Abstract
The exfoliative toxin (ET) is a major virulence factor of Staphylococcus aureus that causes bullous impetigo and its disseminated form, staphylococcal scalded-skin syndrome (SSSS). ET selectively digests one of the intracellular adhesion molecules, desmoglein 1, of epidermal keratinocytes and causes blisters due to intraepidermal cell-cell dissociation. Most S. aureus strains that cause blistering disease produce either ETA or ETB. They are serologically distinct molecules, where ETA is encoded on a phage genome and ETB is enocded on a large plasmid. ETA-producing S. aureus strains are frequently isolated from impetigo patients, and ETB-producing S. aureus strains are isolated from SSSS. ET-induced blister formation can be reproduced with the neonatal mouse. To determine the regulatory mechanism of ET production, we investigated the role of the two-component systems and global regulators for eta or etb expression in vitro and in vivo with the mouse model. Western blot and transcription analyses using a series of mutants demonstrate ETA production was downregulated by sigB, sarS, and sarA, while ETB production was downregulated by sigB and sarA but not by sarS. Production of both toxins is upregulated by saeRS, arlRS, and agrCA. Furthermore, by the in vivo neonatal mouse model, sigB and sarS but not sarA negatively regulate the exfoliation activity of the ETA-producing strain, while sarA negatively regulates the ETB-producing strain. In both strains, saeRS, arlRS, and agrCA positively regulate the exfoliation activity in vivo. The data illustrate similar but distinct regulatory mechanisms for ETA and ETB production in S. aureus in vitro as well as in vivo.
Staphylococcus aureus is a Gram-positive pathogen that causes a wide variety of diseases. It produces a large number of virulence determinants, including proteases, enterotoxins, cytolytic toxins, protein A, clumping factor, and others that may play important roles in establishing and maintaining infections with the bacterium.
Exfoliative toxin (ET) is one of these extracellular proteins and causes blisters in bullous impetigo and, in the disseminated form, staphylococcal scalded-skin syndrome (SSSS) (22). Neonates and young children are primarily affected. Recently, we and others demonstrated three isoforms of ETs (ETA, ETB, and ETD), which are glutamate-specific serine proteases that specifically cleave a single peptide bond in the extracellular region of human and mouse desmoglein 1 (Dsg 1), a desmosomal cadherin-type cell-cell adhesion molecule (1, 2, 40). The exfoliative activity can be assayed monitoring the elicitation of Nikolsky's sign when neonatal mice are injected with the toxin protein or S. aureus strains carrying the ET gene (et) (26, 39). Previous studies show the ETA gene (eta) is carried on the genome of a temperate phage integrated into the S. aureus chromosome (38), whereas the ETB gene (etb) is carried on a large plasmid, pETB (12, 39). Staphylococcus aureus strains carrying eta are frequently isolated from patients with bullous impetigo, whereas those carrying etb are obtained from patients with SSSS (41). The ETD (etd) gene is located on a pathogenicity island where etd-positive strains are primarily isolated from patients with deep pyoderma and not bullous impetigo or SSSS (40).
The production of staphylococcal virulence factors is coordinately modulated by the two-component regulatory systems (TCSs) (e.g., the accessory gene regulator [agr], S. aureus exoprotein expression [saeRS] gene, and autolysis-related locus [arlRS]) and a global regulator (e.g., the staphylococcal accessory regulator family [sarA, sarS, rot, and others] and the alternative sigma factor, sigma B [sigB]) (6, 28). The agr locus has two divergent transcripts, RNA II and RNA III. The RNA II transcript encodes four proteins (AgrB, -D, -C, and -A) that are related to generating a quorum-sensing molecule (AIP) and a two-component regulatory system. The RNA III transcript acts as the effector molecule for agr-specific regulation (29). The SaeRS system was identified as the positive transcriptional regulator of exoprotein independent of agr and SarA (14, 15, 42). Furthermore, saeRS was shown to be an important element for the expression of virulence genes in vivo (17, 31). The ArlRS system was first identified as a regulator involved in biofilm formation, autolysis, and extracellular proteolytic activity (11). In addition, ArlR positively regulates the accessory gene regulator (agr) (23). The sarA and sarS loci are recognized as transcription factors and mediate their effect both directly by binding to the target gene promoters and indirectly via the downstream effect on other regulons (7, 37). Sigma B is one of the three alternative σ factors (σB, σH, and σS RNA polymerases) of S. aureus and has been characterized as a regulator in the general stress response (5, 18, 27, 35). Additionally, regulation of the virulence determinants is shown to be mediated either directly by a σB-dependent promoter or indirectly by additional global regulators, including SarA and SarS, which possess a σB-dependent promoter (3, 7, 10, 21, 37). Furthermore, Oscarsso et al. noted SigB may suppress hla transcription via mechanisms not involving SarA and SarS, suggesting another SigB-dependent factor(s) suppresses hla transcription (30).
Thus, the pathogenicity factors are shown to be modulated by complex regulatory mechanisms in S. aureus, where many other regulatory elements have been identified for other genes (4, 9); however, the regulatory mechanism for ET (et) production has not been extensively studied. Here we investigate the role of the transcription factors sigB, sarS, sarA, saeRS, arlRS, and agrCA involved in the expression of eta and etb in vitro and in vivo by using the neonatal mice model.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are described in Table 1. Staphylococcus aureus and Escherichia coli were grown at 37°C with shaking in Trypticase soy broth (TSB; Becton Dickinson Microbiology Systems, Cockeysville, MD) or Luria-Bertani broth (5 g yeast extract, 10 g polypeptone, 10 g NaCl per liter, pH 7.2), respectively. When necessary, xylose (1%), ampicillin (100 μg/ml), chloramphenicol (10 μg/ml), or tetracycline (3 μg/ml) was added to the medium. The antibiotics were purchased from Sigma Chemical Co., St. Louis, MO. Anti-ETA rabbit serum and anti-ETB serum were prepared as described previously (36, 40). Anti-Hla was a kind gift from T. Tomita (Tohoku University).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype or characteristic(s) | Source or reference |
|---|---|---|
| Strains | ||
| S. aureus | ||
| RN4220 | NCTC8325-4 r− m+ | Kreiswirth et al. (20) |
| TY34 | Clinical isolate (eta+agr type III mecA+) | |
| TY825 | Clinical isolate (etb+agr type IV) | |
| TF5367 | TY34 Δeta::cat | |
| FK101 | TY34 TCS2 MW0199::pFK5 | This work |
| FK102 | TY34 lytSRMW0236::pFK6 | This work |
| FK103 | TY34 graRSMW0621::pFK7 | This work |
