Abstract
Most bacteria with a rod-shaped morphology contain an actin-like cytoskeleton consisting of MreB polymers, which form helical spirals underneath the cytoplasmic membrane to direct peptidoglycan synthesis for the elongation of the cell wall. In contrast, MreB of Streptomyces coelicolor is not required for vegetative growth but has a role in sporulation. Besides MreB, S. coelicolor encodes two further MreB-like proteins, Mbl and SCO6166, whose function is unknown. Whereas MreB and Mbl are highly similar, SCO6166 is shorter, lacking the subdomains IB and IIB of actin-like proteins. Here, we showed that MreB and Mbl are not functionally redundant but cooperate in spore wall synthesis. Expression analysis by semiquantitative reverse transcription-PCR revealed distinct expression patterns. mreB and mbl are induced predominantly during morphological differentiation. In contrast, sco6166 is strongly expressed during vegetative growth but switched off during sporulation. All genes could be deleted without affecting viability. Even a ΔmreB Δmbl double mutant was viable. Δsco6166 had a wild-type phenotype. ΔmreB, Δmbl, and ΔmreB Δmbl produced swollen, prematurely germinating spores that were sensitive to various kinds of stress, suggesting a defect in spore wall integrity. During aerial mycelium formation, an Mbl-mCherry fusion protein colocalized with an MreB-enhanced green fluorescent protein (MreB-eGFP) fusion protein at the sporulation septa. Whereas MreB-eGFP localized properly in the Δmbl mutant, Mbl-mCherry localization depended on the presence of a functional MreB protein. Our results revealed that MreB and Mbl cooperate in the synthesis of the thickened spore wall, while SCO6166 has a nonessential function during vegetative growth.
The peptidoglycan (PG) layer, consisting of long glycan strands cross-linked by short peptides, is a major determinant of bacterial cell shape (51). Most species with a complex, nonspherical morphology contain the actin-like MreB protein, which belongs to the HSP70-actin-sugar kinase (ASHKA) superfamily of proteins (4, 48). MreB was shown to polymerize into a dynamic helical filament underneath the cytoplasmic membrane spanning the long axis of the cells (11, 17, 25, 30, 33). In rod-shaped bacteria, MreB is thought to interact with other proteins to position a cell wall-synthesizing complex at the lateral cell wall (16, 17, 32). The incorporation of new PG at the lateral wall results in cell elongation, thus determining rod-shaped morphology. Gram-negative bacteria seem to have a single mreB gene, usually in an operon with mreC and mreD. Gram-positive bacteria often encode three MreB-like proteins that show a considerable degree of similarity (>50%). All three mreB homologues of Bacillus subtilis, mreB, mbl, and mreBH, have an important role in cell shape determination. Whereas mreB and mbl are essential under normal growth conditions and mutants could survive only after supplementation with 3 mM Mg2+ (18, 25, 27), mreBH inactivation was less severe (8). B. subtilis mutants that are depleted for or defective in a single MreB homologue differed slightly in their phenotype. mreB mutants were straight with an increased diameter, suggesting that MreB controls cell width. mbl mutants were twisted, indicating that Mbl controls the linear axis (27). MreBH probably regulates autolytic activity, since it interacts with the cell wall hydrolase LytE (8). Nevertheless, the three mreB homologues are able to partially complement the defects of the single mutants when overexpressed (27, 44). Also, the three MreB homologues colocalize to the same helical structure in the cell (7, 8). From the phenotype of the mutants and the localization pattern, it was concluded that the three MreB homologues in particular were required to maintain growth and cell shape under stress conditions (27). Besides positioning the lateral wall-synthesizing complex, the MreB cytoskeleton seems to be involved in many other cellular processes, such as the positioning of the DNA replication machinery for proper chromosome segregation during cell division and localizing other proteins to distinct sites (12, 13, 14, 28, 34).
In contrast to the rod-shaped bacteria that depend on MreB proteins to control their cell wall assembly, many Gram-positive bacteria of the phylum Actinobacteria grow in a different way by building their cell walls at the cell poles (10). Corynebacteria and mycobacteria do not contain mreB genes, yet they acquire rod shape by polarized growth, and this depends on the coiled-coil protein DivIVA (35). Similarly, streptomycetes do not divide by binary fission and grow by apical tip extension to form a multiply branching mycelium (22).
Against this background, it was a surprise that an mreB cluster is present in S. coelicolor (6). It was later shown that mreB was dispensable for the apical growth of vegetative hyphae, and loss affected the assembly of the spore wall (37). However, like B. subtilis, S. coelicolor encodes three MreB homologous proteins, MreB (SCO2611), Mbl (SCO2451), and SCO6166, and it could not be excluded that the dispensability of MreB for vegetative growth was due to some redundancy among these proteins. In this work, we characterized the role of the three S. coelicolor MreB-like proteins. We analyzed their expression profile, generated mutants, and localized Mbl and MreB fusion proteins in the wild-type and different mutant backgrounds. We report that the three MreB homologues of S. coelicolor have clearly different functions. Whereas MreB and Mbl cooperate in spore wall synthesis, SCO6166 has a nonessential function during vegetative growth.
MATERIALS AND METHODS
Bacterial strains and media.
Strains and plasmids used in this study are listed in Table 1 . S. coelicolor strains were cultivated on mannitol soya flour (MS) agar plates or in S medium (29). The cultivation of strains and procedures for DNA manipulation were performed as previously described for Escherichia coli (43) and S. coelicolor (29). Oligonucleotides used for reverse transcription-PCR (RT-PCR) and subcloning are given in Tables 2 and 3, respectively.
TABLE 1.
