fisher P. R. 2009. Dictyostelium discoideum—a model for many reasons. Mol. Cell. Biochem. 329:73–91 [DOI] [PubMed] [Google Scholar]
- 5. Bangerth F. 1979. Calcium-related physiological disorders of plants. Annu. Rev. Phytopathol. 17:97–122 [Google Scholar]
- 6. Barry N. P., Bretscher M. S. 2010. Dictyostelium amoebae and neutrophils can swim. Proc. Natl. Acad. Sci. U. S. A. 107:11376–11380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Becker E. L. 1971. Phosphonate inhibition of the accumulation and retention of K+ by rabbit neutrophils in relation to chemotaxis. J. Immunol. 106:689–697 [PubMed] [Google Scholar]
- 8. Becker E. L., Showell H. J. 1972. The effect of Ca2+ and Mg2+ on the chemotactic responsiveness and spontaneous motility of rabbit polymorphonuclear leukocytes. Z. Immunitatsforsch. Exp. Klin. Immunol. 143:466–476 [PubMed] [Google Scholar]
- 9. Beggle D., Durst P. 2001. Managing potassium for crop production. The Pennsylvania State University, College of Agricultural Sciences, University Park, PA: http://cropsoil.psu.edu [Google Scholar]
- 10. Blatt M. R., Beilby M. J. 2007. Mitochondrial sequestration of BCECF after ester loading in the giant alga Chara australis. Protoplasma 232:131–136 [DOI] [PubMed] [Google Scholar]
- 11. Bolourani P., Spiegelman G. B., Weeks G. 2006. Delineation of the roles played by RasG and RasC in cAMP-dependent signal transduction during the early development of Dictyostelium discoideum. Mol. Biol. Cell 17:4543–4550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bonner J. 2009. The social amoebae: the biology of cellular slime molds. Princeton University Press, Princeton, NJ [Google Scholar]
- 13. Booth I. R., Epstein W., Giffard P. M., Rowland G. C. 1985. Roles of the trkB and trkC gene products of Escherichia coli in K+ transport. Biochimie 67:83–89 [DOI] [PubMed] [Google Scholar]
- 14. Bosgraaf L., Van Haastert P. J., Bretschneider T. 2009. Analysis of cell movement by simultaneous quantification of local membrane displacement and fluorescent intensities using Quimp2. Cell Motil. Cytoskeleton 66:156–165 [DOI] [PubMed] [Google Scholar]
- 15. Breitwieser G. E. 2008. Extracellular calcium as an integrator of tissue function. Int. J. Biochem. Cell Biol. 40:1467–1480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Brennan S. C., Conigrave A. D. 2009. Regulation of cellular signal transduction pathways by the extracellular calcium-sensing receptor. Curr. Pharm. Biotechnol. 10:270–281 [DOI] [PubMed] [Google Scholar]
- 17. Brett C. L., Donowitz M., Rao R. 2005. Evolutionary origins of eukaryotic sodium/H+ exchangers. Am. J. Physiol. Cell Physiol. 288:C223–C239 [DOI] [PubMed] [Google Scholar]
- 18. Bright G. R., Whitaker J. E., Haugland R. P., Taylor D. L. 1989. Heterogeneity of the changes in cytoplasmic pH upon serum stimulation of quiescent fibroblasts. J. Cell. Physiol. 141:410–419 [DOI] [PubMed] [Google Scholar]
- 19. Brokaw C. J. 1974. Calcium and flagellar response during the chemotaxis of bracken spermatozoids. J. Cell. Physiol. 83:151–158 [DOI] [PubMed] [Google Scholar]
- 20. Buenemann M., Levine H., Rappel W. J., Sander L. M. 2010. The role of cell contraction and adhesion in Dictyostelium motility. Biophys. J. 99:50–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Case R. M., et al. 2007. Evolution of calcium homeostasis: from birth of the first cell to an omnipresent signalling system. Cell Calcium 42:345–350 [DOI] [PubMed] [Google Scholar]
- 22. Chang A. B., Lin R., Keith Studley W., Tran C. V., Saier M. H., Jr 2004. Phylogeny as a guide to structure and function of membrane transport proteins. Mol. Membr. Biol. 21:171–181 [DOI] [PubMed] [Google Scholar]