| FK104 | TY34 saeRSMW0668::pFK8 | This work |
| FK105 | TY34 TCS6MW1208::pFK9 | This work |
| FK106 | TY34 arlRSMW1305::pFK10 | This work |
| FK107 | TY34 srrABMW1446::pFK11 | This work |
| FK108 | TY34 phoPRMW1637::pFK12 | This work |
| FK109 | TY34 yhcSRMW1790::pFK13 | This work |
| FK110 | TY34 vraSRMW1825::pFK14 | This work |
| FK111 | TY34 agrCAMW1962::pFK15 | This work |
| FK112 | TY34 kdpDEMW2002::pFK16 | This work |
| FK113 | TY34 hssRSMW2282::pFK17 | This work |
| FK114 | TY34 nreCBMW2314::pFK18 | This work |
| FK115 | TY34 TCS16MW2545::pFK19 | This work |
| FK128 | TY34 ΔsarA::cat | This work |
| FK129 | TY34 sarA::catsarS::pFK2 | This work |
| FK130 | TY34 sarS::pFK21 | This work |
| FK131 | TY34 sigB::pFK20 | This work |
| FK132 | TY34 sarA::catsaeRSMW0668::pFK8 | This work |
| FK133 | TY34 sarA::catagrCA MW1962::pFK15 | This work |
| FK134 | TY34 sarS::pFK21 saeRS MW0668::pFK8 | This work |
| FK135 | TY34 sarS::pFK21 agrCAMW1962::pFK15 | This work |
| FK136 | TY34 sigB::pFK20 complemented with pFK22 | This work |
| FK137 | TY34 ΔsarA::cat complemented with pFK23 | This work |
| FK138 | TY34 sarS::pFK21 complemented with pFK24 | This work |
| FK200 | TY825 etb mutant pETB cured | This work |
| FK204 | TY825 saeRSMW0668::pFK8 | This work |
| FK206 | TY825 arlRSMW1305::pFK10 | This work |
| FK211 | TY825 agrCAMW1962::pFK15 | This work |
| FK216 | TY825 sigB::pFK1 | This work |
| FK217 | TY825 sarS::pFK2 | This work |
| FK218 | TY825 ΔsarA::cat | This work |
| E. coli | ||
| DH5α | F− φ80dlacZΔM15 Δ(lacZYA-argF)U169deoRrecA1endA1hsdR17(rK− mK+) phoAsupE44 λ−thi-1gyrA96relA1 | TaKaRa |
| Plasmids | ||
| pUC19 | E. coli cloning vector | TaKaRa |
| pHY300PLK | Shuttle vector between E. coli and S. aureus | TaKaRa |
| pWH1520 | 7.9-kbp xylose-inducible vector; PxylA′xylR Ampr in E. coli, Tetr in S. aureus | Rygus and Hillen (32) |
| pCL52.1 | 8.0-kbp temp-sensitive shuttle vector; Spcr in E. coli, Tetr in S. aureus | Sau et al. (34) |
| pCL15 | 7.0-kbp IPTG-inducible vector; Pspac, Ampr in E. coli, Cmr in S. aureus | Luong and Lee (24) |
| pKFT | 5.7-kbp temp-sensitive shuttle vector; Ampr Tetr in E. coli, Tetr in S aureus | This work |
| pKFC | 5.1-kbp temp-sensitive shuttle vector; Ampr in E. coli, Cmr in S aureus | This work |
| pFK2 | pKFT containing 554-bp fragment of sarS | This work |
| pFK3 | pKFT containing sarA::cat for deletion of the sarA gene | This work |
| pFK4 | pKFT containing 547-bp fragment of MW0018a | This work |
| pFK5 | pKFT containing 658-bp fragment of MW0199a | This work |
| pFK6 | pKFT containing 775-bp fragment of MW0236a | This work |
| pFK7 | pKFT containing 640-bp fragment of MW0621a | This work |
| pFK8 | pKFT containing 545-bp fragment of MW0668a | This work |
| pFK9 | pKFT containing 486-bp fragment of MW1208a | This work |
| pFK10 | pKFT containing 517-bp fragment of MW1305a | This work |
| pFK11 | pKFT containing 552-bp fragment of MW1446a | This work |
| pFK12 | pKFT containing 533-bp fragment of MW1637a | This work |
| pFK13 | pKFT containing 456-bp fragment of MW1790a | This work |
| pFK14 | pKFT containing 510-bp fragment of MW1825a | This work |
| pFK15 | pKFT containing 543-bp fragment of MW1962a | This work |
| pFK16 | pKFT containing 595-bp fragment of MW2002a | This work |
| pFK17 | pKFT containing 534-bp fragment of MW2282a | This work |
| pFK18 | pKFT containing 472-bp fragment of MW2314a | This work |
| pFK19 | pKFT containing 529-bp fragment of MW2545a | This work |
| pFK20 | pKFC containing 503-bp fragment of sigB | This work |
| pFK21 | pKFC containing 554-bp fragment of sarS | This work |
| pFK22 | pWH1520 containing sigB gene | This work |
| pFK23 | pWH1520 containing sarA gene | This work |
| pFK24 | pWH1520 containing sarS gene | This work |
Locus numbers are based on S. aureus strain MW2 (http://www.bio.nite.go.jp). Gene names are based on S. aureus MW2 and according to the first reference where the TCS is described in the text.
DNA procedures.
Routine DNA procedures such as DNA digestion with restriction enzymes, DNA ligations, and gel electrophoresis were performed essentially as described previously (33). The oligonucleotides used in this study are described in Table 2. PCR was performed using the Ex Taq polymerase (TaKaRa Bio) with the appropriate cycling conditions.
TABLE 2.
Oligonucleotide primers used in this study
| Primer | Sequence (5′ to 3′)a |
|---|---|
| Standard sequencing | |
| TsoriF | TACGATGACGTCTTTTGCGCAGTCGGC |
| TsoriR | ATAGACGTCGTGAGAAACAGCGTACAG |
| TetLF | TTATTGCAATGTGGAATTCGGAACGG |
| TetLR | CCGGGAATTCCTGTTATAAAAAAAGG |
| CMF | TTCATATGCCGGCAATAGTTACCCTT |
| CMR | TTCATATGGATCTGGAGCTGTAATAT |
| MW0018F | AAAAGAAGGATACGATGTGTACTGTGC |
| MW0018R | GTAAACGACGAATCGTTACATCGACCG |
| MW0199F | GTAAAATTAAACCATGCTGACCATCGTCTC |
| MW0199R | ATAGCATTTTCTATGAGTGGCTGAAGC |
| MW0236F | GGCCCTTTTGTAGGTCTATTTGTTGGCG |
| MW0236R | CCAATCCTTCTGCAAGTTGACGTTCCAC |
| MW0621F | GACAATACTTTGTTTCAAGAATTGAAA |
| MW0621R | GGTAATAAGAATTTCTAATATAATCATTT |
| MW0668F | TTTGTCAAACCTATTTTGAATATGAAGG |
| MW0668R | GTATATGGACATTCACGGTATTAGCATC |
| MW1208F | TTTCTTTTATAGTGCTTTTGCCGTTCC |
| MW1208R | TATACTATCAATCTCTTCAATAAATGATGG |
| MW1305F | AGCAAGCTTTCTTGAATTGGAACTCAC |
| MW1305R | TTGTTTAAGCTTCACTATTATAACCCC |
| MW1446F | CCATGAAGCAAGTAATGGCCAAGAGGC |
| MW1446R | CACACGATTTAACTTTTCTCTAAGTCG |
| MW1637F | TGGATCCAGTAGATGACGAACATTCAA |
| MW1637R | TAAGCTTATGCTCTCGTAATGACTC |
| MW1790F | GCGTCGAATATCGTCAATTGTTCTCGTA |
| MW1790R | GTCCAACTGGCTTAAGTTGCC |
| MW1825F | GTGAAACGTTAGATTTATACCATACACTCG |
| MW1825R | TTTGTACCGTTTGAATGACGC |
| MW1962F | CACCCTTAAAGAGATGAAATACAAACG |
| MW1962R | TTCTTGGAACAATTCATGAATGCGTGG |
| MW2002F | GAACAAATCACCCTATCGTCCTAAAGGC |
| MW2002R | CCCAGCTTAATGCTTGTTCATGCTGTTG |
| MW2282F | ATTGATGCATACACACAACCAAGTGG |
| MW2282R | ATTTTTTTAATCTTTGGCGTAGTCGC |
| MW2314F | ACATATCAGTTGATTGATCAAGACAGGGG |
| MW2314R | GTACTACTCGATAAACAACCG |
| MW2545F | AAAATGGAATTACGGTGTTATTGTCG |
| MW2545R | AGGCGCGTCATGTTAACAGCTAATGTG |
| sigBF | CAAGAAGCTTAGAATACAGATGCACAGG |
| sigBR | GTTAAAGCTTAATGGTCATCTTGTTGCC |
| sigBFS | TATGACTAGTTATATAAAAAAAGAGCAG |
| sigBRB2 | AATAAAGATCTTCTATTGATGTGCTGC |
| sarAF | ATTTGGGTAGTAAGCTTTGACACAAC |
| sarAR | GGATTTGAAGCTTTGCAACATCAACTAGC |
| sarAFS | ATCGAACTAGTTGCATCAAATAGGGAG |
| sarARB2 | TCATAGATCTCCAAATTGCGCTAAAC |
| sarSF | TATAATCATTGAAGCATATATGTTTCG |
| sarSR | TTATTGAGAGCTCTAACAGTTTGAGGG |
| sarSFS | ATTAAAAACTAGTGCATATACAAGGAG |
| sarSRB2 | CACTTTAGATCTCAGCACACTTGCGT |
| Quantitative RT-PCR | |
| gyrBF | AGGTCTTGGAGAAATGAATG |
| gyrBR | CAAATGTTTGGTCCGCTT |
| etaF | TACAGTTCCGGGAAATTCT |
| etaR | CCCAATACCAACACCATAA |
| etbF | GTGGTAAAGGCGGACAACAT |
| etbR | TCAAATCGTTCCCCAAAGTG |
| saeRF | CTGTAAATGGTCACGAAGT |
| saeRR | GACATTCACGGTATTAGCA |
| agrAF | GCAGTGAAATTCGTAAGCAT |
| agrAR | CGAGTTCTTAATTCTGCTGGA |
| sarAF | CGTAATGAGCATGATGAAAG |
| sarAR | ATTTCGTTGTTTGCTTCAG |
| sarSF | CCACCATAAATACCCTCAAACT |
| sarSR | GTCTTGCTGCGCGTCAT |
Sequence letters in boldface represent restriction sites.