Strains and plasmids
| Strain/plasmid | Relevant characteristic(s) | Reference or source |
|---|---|---|
| E. coli Xl1 blue | F′::Tn10 proA+B+lacIqsupE44 Δ(lacZ)M15 hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | 5 |
| E. coli ET 12567 | GM2929 zjj-202, mutant hsdM, mutant hsdR | 36 |
| E. coli BTH101 | F−cya-99 araD139 galE15 galK16 rpsL1 (Strr) hsdR2 mcrA1 mcrB1 | 26 |
| E. coli BW 25113/pIJ790 | Δ(araD-araB)567 ΔlacZ4787(::rrnB-4) lacIp-4000(lacIq) λ−1rpoS369(Am) rph-1 Δ(rhaD-rhaB)568 hsd514; pIJ790:[oriR101] [repA101(ts)] araBp-gam-beta-exo | 21 |
| S. coelicolor M145 | Prototrophic, SCP1− SCP2− | 29 |
| ΔmreB | mreB replacement mutant of M145 (mreB-IFD) | 37 |
| ΔmreB::pPM6 [mreB-IFDc] | mreB replacement mutant of M145 (mreB-IFD) complemented by the integration of pPM6, aac | 37 |
| Δmbl | mbl replacement mutant of M145 | Present study |
| ΔmreB Δmbl | mreB replacement mutant of M145, mbl replaced by an aac cassette | Present study |
| Δsco6166 | sco6166 replacement mutant of M145 | Present study |
| Δpbp2 | pbp2 replacement mutant of M145 | Kleinschnitz et al., unpublished |
| ΔmreB::pPM4 [SCPM6] | mreB replacement mutant of M145 (mreB-IFD), complemented by the integration of pPM4, aac | 37 |
| Δpbp2::pPM4 | pbp2 replacement mutant of M145 with integration of pPM4, aac | Present study |
| Δmbl::pPM4 | mbl replacement mutant of M145 with integration of pPM4, aac | Present study |
| M145::pAH5 | pK18-mbl-mcherry integrated into mbl of M145, aphII | Present study |
| ΔmreB::pAH5 | mreB replacement mutant of M145 (mreB-IFD), with pAH5 integrated into mbl, aphII | Present study |
| Δpbp2::pAH5 | pbp2 replacement mutant of M145 with integration of pAH5, aphII | Present study |
| pK18 | aphII, lacZα | 40 |
| C24 | aphII, S. coelicolor chromosomal fragment encoding SCO2431-SCO2461 | 41 |
| pTST101 | bla, malE-egfp fusion | J. Altenbuchner, personal communication |
| pTST101-mcherry | bla, malE-mcherry fusion | G. Muth, unpublished |
| pPM4 | pSET152 derivative, mreB-egfp fusion | 37 |
| pUZ8002 | aphII, RP4 transfer region | 29 |
| pCP20 | [repA101(ts)], bla, frt | 9 |
| pIJ773 | bla, aac, oriT | 21 |
| pKT25 | aph, cya-T25 | 26 |
| pUT18c | bla, cya-T18 | 26 |
| pKO6166 | pK18 derivative, aphII, sco6166 deletion vector | Present study |
| pAH4 | pK18, aphII, mbl-egfp fusion | Present study |
| pAH5 | pK18, aphII, mbl-mcherry fusion | Present study |
| M145::pAH5-pPM4 | pAH5 integrated into mbl of M145, integration of pPM4, aphII, aac | Present study |
| pGM202-Mbl | pGM190 derivative, aphII, tsr, PtipA, encoding a Mbl-His6 fusion protein | Present study |
TABLE 2.
Oligonucleotide primers used for RT-PCR
| Primer | Sequence (5′→ 3′) | Fragment size (bp) |
|---|---|---|
| RThrdBleft3 | GGTCGAGGTCATCAACAAGC | 205 |
| RThrdBrig3 | CTCGATGAGGTCACCGAACT | |
| RT-mreBf | GGGGAACTCAATGTCGTTCA | 174 |
| RT-mreBr | CCGATCATCTTCTTCGCTTC | |
| RT-mblleft4 | CATGCTGCGTCATCTGCT | 172 |
| RT-mblright4 | GATGAGCGTGTCGACGAGT | |
| RT-6166f | ATCTCACGGAGGTGGTGCT | 140 |
| RT-6166r | AGCATCGAGGTCGTCATGT |
TABLE 3.
Oligonucleotides used for cloning experiments
| Primer | Sequencea (5′→3′) |
|---|---|
| Ecoupfw | AAGAATTCTGATCTCCTGCGCCTG |
| uprchBgBc | TCATGATCAGATCTCACGGTCATCGTCCG |
| loupchBcBg | GTGAGATCTGATCATGATCCGGTACCGGG |
| Hlorev | GGAAGCTTGACAGCGTGAGAGAC |
| mblupNde | GGCATATGACCGCCAGTACTG |
| mblloBg-gfp | GGAGATCTGTCGGAGTCGGCGTG |
| 6166upNde | GGCATATGACCGTGCTCCGG |
| 6166loBg | CCAGATCTGTGCGGGTGGGTGTC |
| mblredup | GGCCCCGCACGGTCCGCGCTCTCGGGAGGATTCGCCATG GATATCATTCCGGGGATCCGTCGACC |
| mblredrev | TGCGTCAGGCTTCCCCGGTCGCCGCCCGGCGGCGGATCA GATATCTGTAGGCTGGAGCTGCTTC |
| loupchBcBg | GTGAGATCTGATCATGATCCGGTACCGGG |
| Hlorev | GGAAGCTTGACAGCGTGAGAGAC |
| CHSTEup2 | AGGATTCGCCATGGAATTCCACGCCGACTCCGACTG |
| HKHrev2 | CCAAGCTTGGCGGTCGGGTGCAGG |
| mreBXba | AATCTAGAGAACGACATTGAGTTC |
| mreBK | TCGGTACCGATCTACGGGGCGAGGC |
| mblXba | AATCTAGAGTATTGGCAGTACTG |
| mblK | CTGGTACCCAGTCGGAGTCGGCGTG |
| 6166Xba | ACTCTAGAGACCGTGCTCCGGCGG |
| 6166K | AAGGTACCCAGTGCGGGTGGGTGTC |
| rodZXba | ACTCTAGAGTCCAACGGCAAATCC |
| rodZK | TTGGTACCCATCCGACCTGCGGGT |
| mreCXba | ACTCTAGAGAGGGACACGAAAGAG |
| mreCK | AAGGTACCACGGCTCGGGCTGCTC |
| mreDXba | ACTCTAGAGCGCGTCAACCGGATC |
| mreDK | AAGGTACCACAGCCGCTTGACCCC |
| pbp2Xba | ACTCTAGAGACCAACATCCCCGAG |
| pbp2K | AAGGTACCATACGCGGTGCCCTCC |
| sfrXba | ACTCTAGAGACCGGCAACAGCTTC |
| sfrK | AAGGTACCACGCCGACATCGGTCT |
Restriction sites are underlined.
Inactivation of mbl and sco6166.
Mbl was deleted by the PCR targeting (21) of cosmid C24 (41). Following the λ red-mediated replacement of mbl by a PCR-amplified aac(3)IV cassette (primer pair mblredup/mblredrev), the resulting cosmid, C24-aac, was introduced into M145 by intergeneric conjugation, and an mbl replacement mutant, Δmbl-aac (double crossover), was selected.
Subsequently, the aac(3)IV cassette was deleted from cosmid C24-aac by FLP recombination target (FRT) recombination using E. coli BT340 (9), and the resulting cosmid was introduced into the S. coelicolor mutant Δmbl-aac. Finally, apramycin-sensitive Δmbl colonies were selected in which mbl was replaced by a remaining 81-bp scar sequence. The ΔmreB Δmbl double mutant was generated by introducing cosmid C24-aac in the ΔmreB mutant (37) and screening for apramycin-resistant and kanamycin-sensitive colonies. Note that the ΔmreB Δmbl mutant still contains the aac(3)IV gene.
To delete sco6166, a 1.6-kb upstream fragment (primer pair Ecoupfw/uprchBgBc), including the start codon of sco6166, and a 1.6-kb downstream fragment (primer pair loupchBcBg/Hlorev), including the sco6166 stop codon, were amplified by PCR and cloned into pK18, yielding pKO1666. After the transformation of M145 and the integration of the deletion vector pKO6166 by a single crossover (kanamycin resistant), an sco6166 mutant was isolated by selecting for the second crossover (kanamycin sensitive). Correct gene replacement was confirmed by PCR analyses and Southern blotting. Hybridization probes were amplified with primers CHSTEup2/HKHrev2 and loupchBcBg/Hlorev.
Construction of gfp and mcherry fusions.
