Abstract
DET1 (De-etiolated 1) is a chromatin binding protein involved in developmental regulation in both plants and animals. DET1 is largely restricted to multicellular eukaryotes, and here we report the characterization of a DET1 homolog from the social amoeba Dictyostelium discoideum. As in other species, Dictyostelium DET1 is nuclear localized. In contrast to other species, where it is an essential protein, loss of DET1 is nonlethal in Dictyostelium, although viability is significantly reduced. The phenotype of the det1− mutant is highly pleiotropic and results in a large degree of heterogeneity in developmental parameters. Loss of DET1 results in delayed and abnormal development with enlarged aggregation territories. Mutant slugs displayed cell type patterning with a bias toward the prestalk pathway. A number of DET1-interacting proteins are conserved in Dictyostelium, and the apparently conserved role of DET1 in regulatory pathways involving the bZIP transcription factors DimB, c-Jun, and HY5 suggests a highly conserved mechanism regulating development in multicellular eukaryotes. While the mechanism by which DET1 functions is unclear, it appears that it has a key role in regulation of developmental plasticity and integration of information on environmental conditions into the developmental program of an organism.
INTRODUCTION
The social amoeba Dictyostelium discoideum provides a simple model for studying developmental processes. When their food supply is exhausted, Dictyostelium amoebae exit their unicellular proliferative phase and display facultative multicellularity. A few amoebae in the population start emitting cyclic AMP (cAMP) pulses, which act as a chemoattractant for others (7). These will relay the signal, chemotax toward the cAMP source, and aggregate into mounds of approximately 100,000 cells. The amoebae in the mound enter a developmental pathway, differentiating into a number of different cell types to form a multicellular slug. The slug consists of prestalk cells at the anterior fifth and mainly prespore cells, anterior-like cells (ALC), and circulating sentinel cells (10) in the posterior four-fifths, with a small zone of prestalk-like rear guard cells at the posterior extremity. The slugs migrate toward light and away from heat, and the prestalk cells differentiate further into different subtypes (pstO, pstA, pstB, pstAB, and the tip organizer region) (47). Eventually, after a period of migration, the slugs culminate and form a stalk on which a spore head is perched, thus favoring spore dispersal (6, 40).
Aggregation is regulated by cAMP, which induces differentiation of prespore cells. Prespore cells in turn produce a chlorinated polyketide, DIF-1, that induces prestalk cell differentiation (9). While the entire cascade downstream of DIF-1 is not known, DIF-1 ultimately induces the nuclear accumulation of the bZIP transcription factors DimA and DimB, which bind the promoter of the prestalk ecmA gene (51) and regulate its expression. In addition, Myb, MADS-box, Cbl, STAT, and the amoebozoan-specific CudA transcription factors are involved in the regulation of Dictyostelium development (18, 21, 30, 33, 42, 48). Microarray analysis has shown that roughly a third of the genome displays a more than 2-fold change in expression during early development (26).
The Dictyostelium genome encodes several homologs of proteins involved in developmental regulation in plants and animals, including components of a Cul4 (cullin 4)-based ubiquitin ligase complex. Cullins are a small family of evolutionarily conserved proteins that together with substrate adaptors catalyze the conjugation of ubiquitin chains to their targets, resulting in their degradation by the 26S proteasome (25). Cullins have been shown to have critical roles in transducing hormone signals, developmental regulation, environmental signaling, and cell cycle regulation (25, 27). In agreement with this, the loss of either culA or culB in Dictyostelium results in developmental defects (34, 44). Cul4 (CulD in Dictyostelium) has been implicated in the ubiquitination of various chromatin components, including the transcription factor c-Jun, the cell cycle regulator E2F, and the DNA replication licensing factor CTD1 (46), and is involved in developmental regulation in plants, including light response (5, 49, 50). It is also part of the DNA damage repair complex containing DDB1 (RepE in Dictyostelium), which can catalyze the monoubiquitination of histone H2A (12, 29) and/or histones H3 and H4 (45). In Schizosaccharomyces pombe, Cul4 also forms part of a complex with the DDB1 homolog Rik1, which acts to target the H3K9 methyltransferase Clr4 to pericentromeric heterochromatin (24, 28). In plants and animals, DDB1 and Cul4 occur together in a complex with the DET1 protein (39). DET1 was identified in mutant screens in Drosophila (41) and light-signaling mutants in Arabidopsis (11) and tomato (35). DET1 binds to chromatin via the N-terminal tail of histone H2B and acts as a transcriptional repressor at its target genes (3, 4). While DDB1 and Cul4 are highly conserved and appear to be ubiquitous in eukaryotes, DET1 has a much more limited distribution and occurs almost exclusively in multicellular plants and animals. Interestingly a homolog of DET1 appears to be present in the social amoebae Dictyostelium discoideum and Dictyostelium purpureum but not their unicellular relative Entamoeba histolytica. Given its role in developmental regulation in higher eukaryotes and its presence in Dictyostelium, DET1 is a promising candidate for a regulator of development in this species.
MATERIALS AND METHODS
Strains and growth.
For cloning and plasmid isolation, the Escherichia coli strain DH5α was cultured at 37°C in Luria-Bertani (LB) broth or on LB agar plates supplemented with 50 μg ml−1 ampicillin.
The Dictyostelium discoideum strain Ax2-214 (axeA2 axeB2 axeC2) was cultured in petri dishes or shaking culture at 20°C in HL5 medium (ForMedium, Hunstaton, United Kingdom) supplemented with 100 μg ml−1 ampicillin, 100 μg ml−1 amphotericin B, and the appropriate selective agent (10 μg ml−1 Geneticin and/or 10 μg ml−1 blasticidin). Alternatively, Dictyostelium cells were grown on bacterial lawns of Klebsiella aerogenes on SM agar plates.
Cloning.
The N- and C-terminal halves of the DetA (DDB_G0277075) gene were cloned separately with the primers MJD29 (5′-GTCGACAATGATGGATCCATCCACTATATC-3′) and MJD32 (5′-CATGTAAAAGCTTTTTCTCGTGAG-3′) for the N-terminal half and MJD31 (5′-CTCACGAGAAAAAGCTTTTACATG-3′) and MJD30 (5′-CTCGAGTTATGATACTGTATTATTATTATTGTTGTTGTTATTA-3′) for the C-terminal half. After sequencing to confirm the absence of mutations, the N-terminal half was excised with the SalI and HindIII restriction sites, and the C-terminal half was excised with the HindIII and XhoI restriction sites. These fragments were cloned into the green fluorescent protein (GFP)-expressing vector pDneo2a-GFP digested with SalI/XhoI by three-fragment ligation. The C-terminal conserved region was PCR amplified with the primers MJD184 (5′-AGTCGACAACGATGAATTATCAACAAATGTTTGGACAAAC-3′) and MJD30 (5′-CTCGAGTTATGATACTGTATTATTATTATTGTTGTTGTTATTA-3′) and cloned into the vector pDneo2a-3×HA, containing a 3-hemagglutinin (3×HA) tag, by using the SalI and XhoI restriction sites. To create an expression construct for the N-terminal half of DetA, the N-terminal domain described above (amplified with the primers MJD29 and MJD32) was digested with HindIII, blunted with T4 DNA polymerase, and then excised from the cloning vector by SalI digestion. The resulting fragment was cloned into the vector pDneo2a-GFP, which had been XhoI digested, blunted with T4 DNA polymerase, and then digested with SalI.