Isolation of RNA and quantitative RT-PCR analysis of mRNA.
Strains were grown for 6 h in 3 ml TSB medium supplemented with the appropriate antibiotics at 37°C with shaking and subcultured in 30 ml fresh TSB medium adjusted to an initial optical density at 660 nm (OD660) of 0.02. Cultures were then incubated at 37°C with shaking and harvested by centrifugation 2, 3, 4, 5, and 6 h after inoculation. The cells were disrupted with the FastPrep instrument (Qbiogene, Carlsbad, CA) with glass beads. Total RNA was extracted with the FastRNA Pro Blue kit (Qbiogene) according to the manufacturer's protocol. After precipitation with ethanol, RNA was dissolved in diethyl pyrocarbonate (DEPC)-treated water, and the precipitates were washed in 70% ethanol and dried. The resultant RNA preparations were dissolved in DEPC-treated water. The RNA concentrations of the extracts were measured with a Nano Drop (Scrum, Inc.). Ten micrograms of total RNA was treated with reverse transcription (RT)-grade DNase (WaKo) for 30 min at 37°C to digest the remaining DNA, and cDNA synthesis was performed with a Transcriptor first strand cDNA synthesis kit (Roche) in a final volume of 20 μl. The resultant cDNA was diluted 5-fold with Tris-EDTA (TE) buffer and used as the template DNA for the subsequent quantitative PCR. Quantitative RT-PCR analysis was performed with the LightCycler instrument (Roche) using LightCycler FastStart DNA master SYBR green I (Roche) according to the instructions provided by the manufacturer. Gyrase B subunit mRNA (gyrB) was used as an internal standard. The oligonucleotides used in this study are listed in Table 2. The number of copies of each sample transcript was then quantified relative to the internal control, gyrB.
Construction of the temperature-sensitive shuttle vector for gene manipulation of S. aureus.
The temperature-sensitive plasmid in S. aureus, pKFT, was constructed as follows: the tetracycline resistance gene (tetL) was amplified from pHY300PLK with primers TetLF and TetLR and inserted into the SspI site of pUC18. A fragment carrying the p194Ets replicon was amplified from pCL52.1 (34) with the primers TsoriF and TsoriR. The resultant PCR product was digested with AatII and cloned into the AatII site of the resulting plasmid to obtain pKFT.
The temperature-sensitive plasmid in S. aureus pKFC was constructed as follows. A fragment carrying the p194Ets replicon was amplified from pCL52.1 (34) with primers TsoriF and TsoriR. The resultant PCR product was digested with AatII and cloned into the AatII site of pUC18. The chloramphenicol-resistant gene (cat) cassette was amplified from pCL15 (24) with the primers CMF and CMR and inserted into the NdeI site of the resulting plasmid to obtain pKFC.
Construction of mutant S. aureus strains. (i) Construction of the sarA deletion mutant.
A 2,171-bp DNA fragment containing the sarA locus was amplified from TY34 genome DNA with primers sarAF and sarAR. The resultant PCR product was digested with HindIII and subcloned into the same site in pUC19 to obtain pUC19sarA. A 654-bp EcoRV and Eco81I fragment in sarA was replaced by the cat gene cassette, and the resultant 2,332-bp HindIII fragment containing sarA::cat was transferred into the HindIII site of pKFT. The resulting plasmid, pFK3, was first transformed into S. aureus RN4220 (20), and then the modified plasmid was isolated and electroporated into strain TY34. Transformants were selected at 30°C on TSB plates containing chloramphenicol and tetracycline. A double-crossover disrupted mutant was generated by incubation at 42°C, a nonpermissive temperature for the replication of pKFT. The sarA mutant is shown to increase protease production (8); therefore, the mutant can be screened for high protease producers on TSB-containing chloramphenicol agar plates supplemented with 4% skim milk. The transformants were further selected as chloramphenicol-resistant and tetracycline-sensitive colonies. Deletion of the sarA gene was confirmed by PCR. The resultant sarA deletion mutant was designated FK128.
(ii) Construction of sigB, sarS, and TCS mutants by using Campbell-type integration.
Disruption mutants were constructed using a Campbell-type integration as described previously (19). Briefly, DNA fragments containing internal regions of each open reading frame (ORF) were amplified and cloned into the pKFT or pKFC vector. The primers used in this study are described in Table 2. In this step, we cloned the preceding gene of the two-component system (TCS) operon to construct the mutant in which TCS was destroyed. The resulting plasmids were then electroporated into S. aureus RN4220, and then each plasmid was extracted and electroporated into TY34. Transformants were selected at 30°C on TSB plates containing tetracycline or chloramphenicol. Each mutant was generated at 42°C and selected as a tetracycline- or chloramphenicol-resistant colony. The disruption of the target genes was confirmed by PCR. In the same way, sarA sarS (FK129), sarA saeR (FK132), and sarA agrC (FK133) double mutants were constructed from the sarA deletion mutant strain (FK128) (Table 1). Furthermore, sarS saeR (FK134) and sarS agrC (FK135) double mutants were constructed from the sarS mutant strain (FK130) with the pKFT construct.
(iii) Construction of the etb gene null mutant.
The etb gene is carried on a large plasmid, pETB (12, 39); therefore, we constructed the etb null mutant strain in TY825 through plasmid curing. S. aureus strain TY825 was grown in 10 ml TSB medium supplemented with 3 mg/ml ethidium bromide at 37°C with shaking and subcultured into 10 ml of the same fresh medium. The cultures were then plated on a TSB agar plate at 37°C. The pETB contains the etb gene and also a cadmium resistance gene; therefore, the pETB-cured strains were screened for cadmium-sensitive colonies and confirmed by PCR using etb primers. The resultant etb deletion mutant was designated FK200.
(iv) Complementation of isogenic sigB, sarA, and sarS mutants.
To complement the sigB (FK131), sarA (FK128), and sarS (FK130) isogenic mutant strains, we introduced the wild-type sigB, sarA, or sarS gene into the xylose-inducible expression vector pWH1520 vector (32). The plasmid pWH1520 was purchased from MoBiTec (Göttingen, Germany). The sigB, sarA, and sarS genes were amplified by PCR with the following primers: sigBFS and sigBRB2, sarAFS and sarARB2, sarSFS and sarSRB2, respectively (Table 2). These amplified DNA fragments were cut with SpeI and BglII and inserted into the same sites of pWH1520. These resulting plasmids, pFK22, pFK23, and pFK24, containing the sigB, sarA, and sarS genes, respectively, were first transformed into S. aureus RN4220 and selected as a tetracycline-resistant colony, and then modified plasmids were isolated and electroporated into the sigB (FK131), sarA (FK128), and sarS (FK130) isogenic mutant strains, respectively.
Western blot analysis.
Strains were grown with shaking at 37°C for 6 h in 3 ml TSB supplemented with the appropriate antibiotics and subcultured into 3 ml fresh TSB adjusted to an initial OD660 of 0.02. The cultures were then incubated with shaking at 37°C for 15 h, and the culture supernatants were harvested by centrifugation. Equal aliquots from each supernatant sample were electrophoresed with a 12% polyacrylamide gel. SDS-PAGE and Western blotting were performed as described previously (36, 40). Immunodetection of protein was performed with the ECL (enhanced chemiluminescence) Western blot analysis system (Amersham Pharmacia). The intensity of each band was measured with NIH Image 1.59 (National Institutes of Health).