To construct a C-terminal fusion with the enhanced green fluorescent protein (eGFP) or mCherry protein, mbl and sco6166 genes were amplified using the primers mblupNde/mblloBg-gfp and 6166upNde/6166loBg, respectively, cut with NdeI and BglII, and fused to the egfp or mcherry gene in plasmids pTST101 (J. Altenbuchner, personal communication) or pTST101mCherry (G. Muth, unpublished data) cut with NdeI and BamHI. The resulting fusion genes were cut from pTST101 and cloned in the nonreplicative vector pK18, generating pK18-Mbl-eGFP (pAH4), pK18-Mbl-mCherry (pAH5), and pK18-6166-mCherry. The plasmids then were integrated into chromosomal mbl or sco6166 genes of S. coelicolor M145 via homologous recombination.
For colocalization experiments, the plasmid pPM4 (37), carrying an mreB-egfp fusion gene, was integrated into the ΦC31 att site of M145::pAH5.
RNA extraction.
Total RNA was isolated from wild-type M145 grown for 48 and 72 h on LB agar or 48, 72, and 96 h on MS agar overlaid with cellophane discs. Cells were harvested, resuspended in RNAprotect bacteria reagent (Qiagen), centrifuged, and resuspended in Tris-EDTA (TE) buffer. Cell disruption was performed with glass beads (0.45 to 0.5 mm) and a Precellys homogenizer from Peqlab (for substrate mycelium, 2× 6,500 rpm for 30 s; for aerial mycelium and spores, 4× 6,500 rpm for 30 s). RNA was isolated with an RNeasy kit from Qiagen as described previously (46). The RNA samples were resuspended in H20 and quantified spectrophotometrically at 260 nm with a Nanodrop photometer (Peqlab). All RNA isolates had an optical density at 260 nm (OD260)/OD280 ratio between 1.8 and 2.0, indicating clean RNA isolates. The RNA quality also was checked by 1.0% agarose gel electrophoresis and ethidium bromide staining. To exclude the contamination of the RNA with DNA, a control PCR was done with primers for the housekeeping gene hrdB (RNA polymerase principal sigma factor HrdB; SCO5820).
Semiquantitative RT-PCR analysis.
A two-step semiquantitative RT-PCR method was used to measure gene expression during the morphological differentiation of M145. RNA was isolated from two independently grown cultures, and RT-PCR was repeated twice. cDNA synthesis was performed as described in reference 46 using 3 μg of RNA. Sixty ng of cDNA was used for RT-PCR. The housekeeping gene hrdB was used as an internal control. The optimal PCR annealing temperature of all primers was 62°C. Primer sequences are listed in Table 2. PCRs were performed with the Bio-Rad MJ mini personal thermal cycler using 60 ng of cDNA, 400 nM each primer, 200 μM deoxynucleoside triphosphates (dNTPs), 1× polymerase buffer (Qiagen), and 1 U Taq polymerase (Qiagen) in a 25-μl volume. All reactions were done with the same program: 95°C for 2 min, and then 28 cycles of 95°C for 1 min, 62°C for 1 min, and 72°C for 30 s, followed by an additional 7-min extension step at 72°C. The RT-PCR samples were loaded on a 2% agarose gel and stained with ethidium bromide.
Bacterial two-hybrid analyses.
To detect protein interactions, respective S. coelicolor genes were amplified with primers (listed in Table 3) containing XbaI and KpnI sites. Subsequently, PCR fragments were cloned as XbaI/KpnI fragments into plasmids pKT25, pUT18, and pUT18c to generate translational fusions with the catalytic domains of the Bordetella pertussis adenylate cyclase (26). The E. coli cya mutant BTH101 was cotransformed with pUT18c and pKT25 derivatives and spotted on 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) plates. The ability of cotransformants to metabolize X-Gal, resulting in blue color, is based on a functional adenylate cyclase due to the interaction of the fusion proteins.
Phase-contrast microscopy.
For light and fluorescence microscopy of substrate mycelium, aerial mycelium and spore chains cultures were plated on MS agar, and sterile coverslips were inserted at a 45° angle into the agar. Coverslips were removed after 2 to 4 days of incubation at 30°C and mounted on slides coated with phosphate-buffered saline (PBS) with 1% agarose. For the phenotypic characterization of the mutants, pictures were taken with an Olympus system microscope BX60 equipped with an F-view II camera (Olympus). Fluorescent microscopy was done with a Zeiss DM5500B microscope equipped with a Leica DFC360FX camera.
For colocalization studies, the samples were viewed using a Zeiss Axio Imager.Z1 microscope equipped with X-Cite 120 illumination (EXFO Photonic Solution, Inc.), and images were captured and processed using a 9100-02 electron multiplier charge-coupled device camera (Hamamatsu Photonics) and Volocity 3DM software (Improvision).
Spore size measurements.
The wild-type M145 and the mutants Δmbl, ΔmreB Δmbl, and Δsco6166 were inoculated on MS agar. Coverslips were inserted and removed after 4 days of inoculation and observed with an Olympus system BX60 microscope. Average values (length and width, in μm) of 200 spores of M145, Δmbl, ΔmreB, ΔmreB Δmbl, Δsco6166, and Δmbl(pGM202-Mbl) were measured with AnalySIS software (Olympus).
Transmission electron microscopy.
S. coelicolor strains were grown on MS agar for 5 days. Spores were harvested and fixed with 2% glutaraldehyde, embedded in 2% agarose, and treated with 1% OsO4 in 0.1 M phosphate buffer on ice for 30 min. Subsequently, the samples were dehydrated with increasing concentrations of ethanol, infiltrated by ethanol:EPON (2:1 to 1:2 ratio), and embedded in pure EPON. Ultrathin sections were stained with uranylacetate and lead citrate. The samples were examined with a Phillips Tecnai electron microscope at 80 kV.
Analyses of heat, salt, lysozyme, and vancomycin resistance.
Heat resistance was assayed by incubating spores (106/ml) for 30 min at 30 and 60°C. Subsequently, 5 μl of the spore samples was spotted onto LB agar and incubated for 3 days at 30°C. To analyze salt resistance, serial dilutions (H2O) of spore suspensions (titer, 108 to 109/ml) were plated on LB agar with 6 and 0.5% NaCl, respectively. Plates were cultivated for 2 days at 30°C. The survival rate was calculated as the percentage of the colony titer on plates with 6% NaCl divided by the titer on plates with 0.5% NaCl. Resistance to cell wall-damaging agents was assayed by plating spores (105) of the different mutants and the wild type on LB agar and applying filter discs containing 50 μg lysozyme or 5 μg vancomycin. Plates were inoculated for 2 days at 30°C before being photographed.
Bioinformatic analyses.
HHpred (45), a remote homology detection method based on the comparison of profile-hidden Markov models, was used to search the Protein Data Bank (PDB) (3) for homologues of known structure. Searches with MreB and Mbl from S. coelicolor retrieved the rod shape-determining protein MreB from T. maritima (PDB accession no. 1jce) (48) as the closest homolog of known structure. For the MreB-like protein SCO6166, the ethanol utilization protein EutJ from Carboxydothermus hydrogenoformans (3h1q) had the highest score.
Homology models of the three MreB-like proteins of S. coelicolor were generated by Modeler (42) using HHpred alignments to the best hit as a template. The models were superimposed interactively in Swiss-PDB Viewer (20). Molecular structures were rendered in PyMol (www.pymol.org).