RepE (DDB_G0286013) and CulD (DDB_G0292794) were amplified from genomic DNA with a mixture of Pfu/Taq (1:30) polymerases by using the primers: MJD169 (5′-AGTCGACAATGTATAATTTTGTATCAACTGTTCAAAAACC-3′)and MJD170 (5′-TCTCGAGTTAACGAATATATTGCATTAAAGATTCAATTCTTC-3′) for RepE and MJD171 (5′-AGTCGACAATGAATTTTAATAATAACAATAACAATAATAATAATAATAATAA-3′) and MJD172 (5′-TCTCGAGTTAAGCCATGTAATTATATATCATTGCATTTTC-3′) for CulD. After sequencing to confirm the absence of mutations, they were cloned into the vector pDneo2a-3×HA or pDneo2a-GFP (13) by using SalI and XhoI restriction sites introduced during PCR amplification. All accession numbers refer to DictyBase (20).
Disruption of detA.
The N-terminal domain of Dictyostelium detA described above (amplified with the primers MJD29 and MJD32) was cloned into vector pGEM-T Easy (Fermentas). pGEM-T-DetAn was double digested with PstI/SmaI, blunted with T4 polymerase, and then religated with T4 ligase in order to remove the NheI site within the vector backbone. The resulting vector, pGEM-T-DetAn (-NheI), was then digested with NheI (which is present within the detA gene), blunted with T4 polymerase, and treated with alkaline phosphatase. A SmaI-digested fragment from the vector pLPBLP (19) containing a blasticidin cassette flanked by LoxP sites was ligated in, resulting in the vector pGEM-T-detA-LPBLP. The vector was linearized by digestion with EcoRI and used to transform Ax2 cells. Blasticidin-resistant cells were subcloned. Putative disrupted clones were screened with the primers Act15R (5′-CCAACCCAAGTTTTTTTAAACC-3′) and MJD36 (5′-GCAAATAAAGGATTCATTGGGGTG-3′). The disruption of the detA gene was confirmed by Southern blotting, as described in reference 38.
Western blotting.
A total of 107 cells were pelleted and resuspended in 100 μl 1× Laemmli buffer and boiled for 5 min, and 10 μl of the soluble supernatant was run on an 12% SDS–PAGE gel. Proteins were transferred to polyvinylidene difluoride (PVDF) membrane by semidry transfer. Homogeneous loading and transfer of proteins were verified by staining with Ponceau-S dye (0.1% in 5% acetic acid). Excess dye was removed by washing in 1% acetic acid. The membrane was then blocked with 5% bovine serum albumin (BSA) and incubated with the primary antibody overnight at 4°C. After being washed, the membrane was incubated with the appropriate secondary antibody conjugated to alkaline phosphatase (Dianova, Hamburg, Germany) at a dilution of 1:10,000. Staining was done with 0.2 mg ml−1 BCIP (5-bromo-4-chloro-indolylphosphate).
Identification of homologs and alignments.
Dictyostelium homologs of characterized genes were identified in the Dictyostelium genome (16) by using BLAST search (2); a reciprocal BLAST search against the NCBI databank (nonredundant protein sequences) was performed to confirm identity (1).
Immunofluorescence.
Cells were grown overnight on coverslips to approximately 80% confluence and fixed in 4% (wt/vol) paraformaldehyde (PFA) dissolved in 20 mM phosphate buffer (pH 6.7) for 10 min at 22°C. The PFA solution was removed, and cells were permeabilized for 5 min in 0.2% (vol/vol) Triton-X 100 in 20 mM phosphate buffer (pH 6.7). Cells were blocked with 2% (wt/vol) BSA in phosphate-buffered saline (PBS; pH 7.6) for 30 min at room temperature. The coverslips were incubated overnight at 4°C with the primary antibody diluted in 1% (wt/vol) BSA in PBS (pH 7.6). The coverslips were washed 3 times for 5 min each in PBS and then incubated with the secondary antibody diluted in 1% (wt/vol) BSA in PBS (pH 7.6) for 1 h at room temperature. The coverslips were washed 3 times for 5 min each in PBS and then mounted on a slide with a drop of mounting medium: 250 ng ml−1 4′,6-diamidino-2-phenylindole (DAPI) in a mixture of 90% (vol/vol) glycerol, 20 mM Tris-HCl, and 1 μg ml−1 1,4-diazabicyclo[2.2.2]octane (pH 8.3). Cells were examined by fluorescence microscopy.
Coimmunoprecipitation.
A total of 5.5 × 108 cells were harvested, washed with phosphate buffer, and cross-linked in 10 ml phosphate buffer with 5 mM dimethyl 3,3′-dithio-propionimidate dihidrochloride for 30 min. Cells were washed and resuspended in 1 ml lysis buffer (10 mM Tris-Cl [pH 7.5], 150 mM NaCl, 0.5 mM EDTA, 0.5% NP-40, 25 mM MgCl2, 1 tablet of Roche proteinase inhibitor cocktail mini). Sonication was done three times for 15 s each with a Dr. Hielscher UP200S sonifier using the S1 tip and 50% amplitude/50% cycle. After centrifugation (10,000 × g, 15 min), the supernatant was incubated with 20 μl of GFP-Trap (Chromotek, Martinsried, Germany) Sepharose beads for 90 min on a rotating wheel at 4°C. The supernatant was removed, and the beads were washed twice with a mixture of 10 mM Tris-Cl (pH 7.5), 150 mM NaCl, 0.5 mM EDTA, 26 mM MgCl2, and Roche proteinase inhibitor cocktail. Bound protein was boiled off the beads in 25 μl Laemmli buffer. Aliquots were taken from the intermediate steps and compared to bound protein by SDS-PAGE and subsequent Western blotting.
Microscopy.
Images were acquired on a Leica DMIRB inverted microscope equipped with a DC350 camera and IM50 acquisition software (Leica Microsystems; Wetzlar, Germany). Images were prepared for presentation with ImageJ v1.42n-v1.43b (http://rsbweb.nih.gov/ij/).