ET bioassay.
ET activity was assayed with 2-day-old ICR newborn mice as described previously (40). Staphylococcus aureus was grown with shaking at 37°C for 6 h in TSB and washed twice with phosphate-buffered saline (PBS). The cells were suspended in PBS, and 100 μl containing 108 CFU of S. aureus was subcutaneously injected into the back of 2-day-old ICR neonatal mice (5/group). At intervals, the appearance of Nikolsky's sign (peeling of the skin upon slight rubbing) was monitored. These experiments were independently performed twice, and the data are shown as the total value. All of the animal experiments were approved by the Committee of the Institute of Laboratory Animal Science in Hiroshima University (A08-28).
RESULTS
Effect of the two-component signal transduction systems on ETA production in TY34.
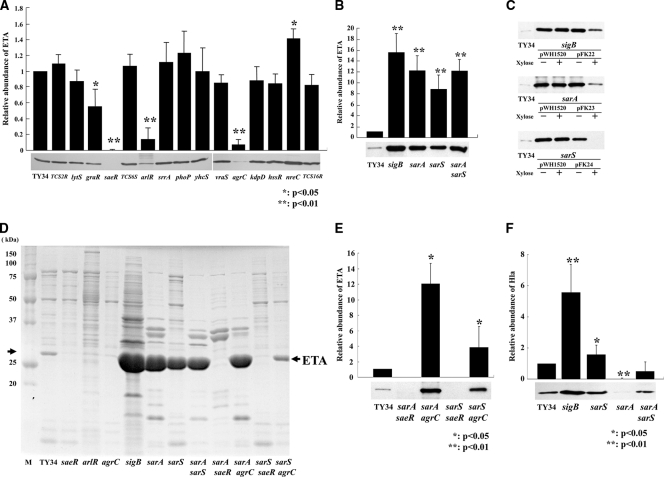
Staphylococcus aureus strain TY34 was isolated from a skin lesion of a patient with impetigo: this strain was eta positive, methicillin-resistant S. aureus (MRSA), and agr type III. To determine the regulation mechanism for ETA production, we constructed isogenic mutants of the two-component systems. Staphylococcus aureus encodes at least 16 two-component signal transduction systems (TCSs) (http://www.bio.nite.go.jp/dogan/project/view/MW2). We hypothesized any of the 16 TCSs identified may be the regulatory element for the ETs production. In particular, saeRS, arlRS, and agrCA were previously identified as global regulatory elements for a number of pathogenic factors in S. aureus (6, 28). In all TCSs, pairs of sensor genes and response regulator genes are arranged on the operon and are tandemly localized; therefore, we constructed gene disruption mutants of TY34 by using integration of the pKFT vector into the preceding gene in the TCS operon. The disrupted genes were TCS2R, lytS, graR, saeR, TCS6S, arlR, srrA, phoP, yhcS, vraS, agrC, kdpD, hssR, nreC, and TCS16R. The resulting isogenic mutants contained insertions in either the sensor or regulator gene, where the downstream gene in the TCS operon is not transcribed. In each mutant, inhibition of downstream gene transcription was confirmed by quantitative PCR. Insertion mutants were obtained for all of the TCSs with the exception of the walK/walR (vicRK) gene, which has been shown to be an essential gene for cell viability (25). The growth rates of the 15 TCS mutants were similar to that of TY34 (not shown). ETA production was not detectible in the saeRS mutant and greatly decreased in arlRS and agrCA mutants (reduced to 1/5 and 1/25 times that of the wild-type, TY34, respectively) (Fig. 1A and D). In the graRS mutant, the production of ETA was 1/2 of that of the wild type (Fig. 1A). The nreCB mutant showed a 1.4-fold increase in ETA production. The other 10 mutants did not show significant effects for ETA production.
FIG. 1.
Comparison of levels of ETA production in wild-type strain TY34 and its regulator mutants. (A) Western immunoblot analysis of ETA production in the wild-type strain, TY34, and its 15 isogenic TCS gene mutants. (B) Western immunoblot analysis of ETA production in the wild-type strain, TY34, and its sigB (FK131), sarA (FK128), sarS (FK130), and sarA sarS (FK129) isogenic mutants. (C) Western immunoblot analysis of ETA production in the wild-type strain, TY34, and the sigB (FK131) mutant containing pWH1520 or pFK22, the sarA (FK128) mutant containing pWH1520 or pFK23, and the sarS (FK130) mutant containing pWH1520 or pFK24. Strains were grown in the presence (+) or absence (−) of 1% xylose. (D) SDS-PAGE analyses of the wild-type strain TY34 extracellular proteins and those of its saeR (FK104), arlR (FK106), agrC (FK111), sigB (FK131), sarA (FK128), sarS (FK130), sarA sarS (FK129), sarA saeR (FK132), sarA agrC (FK133), sarS saeR (FK134), and sarS agrC (FK135) isogenic mutants. M, molecular mass markers. (E) Western immunoblot analysis of ETA production in the wild-type strain, TY34, and its sarA saeR (FK132), sarA agrC (FK133), sarS saeR (FK134), and sarS agrC (FK135) isogenic mutants. (F) Western blot analysis of Hla production of the wild-type strain, TY34, and the sigB (FK131), sarA (FK128), sarS (FK130), and sarA sarS (FK129) mutants. The cells were grown in TSB with shaking at 37°C for 15 h, and culture supernatants were harvested by centrifugation. The concentration of ETA or Hla was detected by Western immunoblotting with antiserum as described in Materials and Methods. The intensity of each band was measured with the program NIH Image and quantified relative to that of wild-type strain TY34. The data are means from four independent experiments. Error bars denote standard deviations.
Effect of sigB, sarA, and sarS mutations on ETA production in TY34.
Staphylococcus aureus expression of various pathogenic factors is known to be coordinately controlled by global regulatory elements: e.g., sigB, sarA, and sarS (4, 9, 28). To study the role of these global regulators affecting ETA production, we constructed a series of sigB, sarA, and sarS single mutants in TY34. The mutant cells showed similar growth rates to the wild type (Fig. 2D). The intensity of the 27-kDa protein corresponding to ETA was the major product in the extracellular protein that greatly increased in the sigB, sarA, and sarS single mutants compared to that in the wild type (Fig. 1D). Furthermore, Western blot analysis demonstrated the amounts of ETA production in the sigB, sarA, and sarS single mutants were approximately 15, 12, and 9 times higher, respectively, than that in the wild-type strain, TY34 (Fig. 1B).
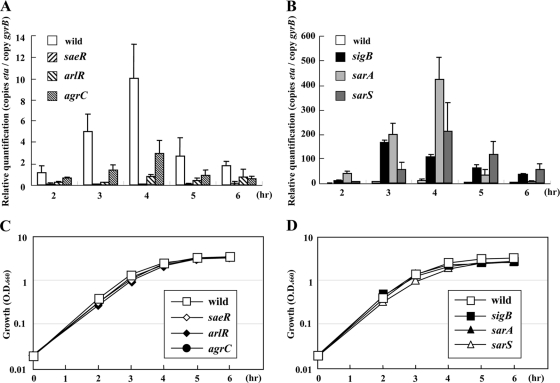
FIG. 2.
Quantitative transcript analysis of eta gene products from TY34 and its isogenic mutants compared to their growth curves. Staphylococcus aureus TY34 and its mutants were grown in TSB with shaking at 37°C. The cell growth was monitored by measuring the turbidity at 660 nm (C and D). The total RNA was extracted from the cultures at the time points shown. The expression level was measured by LightCycler RT-PCR as described in Materials and Methods and quantified relative to the internal control gyrB (A and B). The data presented are mean values from four independent RNA isolations. Error bars denote standard deviations. (A and C) Wild-type TY34 and the saeR (FK104), arlR (FK106), and agrC (FK111) mutants; (B and D) wild-type TY34 and the sigB (FK131), sarA (FK128), and sarS (FK130) mutants.