RESULTS
S. coelicolor has three mreB-like genes.
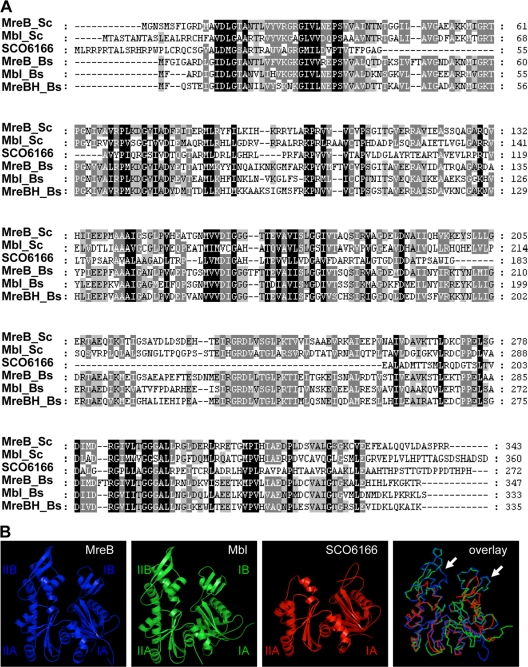
S. coelicolor contains three genes encoding MreB-like proteins. mreB is located in a cluster with the other rod shape-determining genes mreC, mreD, pbp2, and sfr (6). The second mreB homologue, mbl, lies between two genes encoding protein kinases. MreB and Mbl have sizes of 36.5 and 37.5 kDa, respectively, show a similarity of 42% (identical amino acids), and can be aligned over the whole length (Fig. 1A). In contrast, the third MreB-like protein, SCO6166, is shorter and has a molecular size of 28.7 kDa. sco6166 is translationally coupled to sco6165, encoding a 121-amino-acid (aa) protein homologous to the DnaK suppressor DksA of E. coli and TR:O83134 (EMBL:AE001194) from Treponema pallidum. The modeling of SCO6166 on the template of Thermotoga maritima MreB showed that SCO6166 lacks the subdomains IB and IIB of actin-like proteins (Fig. 1B). Whereas homologues of MreB and Mbl are present in all streptomycetes, some strains, such as S. avermitilis (NC_003155.4) or S. viridochromogenes (NZ_ACEZ00000000), do not contain a homologue of SCO6166, indicating a nonessential function for SCO6166.
FIG. 1.
Similarity of the three S. coelicolor MreB homologues. The MreB homologues of S. coelicolor and B. subtilis were compared by ClustalW sequence alignment (A) and homology modeling (B) using MreB of Thermotoga maritima as a template. MreB and Mbl are highly similar to the B. subtilis MreB homologues and have an identical structure; SCO6166 lacks subdomains IB and IIB (arrows). The colors in the superposition (lower right panel) correspond to the colors chosen for the individual homology models. MreB is given in blue, Mbl in green, and SCO6166 in red.
The three mreB-like genes have different expression profiles.
Previous S1 mapping experiments identified three promoters upstream of mreB, one strongly induced during the morphological differentiation of S. coelicolor (6). To characterize the expression profile of the other mreB homologues, we performed semiquantitative RT-PCR experiments with RNA isolated from surface-grown cultures at different time points (Fig. 2). After 48 and 72 h on LB agar, when only substrate mycelium was formed, mreB was expressed but a mbl transcript could not be detected; sco6166 showed the strongest expression. This was in agreement with detailed microarray data from S. coelicolor liquid cultures (38) showing the strong expression of sco6166, intermediate expression of mreB, and only very weak expression of mbl during a period of 40 h (K. Nieselt, personal communication). After 48 h on MS agar, when aerial mycelium already was developed, mreB transcription increased and mbl transcription was detectable at a low level. In contrast, sco6166 expression strongly decreased (Fig. 2). After 72 and 96 h on MS agar, when spore chains have been formed in the aerial mycelium, the transcription of both mreB and mbl further increased, while sco6166 expression was no longer detectable. The expression profile suggested a role for mreB and mbl in sporulation, whereas the third mreB-like gene, sco6166, seems to have a function in vegetatively growing mycelium.
FIG. 2.
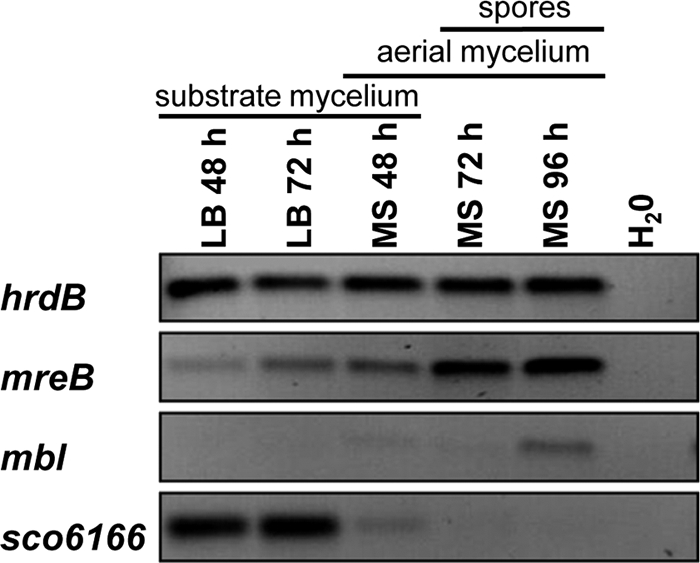
Transcriptional analysis of the mreB-like genes of S. coelicolor by semiquantitative RT-PCR. Cultures of S. coelicolor M145 were grown on cellophane discs on LB agar or MS agar for different time periods. The stage of morphological differentiation is indicated for the different time points. Following RNA isolation and cDNA synthesis, the amounts of transcripts were compared by PCR. The expression of hrdB served as an internal control. The expression of mreB and mbl increased during morphological differentiation. In contrast, sco6166 is highly expressed in vegetative mycelia but no longer is transcribed after the onset of sporulation.
A ΔmreB Δmbl double mutant is viable.
In contrast to B. subtilis, where mreB and mbl are essential under normal growth conditions and mutants require high concentrations of Mg2+ for survival (18, 25), the inactivation of mreB in S. coelicolor was tolerated (37). The phenotype of the ΔmreB (mreB-IFD) mutant suggested a defect in spore wall synthesis. To analyze whether the other mreB-like proteins also are involved in spore wall synthesis, we inactivated mbl and sco6166. mbl was replaced by PCR targeting as described in Materials and Methods and in Fig. S1 in the supplemental material, while sco6166 was deleted from the chromosome by homologous recombination using a nonreplicative deletion vector (see Fig S2). We also deleted mbl in the ΔmreB mutant (37), generating a ΔmreB Δmbl double mutant. All mutants were isolated readily, demonstrating that none of the mreB homologues had an essential role for viability. Even the growth of the ΔmreB Δmbl double mutant was not affected under standard growth conditions.