Developmental assay.
Ax2 or strains of interest were grown in shaking culture to a density of 1 × 106 cells ml−1. A total of 1 × 107 cells were centrifuged for 2 min at 200 × g at 4°C. Cells were washed twice in phosphate buffer and then adjusted to a cell density of 4 × 106 cells ml−1. Fifty microliters of each strain was spotted in triplicate on phosphate agar plates, so that each spot had an area of approximately 1 cm2. Plates were left open in a sterile hood for 30 min to allow excess buffer to evaporate. Plates were examined at regular intervals to examine development.
GFP reporter constructs.
Ax2 or the mutant of interest was transformed with [ecmAO]:GFP or [cotB]:GFP reporter constructs (44) and subcloned. Cells were grown in shaking culture to a density of 1 × 106 cells ml−1. A total of 1 × 107 cells were centrifuged for 2 min at 200 × g at 4°C. Cells were washed 3 times in ice-cold water and then resuspended in a minimal amount (approximately 100 μl) of water and deposited in 3 or 4 small drops on freshly prepared 1% phosphate agar plates. The plates were placed in a dark humid chamber to develop for 18 to 20 h at 20°C. Slugs were examined with a fluorescence microscope, taking care to minimize the exposure of the plates to light beforehand.
LacZ reporter constructs.
Ax2 or the mutant of interest was transformed with a [ecmB]:LacZ, [ecmA]:LacZ, [ecmO]:LacZ, or [ecmAO]:LacZ reporter construct (15), as described above, except that selection was carried out with 50 μg ml−1 Geneticin. β-Galactosidase staining was performed as described in reference 17.
Anti-EB4 staining of developing Dictyostelium.
Staining of developing Dictyostelium with the anti-EB4 antibody was carried out as described for LacZ staining, up to the washing steps with Z buffer. After being washed twice with Z buffer, the cells were incubated with 5% (wt/vol) nonfat milk powder in PBS for 30 min at room temperature. The blocking solution was removed, and the filters were incubated with anti-EB4 antiserum (1:1,000) in 3% (wt/vol) nonfat milk powder in PBS overnight at 4°C. Filters were gently washed with 3× 800 μl PBS and then incubated with alkaline phosphatase-conjugated goat anti-rabbit (1:10,000) in 3% (wt/vol) nonfat milk powder in PBS for 60 min at room temperature. Filters were gently washed with 3× 800 μl PBS and then incubated with development buffer (0.2 mg ml−1 BICP, 100 mM Tris-HCl, 100 mM NaCl, 5 mM MgCl2, 0.05% Tween 20 [pH 9.5]) until blue staining was apparent, at which point the filters were washed twice with PBS.
RESULTS
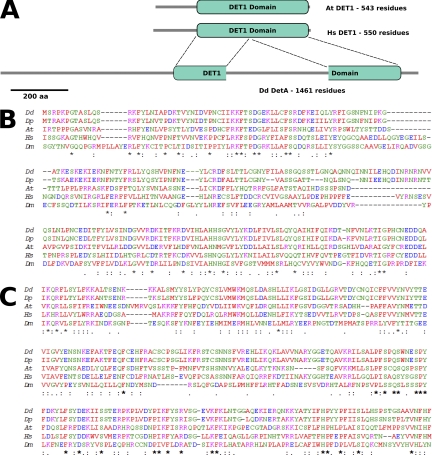
DET1 is a nuclear localized protein of approximately 60 kDa in plants, vertebrates, and Drosophila and has no homology with any other known protein (37). Based on sequence conservation between different species, a DET1 domain of approximately 400 amino acid residues has been defined in the PFAM database (PF09737). A BLAST search against the predicted Dictyostelium proteome using human DET1 as a query sequence revealed one protein (DDB_G0277075) with homology to human DET1 (Fig. 1A). This protein, which we have named DetA, is significantly longer than the other DET1 homologs (1,462 amino acids compared to approximately 550 amino acids for other characterized DET1 proteins). A February 2007 BLAST search of the Swiss-Prot protein database (43) with DetA as the query sequence gave hits against human, mouse (1e−49), and Arabidopsis (1e−45) DET1 (Fig. 1A). A BLAST search of the closely related Dictyostelium purpureum genome with DetA as the query sequence revealed a homolog (1e−130), DPU0059753, while S. pombe and Saccharomyces cerevisiae, as well as the unicellular amoeba Entamoeba histolytica, lack a DET1 homolog. Dictyostelium discoideum detA is located toward the distal end of chromosome 2, has a short intron of 90 bp, and encodes a protein of 169 kDa. Expressed sequence tags (ESTs) (36) span part of the coding region and support the presence of the intron. DetA consists of an asparagine-rich 500-amino-acid region containing homopolymer stretches followed by a region of approximately 200 amino acids, homologous to the N-terminal domain of characterized DET1 proteins (Fig. 1B) and a further asparagine- and glutamine-rich 400-amino-acid stretch of homopolymer sequences followed by another 250-amino-acid region homologous to the C-terminal domain of DET1 (Fig. 1C). The C-terminal domain (CTD) is conserved to a slightly higher degree than the N-terminal domain (NTD) among the different species examined. Dictyostelium appears to be one of the few species outside the metazoan and plant lineages in which a DET1 homolog is present (data not shown; Fig. SA1a in the supplemental material). With the exception of Dictyostelium, DET1 homologs in all other species are small, with a continuous DET1 domain and no additional domains on the protein (data not shown). Though asparagine- and glutamine-rich insertions are frequent in Dictyostelium genes, the presence of a separated NTD and CTD in DetA may suggest that DET1 consists of two separate domains with distinct functions.
Fig. 1.
Putative Dictyostelium DET1 homolog. (A) Schematic presentation of domain organization of DDB_G0277075 (DetA) aligned with DET1 from Homo sapiens and Arabidopsis. (B) Alignment of the N-terminal domain of Dictyostelium DetA (starting at amino acid [aa] 488) with known or putative DET1 homologs from other species. (C) Alignment of the C-terminal domain of DetA (starting at amino acid 1167) with known or putative DET1 homologs from other species. Dd, Dictyostelium discoideum; Dp, Dictyostelium purpureum; At, Arabidopsis thaliana; Hs, Homo sapiens; Dm, Drosophila melanogaster.
DetA localization.