- 23. Chen L., et al. 2007. PLA2 and PI3K/PTEN pathways act in parallel to mediate chemotaxis. Dev. Cell 12:603–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Choi C. H., Patel H., Barber D. L. 2010. Expression of actin-interacting protein 1 suppresses impaired chemotaxis of Dictyostelium cells lacking the Na+-H+ exchanger NHE1. Mol. Biol. Cell 21:3162–3170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cooke D. F., Hallett F. R., Barker C. A. 1976. Motility evaluation of bull spermatozoa by photon correlation spectroscopy. J. Mechanochem. Cell Motil. 3:219–223 [PubMed] [Google Scholar]
- 26. Day J. P., et al. 2008. Identification of two partners from the bacterial Kef exchanger family for the apical plasma membrane V-ATPase of metazoa. J. Cell Sci. 121:2612–2619 [DOI] [PubMed] [Google Scholar]
- 27. De Bary A. 1859. Die Mycetozoen. Ein Beitrag zur Kenntnis der niedersten Tiere. Z. Wiss. Zool. 10:88–175 [Google Scholar]
- 28. Dvorak M. M., et al. 2004. Physiological changes in extracellular calcium concentration directly control osteoblast function in the absence of calciotropic hormones. Proc. Natl. Acad. Sci. U. S. A. 101:5140–5145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Egelhoff T. T., Lee R. J., Spudich J. A. 1993. Dictyostelium myosin heavy chain phosphorylation sites regulate myosin filament assembly and localization in vivo. Cell 75:363–371 [DOI] [PubMed] [Google Scholar]
- 30. Eichinger L., Rivero F. 2006. Methods in molecular biology. Humana Press, Totowa, NJ [Google Scholar]
- 31. Fraser L. R. 1995. Ionic control of sperm function. Reprod. Fertil. Dev. 7:905–925 [DOI] [PubMed] [Google Scholar]
- 32. Fried M., Shapiro R. 1961. Soil-plant relationships in ion uptake. Annu. Rev. Plant Physiol. 12:91–112 [Google Scholar]
- 33. Fujisawa M., Ito M., Krulwich T. A. 2007. Three two-component transporters with channel-like properties have monovalent cation/H+ antiport activity. Proc. Natl. Acad. Sci. U. S. A. 104:13289–13294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Fuller D., et al. 2010. External and internal constraints on eukaryotic chemotaxis. Proc. Natl. Acad. Sci. U. S. A. 107:9656–9659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gilbert O. M., Queller D. C., Strassmann J. E. 2009. Discovery of a large clonal patch of a social amoeba: implications for social evolution. Mol. Ecol. 18:1273–1281 [DOI] [PubMed] [Google Scholar]
- 36. Goldberg E., Broecker W., Gross M., Turekian K. 1971. Marine chemistry, p. 137–146 In Seymour A. H., et al., Radioactivity in the marine environment. National Academy of Sciences, Washington, DC [Google Scholar]
- 37. Guan X. M., Kobilka T. S., Kobilka B. K. 1992. Enhancement of membrane insertion and function in a type IIIb membrane protein following introduction of a cleavable signal peptide. J. Biol. Chem. 267:21995–21998 [PubMed] [Google Scholar]
- 38. Heid P. J., Geiger J., Wessels D., Voss E., Soll D. R. 2005. Computer-assisted analysis of filopod formation and the role of myosin II heavy chain phosphorylation in Dictyostelium. J. Cell Sci. 118:2225–2237 [DOI] [PubMed] [Google Scholar]
- 39. Heid P. J., et al. 2004. The role of myosin heavy chain phosphorylation in Dictyostelium motility, chemotaxis and F-actin localization. J. Cell Sci. 117:4819–4835 [DOI] [PubMed] [Google Scholar]
- 40. Hulo N., et al. 2008. The 20 years of PROSITE. Nucleic Acids Res. 36:D245–D249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Janetopoulos C., Firtel R. A. 2008. Directional sensing during chemotaxis. FEBS Lett. 582:2075–2085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Jin T., et al. 2009. How human leukocytes track down and destroy pathogens: lessons learned from the model organism Dictyostelium discoideum. Immunol. Res. 43:118–127 [DOI] [PubMed] [Google Scholar]