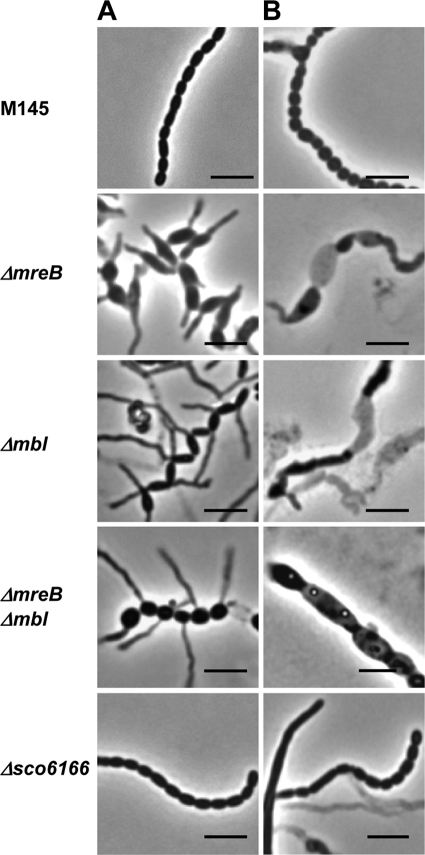
Δmbl and ΔmreB Δmbl had phenotypes very similar to that of the ΔmreB mutant, with swollen, prematurely germinating spores (Fig. 3). Whereas S. coelicolor M145 spores had a size of 1.28 by 0.71 μm, Δmbl and Δmbl ΔmreB spores were 1.72 by 0.95 and 1.7 by 1.15 μm, respectively (Table 4). The mutant phenotype was more severe when the mutants were grown on MS agar supplemented with 10.3% sucrose, as also reported for ftsI and ftsW mutants (2). Under these conditions, ΔmreB, Δmbl, or ΔmreB Δmbl did not sporulate (see Fig. S3 in the supplemental material). Microscopic analyses showed that large areas of the aerial mycelium lysed, and aberrant spore-like particles were formed (Fig. 3B).
FIG. 3.
Aberrant spore phenotype of S. coelicolor mutants defective in mreB-like genes. Phase-contrast microscopy of spore chains from strains grown for 4 days on MS agar (A) and MS agar plus 10.3% sucrose (B), respectively. Whereas the vegetative mycelium was not affected, ΔmreB, Δmbl, and ΔmreB Δmbl formed swollen spores (sizes are given in Table 4) that frequently germinate prematurely. In contrast, Δsco6166 spores had a wild-type appearance. On MS agar containing 10.3% sucrose, large areas of the aerial mycelium of the ΔmreB, Δmbl, and ΔmreB Δmbl mutants were lysed, and aberrant spore-like particles were formed (B). Bar, 4 μm.
TABLE 4.
Spore dimensionsa of S. coelicolor and mutants deficient in mreB homologous genes
| Strain | Length (μm) | Width (μm) |
|---|---|---|
| M145 | 1.28 ± 0.20 | 0.71 ± 0.07 |
| M145 (pGM190) | 1.20 ± 0.15 | 0.64 ± 0.06 |
| ΔmreB [mreB-IFD]b | 1.64 ± 0.24 | 1.16 ± 0.21 |
| ΔmreB::pPM6 [IFDc]b | 1.23 ± 0.27 | 0.84 ± 0.14 |
| Δmbl | 1.72 ± 0.38 | 0.95 ± 0.16 |
| Δmbl (pGM202-Mbl) | 1.31 ± 0.18 | 0.67 ± 0.08 |
| ΔmreB Δmbl | 1.70 ± 0.33 | 1.15 ± 0.11 |
| Δsco6166 | 1.31 ± 0.25 | 0.78 ± 0.08 |
n = 200.
From reference 38.
By immunogold labeling, MreB has been localized to the inner spore wall, suggesting a role in spore wall synthesis (37). This was confirmed by electron microscopic images showing aberrant ΔmreB spores whose spore wall integrity was affected. To demonstrate the involvement of Mbl in spore wall synthesis, we performed electron microscopic studies. These images clearly revealed a distorted spore wall (see Fig. S4 in the supplemental material). Whereas M145 formed uniform round or ellipsoid spores with a confined outer layer, the Δmbl spores were more irregular with a very diffuse outer layer (see Fig. S4 in the supplemental material).
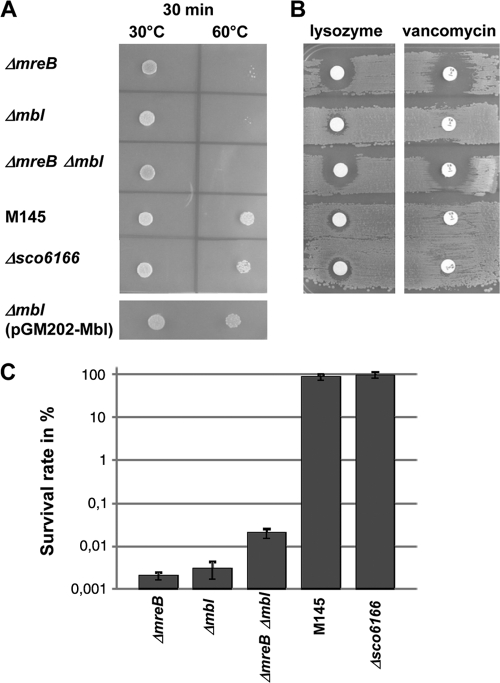
Furthermore, the mutant spores were impaired in their resistance to moderate heat (Fig. 4A), 5% SDS (data not shown), and salt stress (Fig. 4C). Whereas about 100% of M145 spores were able to germinate on LB plus 6% NaCl, only 0.001% of ΔmreB, 0.003% of Δmbl, and 0.02% of ΔmreB Δmbl spores were able to form colonies (mean values from three independent experiments). In contrast, Δsco6166 spores were almost as resistant (89.4% survival) to salt stress as the wild type (Fig. 4C). The transformation of the Δmbl mutant with plasmid pGM202-Mbl, which contains an mbl-his6 fusion gene under the control of the thiostrepton-inducible tipA promoter (Table 1), complemented spore size (Table 4) and heat sensitivity (Fig. 4A), demonstrating that the mutant phenotype was caused solely by mbl inactivation.
FIG. 4.
Sensitivity of mutant spores to moderate heat, cell wall-lytic agents, and salt stress. (A) To assess resistance to moderate heat, spores of M145, ΔmreB, Δmbl, ΔmreB Δmbl, Δsco6166, and Δmbl(pGM202-Mbl) were incubated for 30 min at 30 or at 60°C, spotted onto LB agar, and incubated for 3 days at 30°C. (B) Resistance to cell wall-lytic agents was studied by plating spores onto LB agar, and filter discs with lysozyme (50 μg) or vancomycin (5 μg) were applied. The growth of M145, Δmbl, and Δsco6166 is only slightly affected, while ΔmreB and ΔmreB Δmbl show large inhibition zones. (C) Survival rates of mutant strains exposed to salt stress. Spore dilutions (in H2O) were plated on LB agar plates containing 0.5% NaCl or 6% NaCl. The survival rates (mean values of three independent experiments) represent the percentage of colony titer on LB plus 6% NaCl in relation to the titer on LB plus 0.5% NaCl. In contrast to those of M145 and Δsco6166, the spores of ΔmreB, Δmbl, and ΔmreB Δmbl were impaired in germination and colony formation under high salt concentrations.