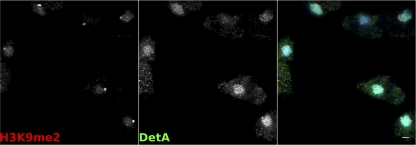
The detA gene was cloned from genomic DNA, inserted into the C-terminal cloning site of pDneo2a-6×MYC or pDneo2a-GFP (13), and used to transform Dictyostelium. Western blotting confirmed that the six-MYC-tagged DetA construct (6×MYC-DetA) was expressed and gave a band of the expected size (180 kDa; see Fig. SA1b in the supplemental material). GFP-tagged DetA localized to the nucleus in both live and fixed cells (see Fig. SA1c and d in the supplemental material), as has been previously described for DET1 in plants and Drosophila (37). Double immunostaining of 6×MYC-DetA-expressing cells with antibodies against H3K9me2 or H3K9me3 showed that the overexpressed fusion protein was largely, but not exclusively, nuclear localized and excluded from the pericentromeric heterochromatin region when examined by wide-field or confocal microscopy (Fig. 2; see Fig. SA1e in the supplemental material).
Fig. 2.
Localization of DetA. Immunofluorescence on fixed cells expressing 6×MYC-DetA with an anti-MYC antibody (green) and an antibody against the heterochromatin marker H3K9me2 (red) counterstained with DAPI. Scale bar, 2 μm.
Knockout of detA.
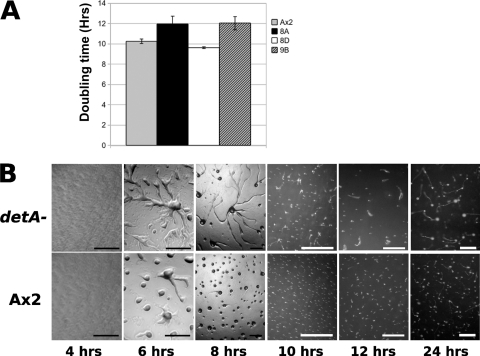
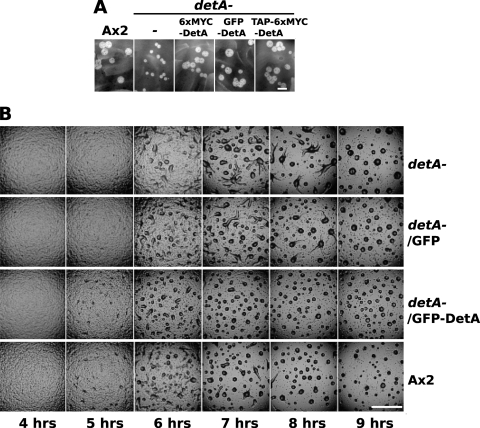
A knockout construct was made by introducing a blasticidin resistance cassette into the 5′ half of the coding sequence of detA (see Fig. SA2a in the supplemental material). This construct was then used to transform Ax2 (wild-type) cells. Potential knockout lines were identified by PCR screening. Southern blotting confirmed that the detA locus was disrupted in three of the putative knockout lines. However, it was also evident that in one of the lines (8D), in addition to the disrupted locus, a second wild-type copy of the gene was present, suggesting that a duplication event had occurred (see Fig. SA2b in the supplemental material). Line 8D was used in further experiments as an internal control since the disruption was compensated for by the gene duplication. The disrupted lines 8A and 9B, but not the duplicated line 8D, displayed an increased doubling time (12 h compared to 10 h for the wild type) when grown in liquid medium, suggesting a mild growth or cell cycle defect (Fig. 3A). When developed on phosphate agar plates, clones 8A and 9B displayed a slight delay in the onset of development (Fig. 3B). In addition to the developmental delay, clones 8A and 9B displayed enlarged aggregation territories, and streaming could still be observed after 10 h. In these experiments, the duplicated line 8D displayed a wild-type phenotype (data not shown). Compared to the wild type, clones 8A and 9B formed fewer and larger slugs. The lower number of slugs appeared to be partly due to the larger aggregation territories and partly because up to 50% of the mounds are arrested at this stage for a considerable period of time without forming tips. The slugs also tended to be thinner than the wild type, a phenotype reminiscent of the dimB− mutant (51). The enlarged aggregation territories in the detA− mutants were only observed when the cells were plated at low densities (2 × 105 cells cm−2) but were not apparent when cells were plated at higher densities (above approximately 5 × 105 cells cm−2) (data not shown). When plated at high density, mutant slugs were often smaller than the wild type (see, for example, Fig. 5). Line 8A and its derivatives were used in all subsequent experiments. To confirm that the observed phenotypes were linked to the disruption of the detA locus, we attempted to complement the detA− knockout. When grown on bacterial lawns, the detA− mutant displayed a growth defect which could be rescued by supertransforming the knockout with a construct expressing 6×MYC-DetA, GFP-DetA, or TAP-6×MYC-DetA (Fig. 4A). The developmental assay was repeated using the wild-type strain (Ax2), the detA− mutant, and the detA− mutant complemented with either GFP alone (empty vector control) or GFP-DetA. In Ax2 and the detA−/GFP-DetA strain, the first mounds were visible at 5 h of development, but not in the detA− strain nor in the detA−/GFP control strain, where the onset of aggregation was delayed approximately 30 min with respect to the wild type (Fig. 4B). In wild-type cells, aggregation was largely complete by 8 h, but it continued until 9 h in the detA− and detA−/GFP strains. In the detA−/GFP-DetA strain, aggregation was even faster than in the wild type and was complete after 7 h. As previously observed, the detA− mutant formed fewer and larger aggregates, many of which arrested at this stage, resulting in fewer fruiting bodies. Except for a slightly accelerated development, the detA−/GFP-DetA strain displayed a wild-type phenotype, confirming that the loss of the DetA protein was responsible for the phenotype observed in the detA− mutant and that epitope-tagged DetA was functional in rescuing the mutant.
Fig. 3.
Phenotype of the detA− mutant. (A) Doubling time of wild-type (Ax2) and putative disruption strains in HL5 shaking culture. Clone 8D contained a gene duplication. (B) Development of the wild type and a disruption strain on phosphate-buffered agar at 22°C. The time after onset of starvation is indicated in hours. Black scale bar, 200 μm; white scale bar, 500 μm.
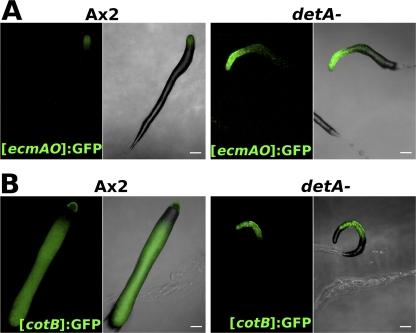
Fig. 5.