- 43. Kay R. R., Langridge P., Traynor D., Hoeller O. 2008. Changing directions in the study of chemotaxis. Nat. Rev. Mol. Cell Biol. 9:455–463 [DOI] [PubMed] [Google Scholar]
- 44. Kilmer V., Younts S., Brady N. 1968. The role of potassium in agriculture. American Society of Agronomy, Madison, WI [Google Scholar]
- 45. King J. S., Insall R. H. 2009. Chemotaxis: finding the way forward with Dictyostelium. Trends Cell Biol. 19:523–530 [DOI] [PubMed] [Google Scholar]
- 46. King J. S., et al. 2009. The mood stabiliser lithium suppresses PIP3 signalling in Dictyostelium and human cells. Dis. Model Mech. 2:306–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kubalski A., Martinac B. 2005. Bacterial ion channels and their eukaryotic homologs. ASM Press, Washington, DC [Google Scholar]
- 48. Laevsky G., Knecht D. A. 2003. Cross-linking of actin filaments by myosin II is a major contributor to cortical integrity and cell motility in restrictive environments. J. Cell Sci. 116:3761–3770 [DOI] [PubMed] [Google Scholar]
- 49. Lawrence G., David M., Bailey S., Shortle W. 1997. Assessment of soil calcium status in red spruce forests in northeastern United States. Biogeochmistry 38:19–39 [Google Scholar]
- 50. Ludtmann M. H., Boeckeler K., Williams R. S. 2011. Molecular pharmacology in a simple model system: implicating MAP kinase and phosphoinositide signalling in bipolar disorder. Semin. Cell Dev. Biol. 22:105–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lusche D. F., Wessels D., Soll D. R. 2009. The effects of extracellular calcium on motility, pseudopod and uropod formation, chemotaxis, and the cortical localization of myosin II in Dictyostelium discoideum. Cell Motil. Cytoskeleton 66:567–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lynn W. S., Mukherjee C. 1978. Motility of human polymorphonuclear leukocytes. Roles of hydroxy fatty acids, other lipids, and cations. Am. J. Pathol. 91:581–594 [PMC free article] [PubMed] [Google Scholar]
- 53. Marchler-Bauer A., et al. 2009. CDD: specific functional annotation with the Conserved Domain Database. Nucleic Acids Res. 37:D205–D210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Maron M. 1982. Numerical analysis: a practical approach. Macmillan, New York, NY [Google Scholar]
- 55. McCann C. P., Kriebel P. W., Parent C. A., Losert W. 2010. Cell speed, persistence and information transmission during signal relay and collective migration. J. Cell Sci. 123:1724–1731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. McLaughlin S. B., Wimmer R. 1999. Tansley review no. 104. Calcium physiology and terrestrial ecosystem processes. Cambridge University Press, Cambridge, United Kingdom [Google Scholar]
- 57. Moores S. L., Sabry J. H., Spudich J. A. 1996. Myosin dynamics in live Dictyostelium cells. Proc. Natl. Acad. Sci. U. S. A. 93:443–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Morris V. L., Chan B. M. 2007. Interaction of epidermal growth factor, Ca2+, and matrix metalloproteinase-9 in primary keratinocyte migration. Wound Repair Regen. 15:907–915 [DOI] [PubMed] [Google Scholar]
- 59. Mozingo N. M., Chandler D. E. 1990. The fluorescent probe BCECF has a heterogeneous distribution in sea urchin eggs. Cell Biol. Int. Rep. 14:689–699 [DOI] [PubMed] [Google Scholar]
- 60. Muller E. G., Muller I. 1980. Sponge cell aggregation. Mol. Cell. Biochem. 29:131–143 [DOI] [PubMed] [Google Scholar]