Since heat and detergent sensitivity is a hallmark for a defective spore wall (37), we studied the effect of lysozyme and vancomycin. Spores of M145 and the mutants were plated onto LB agar, and filter discs soaked with lysozyme or vancomycin were placed on the plates. After incubation at 30°C for 24 h, only very small inhibition zones were formed on M145, Δsco6166, and the Δmbl mutant. However, the ΔmreB mutant and the ΔmreB Δmbl mutant were much more sensitive, and large inhibition zones were observed (Fig. 4B). If the agar plates were incubated overnight to allow spore germination before the use of the lysozyme or vancomycin discs, no inhibition zones were formed (data not shown), demonstrating that the germinating spores were sensitive but substrate mycelium was resistant.
These mutant phenotypes indicated that mreB and mbl are involved in spore wall synthesis, with a less important role for mbl. In contrast, the Δsco6166 mutant behaved like the parent strain M145 in all aspects.
Only MreB, but not Mbl or SCO6166, interacts with RodZ.
RodZ, a remarkable multidomain protein that localizes in a helical manner, recently was shown to interact with MreB, probably linking the cytosolic MreB filaments with the membrane-localized lateral wall-synthesizing machinery (1, 49). To analyze the protein-protein interaction of the MreB-like proteins, we used the bacterial two-hybrid system based on the reconstitution of the Bordetella pertussis adenylate cyclase (26). The three S. coelicolor mreB-like genes, the rodZ homologue sco5751, and the other genes of the mre cluster (mreC, mreD, pbp2, and sfr) were fused to the catalytic domains of cya and analyzed in cotransformation experiments of the cya mutant BTH101 for protein-protein interaction. The MreB homologues were poor interaction partners in these two-hybrid assays, and they did not show the self interaction that would be expected for actin-like proteins. MreB also did not interact with any of the other proteins encoded by the mre cluster (data not shown). However, MreB showed a strong interaction with the scaffold protein RodZ (SCO5751). Neither Mbl nor SCO6166 interacted with RodZ or any of the tested fusion proteins (Fig. 5).
FIG. 5.

Bacterial two-hybrid analysis demonstrating the interaction of MreB with the scaffold protein RodZ (SCO5751). mreB, mbl, sco6166, and further morphogenic genes (see Material and Methods for details) of S. coelicolor were fused to the functional cya domains T25 (pKT25) and T18 (pUT18c) of the Bordetella pertussis adenylate cyclase (26). Cotransformants of the cya mutant BTH101 were spotted onto X-Gal plates. The blue color indicates the reconstitution of a functional adenylate cyclase due to the interaction of the fusion proteins. MreB, but neither Mbl nor SCO6166, interacted with the S. coelicolor RodZ homologue SCO5751. pKT25-zip/pUT18c-zip represent the positive control. As a negative control, BTH101 carrying the empty vector plasmids pKT25 and pUT18c was used.
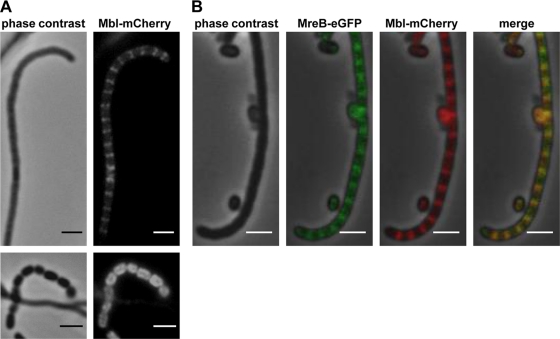
MreB and Mbl colocalize during sporulation.
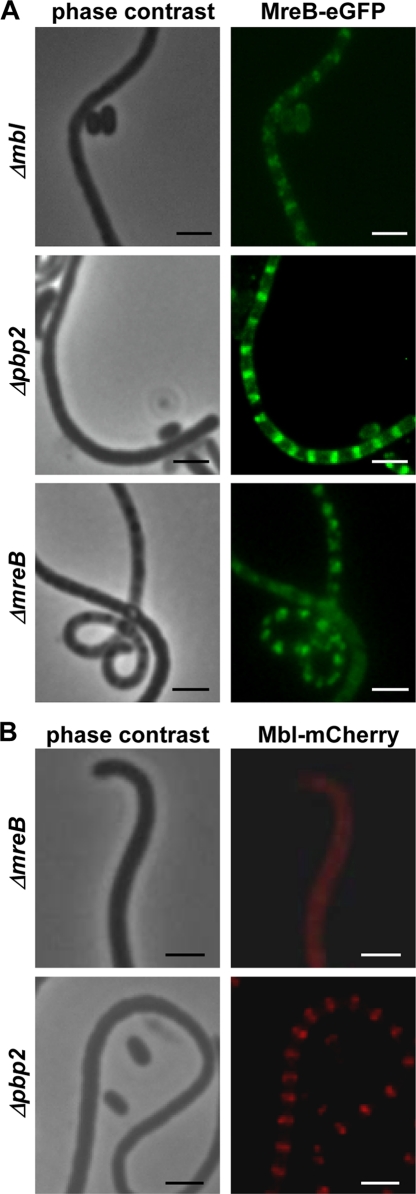
An MreB-eGFP fusion protein showed a dynamic localization pattern during morphological differentiation: disperse distribution in substrate mycelium, localization to the sporulation septa in aerial mycelium, and finally the formation of a shell at the inner spore wall (37). To address whether Mbl and SCO6166 had a similar localization pattern, we generated C-terminal fusions of Mbl and SCO6166 with either eGFP or mCherry. The fusion genes were integrated into the chromosome at the original gene locus via homologous recombination. No fluorescence of Mbl-eGFP or Mbl-mCherry was observed in substrate mycelium, probably reflecting the very low mbl expression, as demonstrated by RT-PCR. However, in aerial mycelium, Mbl localized to the sporulation septa, like MreB. In young spores, Mbl was found underneath the membrane (Fig. 6A). To prove that Mbl really localizes to the same sites as MreB, we performed colocalization studies by integrating plasmid pPM4 (37), which encodes the MreB-eGFP fusion protein, into the ΦC31 attachment site of M145::pAH5. Mbl-mCherry and MreB-eGFP always colocalized (Fig. 6B).
FIG. 6.
Colocalization of Mbl and MreB to putative sporulation septa. (A) Plasmid pAH5 was integrated into the chromosomal mbl gene of S. coelicolor M145 via homologous recombination. The Mbl-mCherry fusion protein forms a ladder-like structure at putative sporulation septa and localizes around the cell periphery in young spores (bottom panels). (B) Colocalization of Mbl-mCherry and MreB-eGFP in the aerial mycelium. Plasmid pPM4 carrying a mreB-egfp fusion gene was integrated into the ΦC31 att site of M145::pAH5, generating M145::pAH5-pPM4. Bar, 2 μm.
In contrast, SCO6166-mCherry showed a dispersed fluorescence of the total substrate mycelium without distinct localization patterns (see Fig. S5 in the supplemental material).
Mbl requires MreB for proper localization.
To analyze whether MreB and Mbl depend on each other for proper localization, fluorescence generated by MreB-eGFP (pPM4) and Mbl-mCherry (pAH5) (Table 1) was analyzed in a ΔmreB or Δmbl background. Mutant Δpbp2 (30a) was used as a control, since it showed the same morphological defects, swollen and prematurely germinating spores, as the ΔmreB or Δmbl mutant. In Δmbl::pPM4, ΔmreB::pPM4, and Δpbp2::pPM4 strains, the characteristic ladder-like fluorescence structure at the sporulation septa of the aerial mycelium (Fig. 7A) and around the spore membrane (see Fig. S6 in the supplemental material) was observed, demonstrating that MreB-eGFP was able to localize independently of Mbl or PBP2.