Developmentally regulated gene expression. (A) Fluorescence microscopy of wild-type (Ax2) or detA− slugs transformed with the prestalk reporter [ecmAO]:GFP 17 h after the onset of starvation; (B) fluorescence microscopy of wild-type or detA− slugs transformed with the prespore reporter [cotB]:GFP 17 h after the onset of starvation. Scale bar, 100 μm.
Fig. 4.
Complementation of the detA− mutant. (A) Growth (plaque size) of wild-type strain Ax2, the detA− mutant strain, and the detA− mutant strain complemented with different DetA expression constructs on bacterial lawns. The pictures were taken after 72 h of growth at 22°C. (B) Development of the Ax2 wild-type strain, the detA− mutant, and the detA− mutant transformed with either GFP-DetA or GFP alone on phosphate-buffered agar at 22°C. Time after onset of starvation is indicated below. Black scale bar, 5 μm; white scale bar, 500 μm.
Role of DetA in cell-type specification.
Given the role of DET1 in developmental regulation in other organisms and the developmental defects observed in the detA− mutant, the role of DetA in cell-type specification was investigated. Wild-type (Ax2) and detA− (clone 8A) cells were transformed with the prespore [cotB]:GFP or prestalk [ecmAO]:GFP reporter construct. Lines expressing the reporter constructs were developed at high density on phosphate agar for 17 h in the dark and observed by fluorescence microscopy. In Ax2, the [cotB]:GFP reporter labeled the prespore cells comprising the posterior 80% of the slug, while the [ecmAO]:GFP reporter labeled the anterior 20% of the slug comprising the prestalk cells. Compared to the wild type, detA− slugs expressed [ecmAO]:GFP more strongly, and the prestalk region was larger (approximately 25 to 30% of the slug) than in Ax2 (Fig. 5A). In agreement with this, the expression level of the endogenous ecmA gene also appeared to be significantly upregulated in the detA− background compared to that in the wild type (see Fig. SA3a in the supplemental material). [cotB]:GFP staining in the detA− mutant was much reduced, and the anterior third of the slug, comprising the enlarged prestalk zone, was free of the label. Surprisingly, staining was also absent in the very posterior of most slugs (Fig. 5B). As [ecmAO]:GFP did not appear to label these posterior cells, their identity remains unknown. We also performed immunostaining on slugs with an antibody against the PsvA prespore markers (EB4 and SP35) (22, 23). In wild-type slugs, the antibody labeled the prespore cells in a rather uniform manner. In contrast, in the detA− mutant, the pattern was quite variable from slug to slug (see Fig. SA3b in the supplemental material), with some cells displaying expression over the entire length of the slug. In many slugs, however, it was largely absent from the anterior third, which corresponded to the enlarged prestalk region, and it was also absent from the posterior of the slug, as was observed for the [cotB]:GFP line.
Analysis of prestalk subtypes.
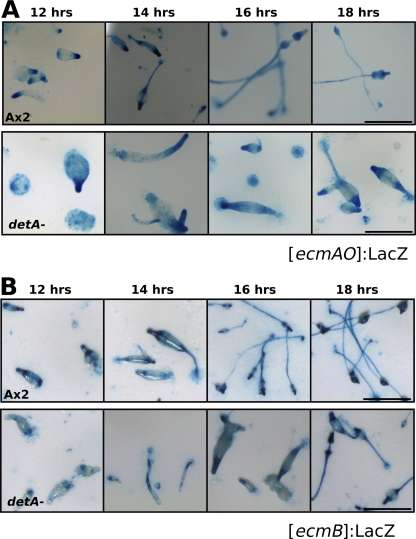
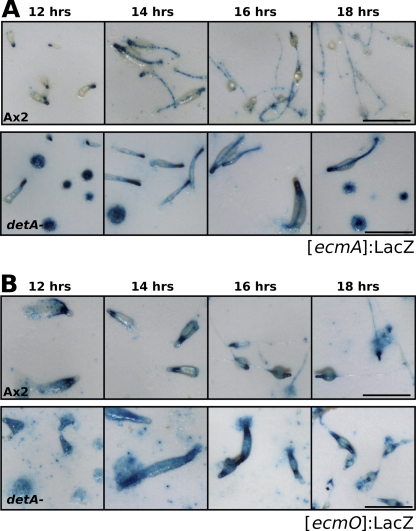
The prestalk region of Dictyostelium is comprised of several zones of different prestalk subtypes (47). To determine the architecture of the enlarged prestalk zone in the detA− mutant, the different subtypes were more closely examined. Ax2 and detA− cells were transformed with a reporter for PstB (prestalk B) cells ([ecmB]:LacZ), PstA cells ([ecmA]:LacZ), or PstO cells ([ecmO]:LacZ) and the reporter [ecmAO]:LacZ (15). Slugs were allowed to develop for 12 to 18 h and then were fixed and stained. The ecmA gene is expressed in almost all prestalk cells, but different cell types express it using either the distal part of the promoter [ecmO] or the proximal part [ecmA]; these cell types are referred to as PstO and PstA, respectively. ecmB is expressed in the PstB cells, which are present at the base of the prestalk zone in migrating slugs and will form the basal disk and lower cup in mature culminants, and also in the rearguard cells. A small region within the PstA zone also expresses ecmB, and this region is called the PstAB zone. The [ecmAO]:LacZ marker labels essentially the entire prestalk region: i.e., the PstA, PstO, and PstAB cells (Fig. 6A). As previously observed for the [ecmAO]:GFP construct, the zone expressing [ecmAO]:LacZ was expanded in the mutant compared to that in the wild type. Although variable, it typically stained the front 25 to 30% of the slug instead of the anterior 20% in the wild type. Staining of the slug posterior and also within the prespore region was also stronger in the detA− mutant. Compared to the wild type, [ecmB]:LacZ staining in the detA− mutant appeared to be slightly weaker after 12 and 14 h of development, but was otherwise similar to that in the wild type (Fig. 6B). It was also apparent that after 16 to 18 h, the mutant has a developmental delay of approximately 4 h compared to the wild type. The [ecmA]:LacZ reporter, which contains the proximal portion of the ecmAO promoter, was much more active in the detA− mutant (Fig. 7A). Some scattered staining was visible well into the prespore zone and at the slug posterior region. Staining of the stalk was strong and homogeneous in the detA− mutant but weak and punctate along the stalk in wild type. Compared to Ax2, [ecmO]:LacZ expression in the detA− mutant appeared to be diffuse, more extended, and possibly weaker (Fig. 7B). Also, the staining did not end abruptly at the prespore region, but continued throughout the slug, suggesting an increased number of anterior-like cells (ALCs), which became more abundant toward the posterior of the slug. Many of the detA− mutant aggregates that arrested at the mound stage showed a rather homogeneous staining of prestalk and prespore markers (Fig. 6 and 7; see Fig. SA3b in the supplemental material), indicating that they were able to differentiate into prestalk and prespore cells. Some of these mounds eventually formed slugs, while others remained arrested 48 h after the onset of development (data not shown).