To complement the sigB, sarA, or sarS mutant strain, we cloned each gene under the control of an xylose-inducible promoter xylA in plasmid pWH1520 and transferred into a sigB, sarA, or sarS mutant strain. When the sigB, sarA, or sarS gene was induced by addition of xylose, ETA production greatly decreased in the sigB, sarA, or sarS mutant strain containing expression vector pFK22, pFK23, or pFK24, respectively, (Fig. 1C). These data further support the conclusion that SigB, SarA, and SarS negatively regulate ETA production.
The sigB mutant showed the highest increase in ETA production comparing these mutants (P < 0.05, sigB mutant versus sarS or sarA mutant). Since sarA and sarS possess a σB-dependent promoter in S. aureus, the negative effect of sigB may be partially attributed to inactivation of sarS and sarA (7, 37). To determine the relationship between these transcription factors, we constructed a sarA sarS double mutant and compared its ETA production to the single mutant. The amount of ETA produced by the sarA sarS double mutant was almost equal to that of the sarA single mutant, suggesting this is not a simple dual-regulation system through two transcriptional regulators, SarA and SarS (Fig. 1B and D).
The relationship between the positive regulators agr and saeR and the negative regulators sarA and sarS.
To evaluate the relationship between positive regulators agr and saeR and negative regulators sarA and sarS, we further constructed saeR sarA, saeR sarS, agr sarA, and agr sarS double mutants and analyzed the amount of ETA production. ETA production was not detectible in the saeR sarA and saeR sarS double mutants, as well as the saeR single mutant (Fig. 1D and E). Surprisingly, agr sarA and agr sarS mutants showed the highest increase in ETA production compared with the agr mutant. In particular, ETA production in the agr sarA mutant was similar level to the sarA single mutants (Fig. 1D and E). Thus, the introduction of sarA or sarS mutation into an agr mutant restored the ability to produce ETA.
Expression of the eta gene in TY34 and its isogenic saeRS, arlRS, agrCA, sigB, sarA, and sarS mutants.
To further verify the data from our Western blot experiments, we examined the transcription level of the eta gene by using LightCycler RT-PCR. Total RNA was extracted from the culture at several growth intervals after inoculation for 2, 3, 4, 5, and 6 h. The wild-type expression of the eta gene was elevated in the exponential phase (up to 4 h) and decreased during the early stationary phase (Fig. 2A). The eta expression was diminished in the saeR, arlR, or agrC mutants at all time points and significantly increased in the sigB, sarA, or sarS mutants (Fig. 2A and B). Using the sigB mutant, the expression level of eta reached a maximum at 3 h. This was 1 h earlier than the peak expression in the wild type or sarA or sarS mutant. Changes in eta expression were consistent with the Western blot data for ETA production (data not shown). These data suggest ETA production was regulated by these regulators at the transcriptional level.
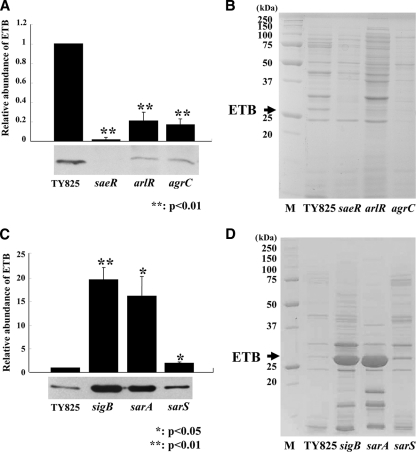
The effect of saeRS, arlRS, agrCA, sigB, sarA, and sarS mutations on ETB production in TY825.
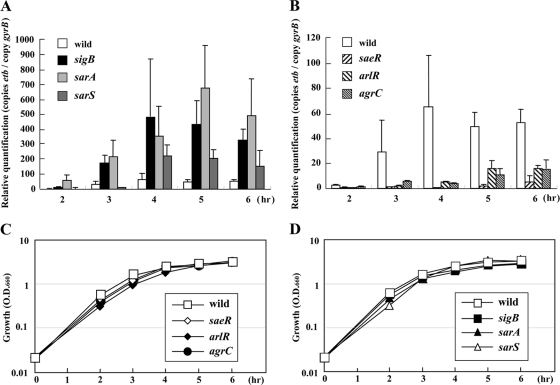
There are at least three serologically distinct exfoliative toxins (ETs), and ETB is one of the three major isoforms of ETs encoded on plasmid pETB (12, 39). To determine the regulatory mechanism for ETB production, we constructed a series of TCSs, sigB, sarS, and sarA mutants, in TY825. Staphylococcus aureus TY825 was isolated from a patient with impetigo. This strain is etb positive, methicillin-susceptible S. aureus, and agr type IV. Among TCS mutants, changes in the production of ETB were found in the saeR, arlR, and agrC mutants (Fig. 3A and B). The other 12 two-component system gene disruption mutants did not show any significant effect on ETB production (data not shown). The amount of ETB production decreased in the saeR, arlR, or agrC mutant at 1/100-, 1/5-, and 1/6-fold, respectively, compared to that in the wild type. We also investigated ETB production in the sigB, sarA, or sarS mutants of TY825. The 27-kDa protein corresponding to ETB is one of the major extracellular protein products that greatly increased in the sigB and sarA single mutants compared to the wild type (Fig. 3D). Western blot analysis demonstrated the levels of production of ETB in the sigB and sarA mutants were 20 times and 16 times, respectively, that of the wild type (Fig. 3C). Of note, unlike with ETA, the mutation in the sarS gene had little effect on ETB production (1.9 times that of the wild type) (Fig. 3C). We analyzed at the transcriptional level for the etb gene and found the expression of the etb gene was consistent with the Western blot data (Fig. 4A and B).
FIG. 3.
ETB production in TY825 and in its regulator mutants. (A and B) Western blot (A) and SDS-PAGE (B) analyses of ETB production in wild-type strain and its saeR (FK204), arlR (FK206), and agrC (FK211) isogenic mutants. M, molecular mass markers. (C and D) Western blot (C) and SDS-PAGE (D) analyses of ETB production in the wild-type strain and its sigB (FK216), sarA (FK218), and sarS (FK217) isogenic mutants. Cells were grown in TSB with shaking at 37°C for 15 h, and culture supernatants were harvested by centrifugation. The concentration of ETB was detected by Western immunoblotting with anti-ETB antiserum, as described in Materials and Methods. The intensity of each band was measured by NIH Image and quantified relative to that of the parental strain, TY825. The data presented are mean values from three independent experiments. Error bars denote standard deviations.
FIG. 4.
Quantitative transcript analysis from the etb gene of TY825 and its isogenic mutants compared to their growth curves. Cells were grown in TSB medium with shaking at 37°C. Cell growth was monitored by measuring turbidity at 660 nm (C and D). Total RNA was extracted from the cultures at the time points shown. The expression level was measured by LightCycler RT-PCR as described in Materials and Methods and quantified relative to that of the internal control gene gyrB (A and B). The data presented are mean values from four independent RNA isolations. Error bars denote standard deviations. (A and C) Wild-type strain TY825 and the saeR (FK204), arlR (FK206), and agrC (FK211) mutants; (B and D) wild-type strain TY825 and the sigB (FK216), sarA (FK218), and sarS (FK217) mutants.
The exfoliative activity of TY34 and TY825 and their derivative mutant strains using the in vivo neonatal mouse model.