FIG. 7.
Mbl-mCherry dependence on MreB for proper localization. The localization of MreB-eGFP and Mbl-mCherry was studied in ΔmreB, Δpbp2, and Δmbl mutants, which have similar morphological defects. (A) MreB-eGFP localizes to the sporulation septa even in the absence of Mbl or PBP2. (B) While the localization of Mbl-mCherry was not affected in the Δpbp2 mutant, it was unable to localize to the sporulation septa in the ΔmreB mutant, suggesting that Mbl depends on the presence of MreB for proper localization. Bar, 2 μm.
Whereas Mbl-mCherry localized properly in the Δpbp2::pAH5 strain, no clear Mbl-mCherry foci were observed in the ΔmreB mutant (Fig. 7B). RT-PCR analyses demonstrated that the transcription of mbl-mcherry was not affected in the ΔmreB mutant (see Fig. S7 in the supplemental material). This clearly suggested that Mbl requires MreB for correct localization.
DISCUSSION
Most rod-shaped bacteria achieve their morphology by incorporating new peptidoglycan at the lateral wall in a helical manner (10, 14, 47), probably directed by cytoplasmic MreB filaments (25, 31). Whereas Gram-negative rods require a single MreB homologue for growth and cell shape determination, Gram-positive ones, like B. subtilis, depend on three MreB homologues (27, 44). The precise role of the three MreB homologues in directing lateral cell wall growth is not known, and some redundancy has been reported (27).
S. coelicolor is a useful model system to study the role of MreB homologues, since none of the three mreB-like genes is essential for viability, demonstrating that they are not involved in vegetative growth by apical tip extension. This provided the unique possibility to analyze the function of the distinct mreB homologues in a defined mutant background. The conservation of mreB in Streptomyces and other sporulating actinomycetes, its mutant phenotype, and its localization to sites where spore wall synthesis occurs demonstrated its role in spore wall synthesis (37). In this report, we provide evidence that Mbl also is involved in sporulation, while the third MreB homologue, SCO6166, has a completely different function. This conclusion is supported by homology modeling, expression analysis, mutant phenotypes, electron microscopy, protein-protein interaction studies, and protein localization experiments.
MreB and Mbl cooperate in the synthesis of a thickened spore wall.
The role of mreB in directing the synthesis of a thickened spore wall has been reported already (37). There are several lines of evidence that suggest a related function for Mbl. The structure of Mbl, deduced from homology modeling, is nearly identical to that of MreB, indicating a very similar function.
Expression analysis by semiquantitative RT-PCR analysis showed the induction of mreB and mbl during morphological differentiation. Whereas mbl expression was not detectable in substrate mycelium but increased considerably during sporulation, mreB also was transcribed in vegetatively growing mycelium. This indicates that besides its function in directing spore wall synthesis, MreB has an additional role in vegetatively growing mycelium under certain conditions. The mreB RT-PCR data are in agreement with previous reports by S1 mapping experiments that identified three promoters, one strongly induced during sporulation (6). Interestingly, the −35 and −10 regions (GGAAC-N17-CGTCCTC) of the mreB P1 promoter (6) closely resemble σE-dependent promoters (GCAAC-N17-CGTCTTC) (23), suggesting the transcription of P1 by SigE and therefore the induction of mreB by cell wall stress. σE is part of a signal transduction system sensing certain changes in the integrity of the cell envelope (24) and transcribing the cwg operon putatively involved in cell wall glycan synthesis (23). The phenotype of a σE mutant, with an altered cell wall structure and increased sensitivity to cell wall-lytic enzymes, resembled somewhat that of the ΔmreB mutant (37, 39). However, whereas the ΔmreB mutant had an impaired spore wall, the sigE mutants suffered from a defective cell wall during vegetative growth (39).
The phenotype of the Δmbl mutant closely resembled that of ΔmreB, with swollen prematurely germinating spores that had lost their resistance to detrimental environmental conditions, such as heat, detergents, and salt stress. This clearly indicated that Mbl and MreB cooperate in directing the synthesis of a stress-resistant spore wall, and that both are necessary for this process. Interestingly, Δmbl and ΔmreB spores showed a different behavior when exposed to cell wall-lytic agents. Whereas spore germination in the ΔmreB strain was highly sensitive to lysozyme and vancomycin, Δmbl spores were almost as resistant as the wild type, revealing a less crucial role for Mbl. The localization of mreB in the same gene cluster as that of pbp2, encoding a monofunctional transpeptidase, allows speculation on the defect leading to the impaired spore wall of the ΔmreB mutant and its increased sensitivity. Due to the missing MreB filaments, PBP2 might not be positioned properly in relation to the spore periphery, causing the spore wall to be less cross-linked.
As previously shown for the ΔmreB mutant (37), Δmbl also had a distorted spore wall. Whereas wild-type spores are ellipsoid with a sharply visible external wall, electron microscopic images showed irregularly shaped Δmbl spores with a more diffuse wall (see Fig. S4 in the supplemental material). Since the Δmbl spores are sensitive to many treatments, we cannot exclude that the appearance of the spore surface is affected by the fixation/staining process.
MreB localized to the sporulation septa and the spore periphery even in the absence of Mbl. It also localized properly in a Δpbp2 mutant, which showed the same morphological defects as those of the ΔmreB and Δmbl mutants (30a). Also, MreB of Caulobacter crescentus was able to form normal helical spirals in MreC-depleted cells (15). In contrast, a distorted MreB localization pattern was observed in E. coli MreC-, MreD-, and RodA-depleted cells (32). Mbl, however, seems to rely on the presence of MreB filaments. While Mbl-eGFP or Mbl-mCherry properly localized at the sporulation septa in M145 or Δpbp2, no clear Mbl-mCherry fluorescence foci were observed in the ΔmreB mutant.
Although MreB and Mbl seem to cooperate in spore wall synthesis, MreB has a more important role than Mbl. This also is supported by the protein-protein interaction studies that showed the strong interaction of MreB with RodZ, which probably represents the main scaffold protein for the peptidoglycan-synthesizing complex (1, 19, 49).
SCO6166 has a nonessential role during vegetative growth.
SCO6166 differs in many aspects from the other MreB homologues. Whereas homology modeling revealed a highly similar structure for MreB and Mbl, SCO6166 lacks subdomains IB and IIB of actin-like proteins. Deviations in the IB and IIB subdomains from MreB and actins also are displayed by ParM, which is involved in plasmid partitioning, and this was proposed to influence the structure of the respective filaments (50).
The localization of sco6166 in an operon with sco6165 and their translational coupling suggest that both proteins cooperate. SCO6165 is homologous to DksA, a suppressor of DnaK. This suggests a function of SCO6165/SCO6166 in heat shock or stress response. Although many Streptomyces strains encode an SCO6165/SCO6166 pair, several Streptomyces strains without an SCO6166 homologue also exist, indicating an ancillary function of SCO6165/SCO6166.