- 61. Nellen W., Silan C., Firtel R. A. 1984. DNA-mediated transformation in Dictyostelium discoideum: regulated expression of an actin gene fusion. Mol. Cell. Biol. 4:2890–2898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Nelson L., McGrady A. V. 1981. Effects of ouabain on spermatozoan function: a review. Arch. Androl. 7:169–176 [DOI] [PubMed] [Google Scholar]
- 63. Nemeth E. 1995. Ca2+ receptor-dependent regulation of cellular functions. News Physiol. Sci. 10:1–5 [Google Scholar]
- 64. Patel H., Barber D. L. 2005. A developmentally regulated Na-H exchanger in Dictyostelium discoideum is necessary for cell polarity during chemotaxis. J. Cell Biol. 169:321–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Pitta T. P., Sherwood E. E., Kobel A. M., Berg H. C. 1997. Calcium is required for swimming by the nonflagellated cyanobacterium Synechococcus strain WH8113. J. Bacteriol. 179:2524–2528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Qian P., Schoenau J. J., King T., Japp M. 2005. Effect of repeated manure application on potassium, calcium and magnesium in soil and cereal crops in Sakatchewan. Can. J. Soil Sci. 85:397–403 [Google Scholar]
- 67. Raper K. B. 1935. Dictyostelium discoideum, a new species of slime mold from decaying forest leaves. J. Agric. Res. 50:135–147 [Google Scholar]
- 68. Rhoads D. B., Waters F. B., Epstein W. 1976. Cation transport in Escherichia coli. VIII. Potassium transport mutants. J. Gen. Physiol. 67:325–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Roberts R. L., Mounessa N. L., Gallin J. I. 1984. Increasing extracellular potassium causes calcium-dependent shape change and facilitates concanavalin A capping in human neutrophils. J. Immunol. 132:2000–2006 [PubMed] [Google Scholar]
- 70. Robertson H. M., Thomas J. H. 2006. The putative chemoreceptor families of C. elegans. WormBook. doi/10.1895/wormbook.1.66.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Sathe S., et al. 2010. Genetic heterogeneity in wild isolates of cellular slime mold social groups. Microb. Ecol. 60:137–148 [DOI] [PubMed] [Google Scholar]
- 72. Scott D. A., Docampo R., Benchimol M. 1998. Analysis of the uptake of the fluorescent marker 2′,7′-bis-(2-carboxyethyl)-5(and-6)-carboxyfluorescein (BCECF) by hydrogenosomes in Trichomonas vaginalis. Eur. J. Cell Biol. 76:139–145 [DOI] [PubMed] [Google Scholar]
- 73. Shelden E., Knecht D. A. 1996. Dictyostelium cell shape generation requires myosin II. Cell Motil. Cytoskeleton 35:59–67 [DOI] [PubMed] [Google Scholar]
- 74. Slayman C. W., Tatum E. L. 1964. Potassium transport in Neurospora. I. Intracellular sodium and potassium concentrations, and cation requirements for growth. Biochim. Biophys. Acta 88:578–592 [PubMed] [Google Scholar]
- 75. Soll D., Voss E. 1998. Two and three dimensional computer systems for analyzing how cells crawl, p. 25–52 In Soll D., Wessels D. (ed.), Motion analysis of living cells. John Wiley, Inc., Hoboken, NJ [Google Scholar]
- 76. Soll D. R. 1979. Timers in developing systems. Science 203:841–849 [DOI] [PubMed] [Google Scholar]
- 77. Soll D. R. 1995. The use of computers in understanding how animal cells crawl. Int. Rev. Cytol. 163:43–104 [PubMed] [Google Scholar]
- 78. Soll D. R., Wessels D., Kuhl S., Lusche D. F. 2009. How a cell crawls and the role of cortical myosin II. Eukaryot. Cell 8:1381–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Sparks D., Huang P. 1985. Physical chemistry of soil potassium. In Munson R. (ed.), Potassium in agriculture. American Society of Agronomy, Madison, WI [Google Scholar]
- 80. Suki W. N. 1976. Disposition and regulation of body potassium: an overview. Am. J. Med. Sci. 272:31–41 [DOI] [PubMed] [Google Scholar]
- 81. Sussman M. 1987. Cultivation and synchronous morphogenesis of Dictyostelium under controlled experimental conditions. Methods Cell Biol. 28:9–29 [DOI] [PubMed] [Google Scholar]