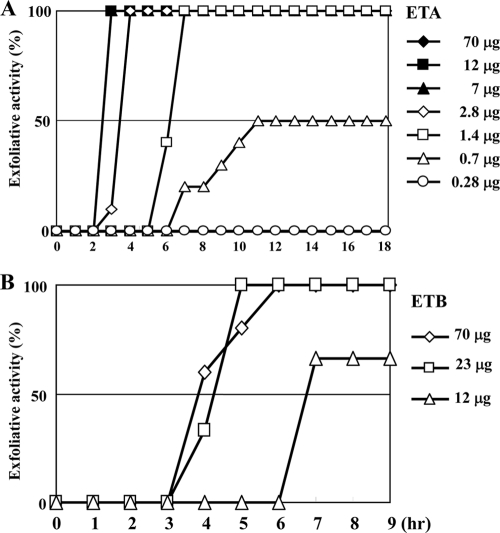
To further assess the effect of saeRS, arlRS, agrCA, sigB, sarS, and sarA on ETA or ETB production in a clinical setting, we used the neonatal mouse model. In S. aureus, the exfoliative toxin is the only toxin causing exfoliation of neonatal mouse skin. We first assessed the exfoliative activity of purified ETA and ETB by subcutaneous injection of the toxin at various concentrations into neonatal mice (Fig. 5). For purified ETA, concentrations of toxin over 1.4 μg caused exfoliation in all tested mice, and the onset of exfoliation was dose dependent (Fig. 5A). It took at least 3 h postinjection for the mice to show exfoliation even at the highest dose (70 μg). In contrast, purified ETB showed significantly lower exfoliation activity (Fig. 5B). At least 23 μg of toxin was necessary to cause exfoliation of neonatal mice skin.
FIG. 5.
Exfoliative activity with in vivo injection of S. aureus purified exfoliative toxin into neonatal mice. Two-day-old ICR neonatal mice (10 mice/group) were subcutaneously injected in the back with 100 μl PBS containing various concentrations of ETA (A) or ETB (B). At the intervals shown, the occurrence of Nikolsky's sign was monitored.
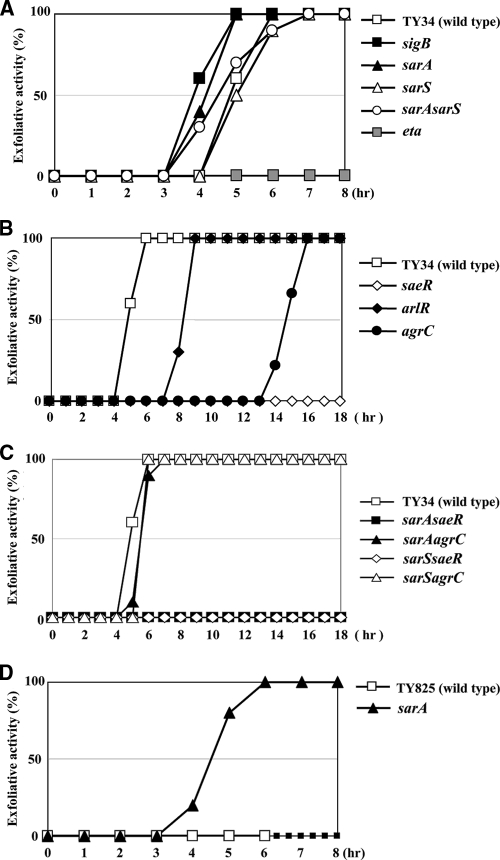
We administered approximately 108 CFU of S. aureus TY34 or its derivative mutants by subcutaneous injection into the skin of the back of neonatal mice and monitored exfoliation of the epidermis. All tested mice did not die during the experiment. Using the wild-type strain, TY34, the mice started to show exfoliation 5 h after injection, and all mice were positive after 7 h (Fig. 6A to C). Whereas, mice injected with the eta null mutant of TY34 did not show any exfoliation until 18 h postinjection (Fig. 6A). As expected from the in vitro experiments, exfoliation started 1 h earlier in mice injected with the sigB or sarS mutant compared to the wild type (Fig. 6A). Unexpectedly, the timing of exfoliation in mice injected with the sarA mutant was identical to that of the wild type, although ETA production in the sarA mutant was greatly increased in vitro. In mice injected with the saeR, arlR, or agrC mutant, exfoliation of the epidermis was markedly delayed, in good agreement with attenuated ETA production seen in the in vitro experiments (Fig. 6B). In particular, mice injected with the saeR mutant did not show any exfoliation during the test period. Similarly, mice injected with the sarA saeR or sarS saeR double mutant also did not show any exfoliation until 18 h. Whereas, mice injected with the sarA agrC or sarS agrC double mutant were similar to the wild type (Fig. 6C). We concluded ETA production was upregulated by saeRS, arlRS, and agrCA and was downregulated by sigB and sarS by the in vivo mouse model.
FIG. 6.
Exfoliative activity in vivo after injection of S. aureus mutants into neonatal mice. Two-day-old ICR neonatal mice (10 mice/group) were subcutaneously injected in the back with 100 μl PBS containing 108 CFU of S. aureus TY34 or the saeR (FK104), arlR (FK106), and agrC (FK111) isogenic mutants (A); the eta (TF5367), sigB (FK131), sarA (FK128), sarS (FK130), and sarA sarS (FK129) isogenic mutants (B); or the sarA saeR (FK132), sarA agrC (FK133), sarS saeR (FK134), and sarS agrC (FK135) isogenic mutants (C). Alternatively, mice were injected with 100 μl PBS containing 108 CFU of S. aureus TY825 and its sarA (FK218) isogenic mutant (D). At the intervals shown, the occurrence of Nikolsky's sign was monitored.
We further investigated the exfoliation activity of ETB-producing S. aureus. Injection of ETB-producing S. aureus TY825 did not induce exfoliation of the skin but caused death. The mice started to die 7 h after inoculation. The pETB-cured etb null mutant of TY825 also showed a similar symptom (Table 3). Mice injected with the sigB or sarS mutant started to die earlier, with symptoms of exfoliation of the skin at 5 to 6 h (Table 3). Of note, the mice injected with the sarA mutant of TY825 did not die; furthermore, the mice showed exfoliation of the skin after 4 h. After 6 h, all mice showed exfoliation of the skin (Fig. 6D). Mice injected with the saeR or agrC mutant did not die, and those injected with the saeR, arlR, or agrC mutant did not show symptoms of exfoliation of the skin (Table 3).
TABLE 3.
ET bioassay results
| Time (h) | No. of mice (n = 5/group) with result after injection witha: |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TY825 (wild type) |
etb mutant |
saeR mutant |
arlR mutant |
agrC mutant |
sigB mutant |
sarA mutant |
sarS mutant |
|||||||||
| Activity | Death | Activity | Death | Activity | Death | Activity | Death | Activity | Death | Activity | Death | Activity | Death | Activity | Death | |
| 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 4 | 0 | 0 | 0 |
| 6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 2 | 5 | 0 | 0 | 2 |
| 7 | 0 | 1 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 3 | 5 | 0 | 1 | 5 |
| 8 | 0 | 3 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | 0 | 2 | 3 | 5 | 0 | 1 | 5 |
Aliquots of bacterial cell suspension (108 bacteria ml−1) were administered subcutaneously to groups of 2-day-old ICR neonatal mice, and the occurrence of Nikolsky's sign (exfoliation activity) was monitored.
DISCUSSION
ETA production was upregulated by the SaeRS, ArlRS, and Agr system.
TCSs sense stimuli and respond to environmental conditions; therefore, the TCSs may be central elements for regulation of ETA production. We demonstrated ETA production was upregulated by the SaeRS, ArlRS, and Agr system by Western blotting and quantitative RT-PCR (Fig. 1A and 2A). Transcription analysis further showed the eta transcription was at the maximum level during the exponential phase and diminished in the early stationary phase. This indicates some regulatory elements may be involved in the activation of eta transcription to function primarily in the exponential growth phase (2 to 4 h).
The agr locus is known to play a central role in the regulation of pathogenic factors (29). We demonstrated that agr upregulated eta expression (Fig. 1A and 2A). However, eta expression started to decline during the early stationary phase (5 to 6 h), even though agrA expression was increasing (not shown). Our data suggest the production of ETA requires the agr system; however, full expression of agr is not necessary for eta transcription.
ETA production was strongly downregulated by SigB, SarA, and SarS.