Fig. 6.
Effect of DET1 on the formation of prestalk cell subtypes. (A) Wild-type or detA− slugs transformed with the prestalk subtype reporter [ecmB]:LacZ were fixed and stained for β-galactosidase at the indicated times after the onset of starvation. (B) Wild-type or detA− slugs transformed with the prestalk subtype reporter [ecmAO]:LacZ were fixed and stained for β-galactosidase at the indicated times after the onset of starvation. All micrographs are of identical magnification. Scale bar, 500 μm.
Fig. 7.
Effect of DET1 on the formation of prestalk cell subtypes. (A) Wild-type or detA− slugs transformed with the prestalk subtype reporter [ecmA]:LacZ were fixed and stained for β-galactosidase at the indicated times after the onset of starvation. (B) Wild-type or detA− slugs transformed with the prestalk subtype reporter [ecmO]:LacZ were fixed and stained for β-galactosidase at the indicated times after the onset of starvation. All micrographs are of identical magnification. Scale bar, 500 μm.
Possible DetA-interacting partners.
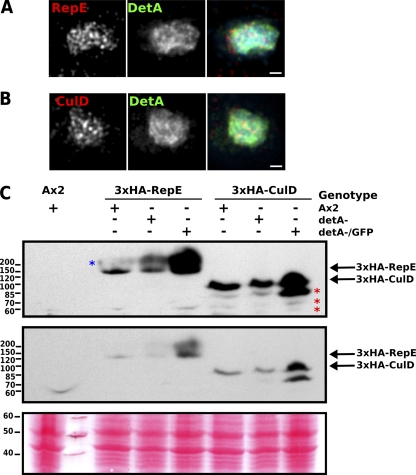
The Dictyostelium genome encodes homologs of the DET1-associated proteins Cul4 (CulD; DDB_G0292794), RepE (DDB1; DDB_G0286013), and COP1 (for constitutive photomorphogenesis 1; DDB_G0288453). These were cloned and expressed as GFP or 3×HA epitope-tagged fusions. All three were nuclear localized and displayed a localization pattern similar to DetA (Fig. 8A and B) (data not shown). When they were expressed in the detA− mutant background, no noticeable changes in their distribution were observed, suggesting that DetA is not primarily responsible for their nuclear localization (data not shown). DET1 is known to bind directly to DDB1 (14, 39). While DET1 and Cul4 can also be detected in the same complex (5, 46), the interaction appears to be weaker and occur via DDB1. The 3×HA-tagged CulD and RepE were both expressed at the expected size (Fig. 8C), although for CulD, a number of bands probably representing degradation products were observed, while for RepE, higher-molecular-weight species were apparent, which may correspond to ubiquitinated RepE. Loss of DetA appeared to result in a relative increase in the higher-molecular-weight species of RepE, while epitope-tagged CulD was unaffected (Fig. 8C). Overexpression of GFP-DetA in the detA− mutant background resulted in a significant increase in the levels of both CulD and RepE (Fig. 8C).
Fig. 8.
Putative DetA interaction partners. (A) Immunofluorescence on fixed detA−/GFP-DetA cells supertransformed with a 3×HA-RepE construct; (B) Immunofluorescence on fixed detA−/GFP-DetA cells supertransformed with a 3×HA-CulD expression construct; (C) Western blot showing Ax2, detA− or detA−/GFP-DetA cells supertransformed with the expression construct 3×HA-RepE or 3×HA-CulD. The middle panel is a shorter exposure of the top panel, and the lower panel shows the Ponceau-S-stained membrane to confirm equal loading. The red asterisks indicate putative degradation products of 3×HA-CulD, and the blue asterisk indicates a species of 3×HA-RepE with a higher than predicted Mr, possibly due to ubiquitination or another posttranslational modification. Scale bar, 1 μm.
In order to more directly test the interaction between DetA, RepE, and CulD, coimmunoprecipitations were performed with detA−/GFP-DetA cells supertransformed with 3×HA-CulD or 3×HA-RepE. As shown in Fig. 9, HA-tagged RepE was found on the GFP-Trap Sepharose beads. The predicted indirect interaction of DetA via RepE with CulD was too weak to be seen in the reproduction of the Western blot.
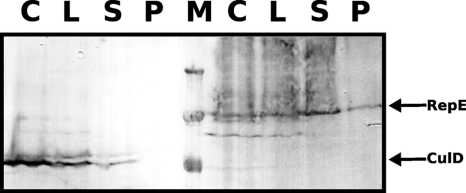
Fig. 9.
RepE interacts with DetA. DetA−/GFP-DetA cells expressing 3×HA-RepE (left) or 3×HA-CulD (right) were processed for coimmunoprecipitation. The anti-HA-stained protein bands are indicated by arrows. C, cells before processing; L, lysate; S, supernatant from GFP-Trap Sepharose beads; P, protein bound to the beads. Lanes C, L, and S contain aliquots equivalent to 1 × 107 cells; lane P contains protein from 5 × 108 cells bound to the beads; and lane M contains 250-, 130-, and 100-kDa markers. Washes of the beads (not shown) do not contain any RepE or CulD protein.
DISCUSSION
We report here the identification and characterization of DetA, the Dictyostelium homolog of DET1, a chromatin binding protein involved in developmental regulation in both plants and animals. DET1 homologs are almost exclusively restricted to multicellular organisms. Consistent with this, among the amoebozoa, a DET1 homolog is present in the multicellular species Dictyostelium discoideum and Dictyostelium purpureum but not in the obligatory unicellular species Entamoeba histolytica.
In contrast to all other known DET1 homologs, Dictyostelium DetA is split by a large homopolymer stretch. It is thus significantly larger than DET1 proteins from other organisms. This organization, together with the observation that both parts were independently able to localize to the nucleus (data not shown), suggests that DET1 may consist of two separate domains.
Loss of DetA resulted in a slight increase in doubling time in vegetative cells, delayed aggregation and enlarged aggregation territories suggesting a defect in quorum sensing. Up to 50% of the mounds had arrested development, but some resumed development later, indicating a role of DetA in the transition from mounds to slugs. The prespore cotB promoter showed a reduced zone of expression in detA− slugs, while the prestalk zone labeled by [ecmAO]:GFP was extended. The PstB zone was rather unaffected, but the PstA zone was enlarged, and expression of [ecmA]:LacZ was stronger in the mutant. DetA− cells thus have an altered prespore/prestalk ratio and a misproportioning within the prestalk population. As the posterior of the detA− slugs failed to express [cotB]:GFP or stain with an antibody against the SP35 protein (PsvA/EB4), these cells are not well defined: they are probably prestalk like, with at least some PstO characteristics. Since DET1 appears to act as a transcriptional repressor, it seems probable that the increase in the percentage of prestalk cells in the detA− mutant is due to the inability to repress the prestalk genes.