- 82. Swaney K. F., Huang C. H., Devreotes P. N. 2010. Eukaryotic chemotaxis: a network of signaling pathways controls motility, directional sensing, and polarity. Annu. Rev. Biophys. 39:265–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Teany L. E. 2004. Phosphorus losses from simulated dairy management intensive grazing forage system. Virginia Polytechnic Institute and State University, Blacksburg, VA [Google Scholar]
- 84. Telo W., Pieragnoli E. 1955. Study of tissue metabolism. I. Means of investigating muscular potassium; critical review and personal contribution. Arch. Sci. Med. 99:260–273 (In Italian.) [PubMed] [Google Scholar]
- 85. Thomas J. H., Robertson H. M. 2008. The Caenorhabditis chemoreceptor gene families. BMC Biol. 6:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Urushihara H. 2009. The cellular slime mold: eukaryotic model microorganism. Exp. Anim. 58:97–104 [DOI] [PubMed] [Google Scholar]
- 87. Van Haastert P. J. 2010. A model for a correlated random walk based on the ordered extension of pseudopodia. PLoS Comput. Biol. 6:e1000874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Varnum B., Edwards K. B., Soll D. R. 1986. The developmental regulation of single-cell motility in Dictyostelium discoideum. Dev. Biol. 113:218–227 [DOI] [PubMed] [Google Scholar]
- 89. Varnum B., Soll D. R. 1984. Effects of cAMP on single cell motility in Dictyostelium. J. Cell Biol. 99:1151–1155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Wakabayashi S., Pang T., Su X., Shigekawa M. 2000. A novel topology model of the human Na(+)/H(+) exchanger isoform 1. J. Biol. Chem. 275:7942–7949 [DOI] [PubMed] [Google Scholar]
- 91. Wang W. H., Giebisch G. 2009. Regulation of potassium (K) handling in the renal collecting duct. Pflugers Arch. 458:157–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Weijer C. J. 2009. Collective cell migration in development. J. Cell Sci. 122:3215–3223 [DOI] [PubMed] [Google Scholar]
- 93. Wessels D., et al. 2004. RasC plays a role in transduction of temporal gradient information in the cyclic-AMP wave of Dictyostelium discoideum. Eukaryot. Cell 3:646–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Wessels D., Lusche D. F., Kuhl S., Heid P., Soll D. R. 2007. PTEN plays a role in the suppression of lateral pseudopod formation during Dictyostelium motility and chemotaxis. J. Cell Sci. 120:2517–2531 [DOI] [PubMed] [Google Scholar]
- 95. Wessels D., et al. 1988. Cell motility and chemotaxis in Dictyostelium amoebae lacking myosin heavy chain. Dev. Biol. 128:164–177 [DOI] [PubMed] [Google Scholar]
- 96. Wessels D. J., Kuhl S., Soll D. R. 2009. Light microscopy to image and quantify cell movement. Methods Mol. Biol. 571:455–471 [DOI] [PubMed] [Google Scholar]
- 97. Wickner W., Schekman R. 2005. Protein translocation across biological membranes. Science 310:1452–1456 [DOI] [PubMed] [Google Scholar]
- 98. Williams R. S., et al. 2006. Towards a molecular understanding of human diseases using Dictyostelium discoideum. Trends Mol. Med. 12:415–424 [DOI] [PubMed] [Google Scholar]
- 99. Young L. G., Nelson L. 1974. Calcium ions and control of the motility of sea urchin spermatozoa. J. Reprod. Fertil. 41:371–378 [DOI] [PubMed] [Google Scholar]
- 100. Yumura S., Furuya K., Takeuchi I. 1996. Intracellular free calcium responses during chemotaxis of Dictyostelium cells. J. Cell Sci. 109(Pt 11):2673–2678 [DOI] [PubMed] [Google Scholar]
- 101. Zigmond S. H. 1977. Ability of polymorphonuclear leukocytes to orient in gradients of chemotactic factors. J. Cell Biol. 75:606–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.