We demonstrated ETA production was strongly downregulated by SigB, SarA, and SarS in TY34 (Fig. 1B and C and 2B). The eta transcript level of the sigB mutant was similar to that in the sarA or sarS mutants (Fig. 2B). Transcription analysis demonstrated the eta expression in the sigB mutant reached a maximum at 3 h, 1 h earlier than that in the wild type. At 3 h, the expression of saeR also reached a maximum where the sigB mutation increased the expression of saeR (not shown). In the sigB mutant, the expression of sarS was significantly attenuated at all times, whereas sarA expression was not affected until 3 h (not shown). Taken together, the data suggest the negative effect of SigB on eta expression was mediated by upregulation of sarS and downregulation of saeRS that was most apparent at 3 h. Conversely, the sarS mutation did not affect the transcription of any of the other regulators (e.g., saeRS, agr, and sarA) (data not shown). Additionally, the mutation in sarS did not cause a change in the exoprotein synthesis pattern except for ETA (Fig. 1D). The data suggest SarS directly regulates eta transcription.
Our data demonstrate the regulatory network for eta transcription by sigB, sarA, and sarS is very similar to hla expression (30). However, Western blot analysis demonstrated Hla production increased in the sigB and sarS mutants but is completely diminished in the sarA mutant (Fig. 1F). This suggests SarA controls the production of ETA and Hla through different mechanisms, and SigB, SarA, and SarS preferentially downregulate ETA production more strongly than other pathogenic factors such as Hla in TY34, although the eta gene is exogenously acquired on the genome of a temperate phage (38).
Relationship between the positive regulators agr and saeR and the negative regulators sarA and sarS.
We further constructed these saeR sarA, saeR sarS, agr sarA, and agr sarS double mutants and analyzed the concentration of ETA produced. ETA production in the saeR sarA or saeR sarS double mutant was not detectible as well as that in the saeR mutant, although the sarA or sarS mutant produces a large amount of ETA (Fig. 1D and E). These data strongly suggest saeRS is an essential TCS for ETA production as a major positive regulator. Conversely, ETA production in the agr sarA or agr sarS mutant is very similar to that in the sarA or sarS mutant, respectively, suggesting agr may not effectively function in the sarA or sarS mutant (Fig. 1D and E).
In vivo neonatal mouse model.
We investigated the regulation mechanism of eta by using the in vivo mouse model and demonstrated the difference between the in vitro and in vivo environments. Using the in vivo exfoliation assay, ETA production was suggested to be upregulated by saeR, arlR, and agrC and downregulated by sigB and sarS. This is in good agreement with the in vitro experiments (Fig. 1). In the in vivo mouse model, particularly saeR and -S are necessary TCSs for the production of ETA (Fig. 6B and C). An unknown key signal may induce ETA production via sensing by using these TCSs in vivo. The saeRS mutant completely lost exfoliative activity even in the sarA or sarS mutation genetic background. Reports show the regulation by saeRS is important in several animal models: e.g., the murine model for hematogenous pyelonephritis (16, 17, 31), where the promoter of saeRS is activated by H2O2 and subinhibitory concentrations of α-defensins (13). However, it is not known what signaling molecule(s) is actually involved in the regulation of ETA production in vivo. Further investigations are needed to understand the regulatory mechanism of ETA production. Unexpectedly, the sarA mutant showed identical exfoliative activity to that of the wild-type strain TY34, although it produced a large amount of ETA in in vitro experiments. This suggests sarA in TY34 may not work in vivo or some additional factors produced in the sarA mutant may inhibit the exfoliative activity.
The regulatory network for etb in vitro and in vivo.
We suggest the regulatory pathway for ETB production is similar to but distinct from that for ETA production. ETB production was upregulated by saeRS, arlRS, and agr and downregulated by SigB and SarA in TY825 in vitro (Fig. 3 and 4). Interestingly, the inactivation of sarS had little effect on ETB production, although etb expression increased in the sarS mutant (Fig. 3C and 4A). Thus, the negative effect of sigB on etb expression was predicted to be mediated primarily by sarA in TY825. We attempted to investigate the regulatory mechanism of etb by using the in vivo neonatal mouse model; however, administration of TY825 did not show exfoliation of the epidermis, possibly due to the low specific activity of ETB (Fig. 5B). Furthermore, the toxic effect of TY825 made it difficult to interpret the exfoliative activity of the mutant strains in vivo. The sarA mutant showed a significantly stronger exfoliative activity and didn't show a toxic effect (Fig. 6D and Table 3). Thus, in the in vivo environment, sarA appears to primarily downregulate ETB production. Conversely, in mice injected with the saeR, arlR, or agrC mutant, the ET did not cause death or exfoliation of the epidermis (Table 3). The data suggest saeRS, arlRS, and agr positively regulate ETB production, and the lethal factor(s) is also positively regulated by saeRS, arlRS, or agr and is not located on plasmid pETB.
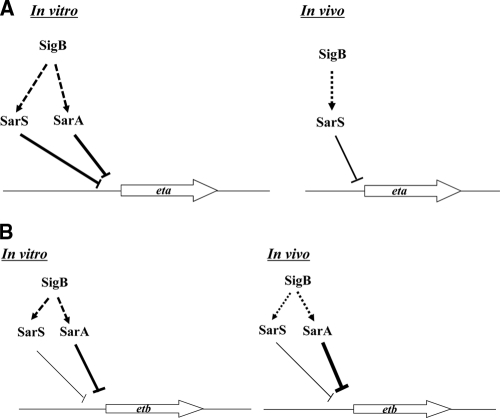
In conclusion, we examined the regulation pathway of eta and etb both in vitro and in vivo, observing the regulators saeRS, arlRS, agr, sigB, sarA, and sarS. The expression of eta and etb was positively regulated by common regulators saeR, arlR, and agrC, both in vitro and in vivo. However, they are downregulated differently. The expression of eta is negatively regulated by sarA and sarS, whereas etb expression is negatively regulated by sarA and slightly negatively regulated by sarS in vitro (Fig. 7A and B). In vivo, eta is negatively regulated by sarS, while etb is negatively regulated by sarA. Whether or not this difference in the regulation mechanisms of these ET genes' expression may affect virulence of S. aureus in blistering diseases remains to be determined.
FIG. 7.
Regulation model of SarA, SarS, and SigB for eta (A) and etb (B) expression in vitro and in vivo. Arrows indicate activation of gene expression. Repression is noted as bars. Dotted arrows and bars indicate presumed regulation.
Acknowledgments
We thank Tamaki Fujiwara for providing the eta null mutant strain. We also thank Jim Nelson for editorial assistance.
This study was supported in part by Grants-in-Aid for Scientific Research (F.K.) and Grants-in-Aid for Scientific Research for Priority Areas of “Applied Genomics” (M.S.) from the Ministry of Education, Culture, Sports, Sciences, and Technology of Japan.
Editor: J. B. Bliska
Footnotes
Published ahead of print on 31 January 2011.