The DET1-associated proteins RepE, CulD, and Cop1 are conserved in Dictyostelium and localized to the nucleus in a manner similar to DetA. No significant change in their localization was observed in the detA− mutant, but substantial overexpression of RepE and CulD in the GFP-DetA overexpression mutant pointed to at least regulatory interactions. The binding of DetA to RepE could be shown by coimmunopreciptitation, while the indirect interaction with CulD was only barely detectable.
Plant development is highly plastic in order to adapt to changing environmental conditions (1). While DET1 was originally described as a negative regulator of photomorphogenesis, however, further analysis revealed extensive and rather variable organ patterning defects, somewhat reminiscent of polycomb mutants (32). Likewise, loss of the Drosophila DET1 homolog ABO1 also results in extensive but highly variable patterning defects during embryogenesis (41). Developmental patterning of most animals displays a certain degree of plasticity patterning. For example, cell fate been specified in different segments of Drosophila embryos by morphogen gradients, while mammalian embryos go through a number of cell divisions before cell fate is specified (31). An interesting exception is Caenorhabditis elegans, which relies on asymmetry that exists prior to the first cell division to establish polarity. In contrast to most other organisms, it exhibits almost no developmental plasticity, always going on to form an organism with exactly the same number of cells (1,031 for males) and the same proportion of cell types (8). C. elegans is one of the few multicellular animals that lacks a DET1 homolog. It is tempting to speculate that the loss of DET1 in this organism is linked to the lack of plasticity in its developmental program. In contrast, Dictyostelium development is highly plastic, with cell fate only been specified late in development after the individual amoebae have aggregated into a mound and in response to morphogen gradients such as DIF-1 (40). Being among the most simple organisms that harbor a DET1 homolog, Dictyostelium provides an ideal system to study developmental plasticity and the functions of this evolutionarily conserved chromatin regulator.
Supplementary Material
ACKNOWLEDGMENTS
We thank Rick Firtel for providing the GFP reporter constructs and the Dictyostelium Stock Center for the LacZ reporter constructs and bioinformatics resources. We thank Markus Maniak for helpful comments and critical reading of the manuscript.
This work was supported by grants (Ne285/10 and Ne285/12) from the Deutsche Forschungsgemeinschaft to W.N.
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
Published ahead of print on 30 December 2010.
REFERENCES
- 1. Alabadí D., Blázquez M. A. 2009. Molecular interactions between light and hormone signaling to control plant growth. Plant Mol. Biol. 69:409–417 [DOI] [PubMed] [Google Scholar]
- 2. Altschul S. F., et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Benvenuto G., Formiggini F., Laflamme P., Malakhov M., Bowler C. 2002. The photomorphogenesis regulator DET1 binds the amino-terminal tail of histone H2B in a nucleosome context. Curr. Biol. 12:1529–1534 [DOI] [PubMed] [Google Scholar]
- 4. Berloco M., Fanti L., Breiling A., Orlando V., Pimpinelli S. 2001. The maternal effect gene, abnormal oocyte (abo), of Drosophila melanogaster encodes a specific negative regulator of histones. Proc. Natl. Acad. Sci. U. S. A. 98:12126–12131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bernhardt A., et al. 2006. CUL4 associates with DDB1 and DET1 and its downregulation affects diverse aspects of development in Arabidopsis thaliana. Plant J. 47:591–603 [DOI] [PubMed] [Google Scholar]
- 6. Bonner J. 1950. Observations on polarity in the slime mold Dictyostelium discoideum. Biol. Bull. 99:143–151 [DOI] [PubMed] [Google Scholar]
- 7. Bonner J. T. 1970. Induction of stalk cell differentiation by cyclic AMP in the cellular slime mold dictyostelium discoideum. Proc. Natl. Acad. Sci. U. S. A. 65:111–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Brenner S. 1974. The genetics of Caenorhabditis elegans. Genetics 77:71–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Brookman J. J., Jermyn K. A., Kay R. R. 1987. Nature and distribution of the morphogen DIF in the Dictyostelium slug. Development 100:119–124 [DOI] [PubMed] [Google Scholar]
- 10. Chen G., Zhuchenko O., Kuspa A. 2007. Immune-like phagocyte activity in the social amoeba. Science 317:678–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chory J., Peto C., Feinbaum R., Pratt L., Ausubel F. 1989. Arabidopsis thaliana mutant that develops as a light-grown plant in the absence of light. Cell 58:991–999 [DOI] [PubMed] [Google Scholar]
- 12. Dai Q., Wang H. 2006. Cullin 4 makes its mark on chromatin. Cell Division 1:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dubin M., Nellen W. 2010. A versatile set of tagged expression vectors to monitor protein localisation and function in Dictyostelium. Gene 465:1–8 [DOI] [PubMed] [Google Scholar]
- 14. Dubin M. J., Bowler C., Benvenuto G. 2008. A modified Gateway cloning strategy for overexpressing tagged proteins in plants. Plant Methods 4:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Early A. E., Gaskell M. J., Traynor D., Williams J. G. 1993. Two distinct populations of prestalk cells within the tip of the migratory Dictyostelium slug with differing fates at culmination. Development 118:353–362 [DOI] [PubMed] [Google Scholar]
- 16. Eichinger L., et al. 2005. The genome of the social amoeba Dictyostelium discoideum. Nature 435:43–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Escalante R., Sastre L. 2006. Investigating gene expression: in situ hybridization and reporter genes. Methods Mol. Biol. 346:247–260 [DOI] [PubMed] [Google Scholar]
- 18. Escalante R., Vicente J. J., Moreno N., Sastre L. 2001. The MADS-box gene srfA is expressed in a complex pattern under the control of alternative promoters and is essential for different aspects of Dictyostelium development. Dev. Biol. 235:314–329 [DOI] [PubMed] [Google Scholar]
- 19. Faix J., Kreppel L., Shaulsky G., Schleicher M., Kimmel A. R. 2004. A rapid and efficient method to generate multiple gene disruptions in Dictyostelium discoideum using a single selectable marker and the Cre-loxP system. Nucleic Acids Res. 32:e143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fey P., et al. 2009. dictyBase—a Dictyostelium bioinformatics resource update. Nucleic Acids Res. 37:D515–D519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fukuzawa M., Zhukovskaya N. V., Yamada Y., Araki T., Williams J. G. 2006. Regulation of Dictyostelium prestalk-specific gene expression by a SHAQKY family MYB transcription factor. Development 133:1715–1724 [DOI] [PubMed] [Google Scholar]
- 22. Hildebrandt M., Humbel B. M., Nellen W. 1991. The Dictyostelium discoideum EB4 gene product and a truncated mutant form of the protein are localized in prespore vesicles but absent from mature spores. Dev. Biol. 144:212–214 [DOI] [PubMed] [Google Scholar]
- 23. Hildebrandt M., Nellen W. 1992. Differential antisense transcription from the Dictyostelium EB4 gene locus: implications on antisense-mediated regulation of mRNA stability. Cell 69:197–204 [DOI] [PubMed] [Google Scholar]
- 24. Hong E. E., Villén J., Gerace E. L., Gygi S. P., Moazed D. 2005. A cullin E3 ubiquitin ligase complex associates with Rik1 and the Clr4 histone H3-K9 methyltransferase and is required for RNAi-mediated heterochromatin formation. RNA Biol. 2:106–111 [DOI] [PubMed] [Google Scholar]
- 25. Hotton S. K., Callis J. 2008. Regulation of cullin RING ligases. Annu. Rev. Plant Biol. 59:467–489 [DOI] [PubMed] [Google Scholar]
- 26. Iranfar N., Fuller D., Loomis W. F. 2003. Genome-wide expression analyses of gene regulation during early development of Dictyostelium discoideum. Eukaryot. Cell 2:664–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jackson S., Xiong Y. 2009. CRL4s: the CUL4-RING E3 ubiquitin ligases. Trends Biochem. Sci. 34:562–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jia S., Kobayashi R., Grewal S. I. S. 2005. Ubiquitin ligase component Cul4 associates with Clr4 histone methyltransferase to assemble heterochromatin. Nat. Cell Biol. 7:1007–1013 [DOI] [PubMed] [Google Scholar]
- 29. Kapetanaki M. G., et al. 2006. The DDB1-CUL4-DDB2 ubiquitin ligase is deficient in xeroderma pigmentosum group E and targets histone H2A at UV-damaged DNA sites. Proc. Natl. Acad. Sci. U. S. A. 103:2588–2593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Langenick J., Araki T., Yamada Y., Williams J. G. 2008. A Dictyostelium homologue of the metazoan Cbl proteins regulates STAT signalling. J. Cell Sci. 121:3524–3530 [DOI] [PubMed] [Google Scholar]
- 31. Lu C. C., Brennan J., Robertson E. J. 2001. From fertilization to gastrulation: axis formation in the mouse embryo. Curr. Opin. Genet. Dev. 11:384–392 [DOI] [PubMed] [Google Scholar]
- 32. Mayer R., Raventos D., Chua N. H. 1996. det1, cop1, and cop9 mutations cause inappropriate expression of several gene sets. Plant Cell 8:1951–1959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mohanty S., et al. 1999. Evidence that the Dictyostelium Dd-STATa protein is a repressor that regulates commitment to stalk cell differentiation and is also required for efficient chemotaxis. Development 126:3391–3405 [DOI] [PubMed] [Google Scholar]
- 34. Mohanty S., et al. 2001. Regulated protein degradation controls PKA function and cell-type differentiation in Dictyostelium. Genes Dev. 15:1435–1448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mustilli A. C., Fenzi F., Ciliento R., Alfano F., Bowler C. 1999. Phenotype of the tomato high pigment-2 mutant is caused by a mutation in the tomato homolog of DEETIOLATED1. Plant Cell 11:145–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Parikh A., et al. 2010. Conserved developmental transcriptomes in evolutionarily divergent species. Genome Biol. 11:R35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pepper A., Delaney T., Washburn T., Poole D., Chory J. 1994. DET1, a negative regulator of light-mediated development and gene expression in arabidopsis, encodes a novel nuclear-localized protein. Cell 78:109–116 [DOI] [PubMed] [Google Scholar]
- 38. Sambrook J., Fritsch E. F., Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 39. Schroeder D. F., et al. 2002. De-etiolated 1 and damaged DNA binding protein 1 interact to regulate Arabidopsis photomorphogenesis. Curr. Biol. 12:1462–1472 [DOI] [PubMed] [Google Scholar]
- 40. Strmecki L., Greene D. M., Pears C. J. 2005. Developmental decisions in Dictyostelium discoideum. Dev. Biol. 284:25–36 [DOI] [PubMed] [Google Scholar]
- 41. Tomkiel J., et al. 1995. Developmental genetical analysis and molecular cloning of the abnormal oocyte gene of Drosophila melanogaster. Genetics 140:615–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Tsujioka M., et al. 2007. Dictyostelium Myb transcription factors function at culmination as activators of ancillary stalk differentiation? Eukaryot. Cell 6:568–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. UniProt Consortium 2010. The Universal Protein Resource (UniProt) in 2010. Nucleic Acids Res. 38:D142–D148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wang B., Kuspa A. 2002. CulB, a putative ubiquitin ligase subunit, regulates prestalk cell differentiation and morphogenesis in Dictyostelium spp. Eukaryot. Cell 1:126–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wang H., et al. 2006. Histone H3 and H4 ubiquitylation by the CUL4-DDB-ROC1 ubiquitin ligase facilitates cellular response to DNA damage. Mol. Cell 22:383–394 [DOI] [PubMed] [Google Scholar]
- 46. Wertz I. E., et al. 2004. Human De-etiolated-1 regulates c-Jun by assembling a CUL4A ubiquitin ligase. Science 303:1371–1374 [DOI] [PubMed] [Google Scholar]
- 47. Williams J. G. 2006. Transcriptional regulation of Dictyostelium pattern formation. EMBO Rep. 7:694–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yamada Y., Wang H. Y., Fukuzawa M., Barton G. J., Williams J. G. 2008. A new family of transcription factors. Development 135:3093–3101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhang K., Mosch K., Fischle W., Grewal S. I. 2008. Roles of the Clr4 methyltransferase complex in nucleation, spreading and maintenance of heterochromatin. Nat. Struct. Mol. Biol. 15:381–388 [DOI] [PubMed] [Google Scholar]
- 50. Zhang Y., et al. 2008. Arabidopsis DDB1-CUL4 ASSOCIATED FACTOR1 forms a nuclear E3 ubiquitin ligase with DDB1 and CUL4 that is involved in multiple plant developmental processes. Plant Cell 20:1437–1455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhukovskaya N. V., Fukuzawa M., Yamada Y., Araki T., Williams J. G. 2006. The Dictyostelium bZIP transcription factor DimB regulates prestalk-specific gene expression. Development 133:439–448 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.