REFERENCES
- 1.Amagai, M., N. Matsuyoshi, Z. H. Wang, C. Andl, and J. R. Stanley. 2000. Toxin in bullous impetigo and staphylococcal scalded-skin syndrome targets desmoglein 1. Nat. Med. 6:1275-1277. [DOI] [PubMed] [Google Scholar]
- 2.Amagai, M., et al. 2002. Staphylococcal exfoliative toxin B specifically cleaves desmoglein 1. J. Invest. Dermatol. 118:845-850. [DOI] [PubMed] [Google Scholar]
- 3.Bischoff, M., et al. 2004. Microarray-based analysis of the Staphylococcus aureus σB regulon. J. Bacteriol. 186:4085-4099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bronner, S., H. Monteil, and G. Prevost. 2004. Regulation of virulence determinants in Staphylococcus aureus complexity and applications. FEMS Microbiol. Rev. 28:183-200. [DOI] [PubMed] [Google Scholar]
- 5.Chan, P. F., S. J. Foster, E. Ingham, and M. O. Clements. 1998. The Staphylococcus aureus alternative sigma factor σB controls the environmental stress response but not starvation survival or pathogenicity in a mouse abscess model. J. Bacteriol. 180:6082-6089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheung, A. L., A. S. Bayer, G. Zhang, H. Gresham, and Y.-Q. Xiong. 2004. Regulation of virulence determinants in vitro and in vivo in Staphylococcus aureus. FEMS Immunol. Med. Microbiol. 40:1-9. [DOI] [PubMed] [Google Scholar]
- 7.Cheung, A. L., Y.-T. Chien, and A. S. Bayer. 1999. Hyperproduction of alpha-hemolysin in a sigB mutant is associated with elevated SarA expression in Staphylococcus aureus. Infect. Immun. 67:1331-1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheung, A. L., J. M. Koomey, C. A. Butler, S. Projan, and V. A. Fischetti. 1992. Regulation of exoprotein expression in Staphylococcus aureus by a locus (sar) distinct from agr. Proc. Natl. Acad. Sci. U. S. A. 89:6462-6466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheung, A. L., K. A. Nishina, M. P. T. Pous, and S. Tamber. 2008. The SarA protein family of Staphylococcus aureus. Int. J. Biochem. Cell Biol. 40:355-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deora, R., T. Tseng, and T. K. Misra. 1997. Alternative transcription factor σSB of Staphylococcus aureus: characterization and role in transcription of the global regulatory locus sar. J. Bacteriol. 179:6355-6359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fournier, B., and D. C. Hooper. 2000. A new two-component regulatory system involved in adhesion, autolysis, and extracellular proteolytic activity of Staphylococcus aureus. J. Bacteriol. 182:3955-3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Freer, J. H., and J. P. Arbuthnott. 1982. Toxins of Staphylococcus aureus. Pharmacol. Ther. 19:55-106. [DOI] [PubMed] [Google Scholar]
- 13.Geiger, T., C. Goerke, M. Mainiero, D. Kraus, and C. Wolz. 2008. The virulence regulator Sae of Staphylococcus aureus promoter activities and response to phagocytosis-related signals. J. Bacteriol. 190:3419-3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Giraudo, A. T., A. L. Cheung, and R. Nagel. 1997. The sae locus of Staphylococcus aureus controls exoprotein synthesis at the transcriptional level. Arch. Microbiol. 168:53-58. [DOI] [PubMed] [Google Scholar]
- 15.Giraudo, A. T., C. G. Raspanti, A. Calzolari, and R. Nagel. 1994. Characterization of a Tn551-mutant of Staphylococcus aureus defective in the production of several exoproteins. Can. J. Microbiol. 40:677-681. [DOI] [PubMed] [Google Scholar]
- 16.Goerke, C., et al. 2005. Role of Staphylococcus aureus global regulators sae and σB in virulence gene expression during device-related infection. Infect. Immun. 73:3415-3421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goerke, C., U. Fluckiger, A. Steinhuber, W. Zimmerli, and C. Wolz. 2001. Impact of the regulatory loci agr, sarA and sae of Staphylococcus aureus on the induction of α-toxin during device-related infection resolved by direct quantitative transcript analysis. Mol. Microbiol. 40:1439-1447. [DOI] [PubMed] [Google Scholar]
- 18.Horsburgh, M. J., et al. 2002. σB modulates virulence determinant expression and stress resistance: characterization of a functional rsbU strain derived from Staphylococcus 8325-4. J. Bacteriol. 184:5457-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Komatsuzawa, H., et al. 2004. The gate controlling cell wall synthesis in Staphylococcus aureus. Mol. Microbiol. 53:1221-1231. [DOI] [PubMed] [Google Scholar]
- 20.Kreiswirth, N., et al. 1983. The toxic syndrome exotoxin structural gene is not detectably transmitted by a phage. Nature 305:704-712. [DOI] [PubMed] [Google Scholar]
- 21.Kullik, I., P. Giachino, and T. Fuchs. 1998. Deletion of the alternative sigma factor σB in Staphylococcus aureus reveals its function as a global regulator of virulence genes. J. Bacteriol. 180:4814-4820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ladhani, S., C. L. Joannou, D. P. Lochrie, R. W. Evans, and S. M. Poston. 1999. Clinical, microbial, and biochemical aspects of the exfoliative toxins causing staphylococcal scalded-skin syndrome. Clin. Microbiol. Rev. 12:224-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liang, X., et al. 2005. Global regulation of gene expression by ArlRS, a two-component signal transduction regulatory system of Staphylococcus aureus. J. Bacteriol. 187:5486-5492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luong, T. T., and C. Y. Lee. 2006. The arl locus regulates Staphylococcus aureus type 5 capsule via mgr dependent and independent pathways. Microbiology 152:3123-3131. [DOI] [PubMed] [Google Scholar]
- 25.Martin, P. K., T. Li, D. Sun, D. P. Biek, and M. B. Schmid. 1999. Role in cell permeability of an essential two-component system in Staphylococcus aureus. J. Bacteriol. 181:3666-3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Melish, M. E., and L. A. Glasgow. 1970. The staphylococcal scalded-skin syndrome. N. Engl. J. Med. 282:1114-1119. [DOI] [PubMed] [Google Scholar]
- 27.Morikawa, K., et al. 2003. A new staphylococcal sigma factor in the conserved gene cassette: functional significance and implication for the evolutionary processes. Genes Cells 8:699-712. [DOI] [PubMed] [Google Scholar]
- 28.Novick, R. N. 2003. Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol. Microbiol. 48:1429-1449. [DOI] [PubMed] [Google Scholar]
- 29.Novick, R. N., and E. Geisinger. 2008. Quorum sensing in staphylococci. Annu. Rev. Genet. 42:541-564. [DOI] [PubMed] [Google Scholar]
- 30.Oscarsson, J., A. Kanth, K. Tegmark-Wisell, and S. Arvidson. 2006. SarA is a repressor of hla (α-hemolysin) transcription in Staphylococcus aureus: its apparent role as an activator of hla in the prototype strain NCTC 8325 depends on reduced expression of sarS. J. Bacteriol. 188:8526-8533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rampone, H., G. L. Martinez, A. T. Giraudo, A. Calzolari, and R. Nagel. 1996. In vivo expression of exoprotein synthesis with a Sae mutant of Staphylococcus aureus. Can. J. Vet. Res. 60:237-240. [PMC free article] [PubMed] [Google Scholar]
- 32.Rygus, T., and W. Hillen. 1991. Inducible high-level expression of heterologous genes in Bacillus megaterium using the regulatory elements of the xylose-utilization operon. Appl. Microbiol. Biotechnol. 35:594-599. [DOI] [PubMed] [Google Scholar]
- 33.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 34.Sau, S., S. Jiwen, and C. Y. Lee. 1997. Molecular characterization and transcriptional analysis of type 8 capsule genes in Staphylococcus aureus. J. Bacteriol. 179:1614-1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shaw, L. N., et al. 2008. Identification and characterization of sigma, a novel component of the Staphylococcus aureus and virulence responses. PLoS One 3:e3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sugai, M., et al. 1990. Purification of staphylococcal exfoliative toxin by high pressure liquid chromatography. Zentralbl. Bakteriol. 273:5-11. [DOI] [PubMed] [Google Scholar]
- 37.Tegmark, K., A. Karisson, and S. Arvidson. 2000. Identification and characterization of SarH1, a new global regulator of virulence gene expression in Staphylococcus aureus. Mol. Microbiol. 37:398-409. [DOI] [PubMed] [Google Scholar]
- 38.Yamaguchi, T., et al. 2000. Phage conversion of exfoliative toxin A production in Staphylococcus aureus. Mol. Microbiol. 38:694-705. [DOI] [PubMed] [Google Scholar]
- 39.Yamaguchi, T., et al. 2001. Complete nucleotide sequence of a Staphylococcus aureus exfoliative toxin B plasmid and identification of a novel ADP-ribosyltransferase, EDIN-C. Infect. Immun. 69:7760-7771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yamaguchi, T., et al. 2002. Identification of the Staphylococcus aureus etd pathogenicity island which encodes a novel exfoliative toxin, ETD, and EDIN-B. Infect. Immun. 70:5835-5845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yamasaki, O., et al. 2005. Clinical manifestations of staphylococcal scalded-skin syndrome depend on serotypes of exfoliative toxins. J. Clin. Microbiol. 43:1890-1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yamazaki, K., F. Kato, F. Kamio, and J. Kaneko. 2006. Expression of γ-hemolysin regulated by sae in Staphylococcus aureus strain Smith5R. FEMS Microbiol. Lett. 259:174-180. [DOI] [PubMed] [Google Scholar]