In contrast to mreB and mbl, which are induced during morphological differentiation, sco6166 had a completely different expression profile, being highly expressed in vegetative mycelium but completely switched off during sporulation. The RT-PCR experiments are in agreement with fluorescence microscopy studies, which detected SCO6166-mCherry only in substrate mycelium and not in aerial mycelium or spores. This clearly indicates a role of SCO6166 during vegetative growth and not in sporulation.
In summary, we showed that the three MreB proteins of S. coelicolor have distinct functions. MreB and Mbl are involved in the synthesis of a stress-resistant spore wall, with MreB probably providing a scaffold for the assembly of Mbl filaments. The role of SCO6166 during vegetative growth still is unclear and remains to be elucidated.
Supplementary Material
Acknowledgments
We thank D. Ladant for providing the bacterial two-hybrid system, C. Menzel for help with TEM analyses, and K. Nieselt for providing microarray data.
The DFG (SFB766) provided financial support.
Footnotes
Published ahead of print on 21 January 2011.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Alyahya, S. A., et al. 2009. RodZ, a component of the bacterial core morphogenic apparatus. Proc. Natl. Acad. Sci. U. S. A. 106:1239-1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bennett, J. A., et al. 2009. Medium-dependent phenotypes of Streptomyces coelicolor with mutations in ftsI or ftsW. J. Bacteriol. 191:661-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berman, H. M., et al. 2000. The Protein Data Bank. Nucleic Acids Res. 28:235-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bork, P., C. Sander, and A. Valencia. 1992. An ATPase domain common to prokaryotic cell cycle proteins, sugar kinases, actin, and hsp70 heat shock proteins. Proc. Natl. Acad. Sci. U. S. A. 89:7290-7294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bullock, W. O., J. M. Fernandez, and M. J. Short. 1987. X-L1blue, a high efficiency plasmid transforming recA Escherichia coli strain with beta galactosidase selection. Biotechniques 5:376-378. [Google Scholar]
- 6.Burger, A., K. Sichler, G. Kelemen, M. Buttner, and W. Wohlleben. 2000. Identification and characterization of the mre gene region of Streptomyces coelicolor A3(2). Mol. Gen. Genet. 263:1053-1060. [DOI] [PubMed] [Google Scholar]
- 7.Carballido-López, R. 2006. The bacterial actin-like cytoskeleton. Microbiol. Mol. Biol. Rev. 70:888-909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carballido-López, R., et al. 2006. Actin homolog MreBH governs cell morphogenesis by localization of the cell wall hydrolase LytE. Dev. Cell 11:399-409. [DOI] [PubMed] [Google Scholar]
- 9.Cherepanov, P. P., and W. Wackernagel. 1995. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158:9-14. [DOI] [PubMed] [Google Scholar]
- 10.Daniel, R. A., and J. Errington. 2003. Control of cell morphogenesis in bacteria: two distinct ways to make a rod-shaped cell. Cell 113:767-776. [DOI] [PubMed] [Google Scholar]
- 11.Defeu Soufo, H. J., and P. L. Graumann. 2004. Dynamic movement of actin-like proteins within bacterial cells. EMBO Rep. 5:789-794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Defeu Soufo, H. J., and P. L. Graumann. 2005. Bacillus subtilis actin-like protein MreB influences the positioning of the replication machinery and requires membrane proteins MreC/D and other actin-like proteins for proper localization. BMC Cell Biol. 6:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Defeu Soufo, H. J., et al. 2010. Bacterial translation elongation factor EF-Tu interacts and colocalizes with actin-like MreB protein. Proc. Natl. Acad. Sci. U. S. A. 107:3163-3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Divakaruni, A. V., C. Baida, C. L. White, and J. W. Gober. 2007. The cell shape proteins MreB and MreC control cell morphogenesis by positioning cell wall synthetic complexes. Mol. Microbiol. 66:174-188. [DOI] [PubMed] [Google Scholar]
- 15.Dye, N. A., Z. Pincus, J. A. Theriot, L. Shapiro, and Z. Gitai. 2005. Two independent spiral structures control cell shape in Caulobacter. Proc. Natl. Acad. Sci. U. S. A. 102:18608-18613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Errington, J. 2003. Dynamic proteins and a cytoskeleton in bacteria. Nat. Cell Biol. 5:175-178. [DOI] [PubMed] [Google Scholar]
- 17.Figge, R. M., A. V. Divakaruni, and J. W. Gober. 2004. MreB, the cell shape-determining bacterial actin homologue, co-ordinates cell wall morphogenesis in Caulobacter crescentus. Mol. Microbiol. 51:1321-1332. [DOI] [PubMed] [Google Scholar]
- 18.Formstone, A., and J. Errington. 2005. A magnesium-dependent mreB null mutant: implications for the role of mreB in Bacillus subtilis. Mol. Microbiol. Mar;. 55:1646-1657. [DOI] [PubMed] [Google Scholar]
- 19.Gerdes, K. 2009. RodZ, a new player in bacterial cell morphogenesis. EMBO J. 28:171-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guex, N., and M. C. Peitsch. 1997. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18:2714-2723. [DOI] [PubMed] [Google Scholar]
- 21.Gust, B., G. L. Challis, K. Fowler, T. Kieser, and K. F. Chater. 2003. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc. Natl. Acad. Sci. U. S. A. 100:1541-1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hempel, A. M., S. B. Wang, M. Letek, J. A. Gil, and K. Flardh. 2008. Assemblies of DivIVA mark sites for hyphal branching and can establish new zones of cell wall growth in Streptomyces coelicolor. J. Bacteriol. 190:7579-7583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hong, H. J., M. S. Paget, and M. J. Buttner. 2002. A signal transduction system in Streptomyces coelicolor that activates the expression of a putative cell wall glycan operon in response to vancomycin and other cell wall-specific antibiotics. Mol. Microbiol. 44:1199-1211. [DOI] [PubMed] [Google Scholar]
- 24.Hutchings, M. I., H. J. Hong, E. Leibovitz, I. C. Sutcliffe, and M. J. Buttner. 2006. The sigma(E) cell envelope stress response of Streptomyces coelicolor is influenced by a novel lipoprotein, CseA. J. Bacteriol. 188:7222-7229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jones, L. J., R. Carballido-Lopez, and J. Errington. 2001. Control of cell shape in bacteria: helical, actin-like filaments in Bacillus subtilis. Cell 104:913-922. [DOI] [PubMed] [Google Scholar]
- 26.Karimova, G., J. Pidoux, A. Ullmann, and D. Ladant. 1998. A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc. Natl. Acad. Sci. U. S. A. 95:5752-5756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kawai, Y., K. Asai, and J. Errington. 2009. Partial functional redundancy of MreB isoforms, MreB, Mbl and MreBH, in cell morphogenesis of Bacillus subtilis. Mol. Microbiol. 73:719-731. [DOI] [PubMed] [Google Scholar]
- 28.Kawai, Y., R. A. Daniel, and J. Errington. 2009. Regulation of cell wall morphogenesis in Bacillus subtilis by recruitment of PBP1 to the MreB helix. Mol. Microbiol. 71:1131-1144. [DOI] [PubMed] [Google Scholar]
- 29.Kieser, T., M. J. Bibb, M. J. Buttner, K. F. Chater, and D. A. Hopwood. 2000. Practical Streptomyces genetics. The John Innes Foundation, Norwich, CT.
- 30.Kim, S. Y., Z. Gitai, A. Kinkhabwala, L. Shapiro, and W. E. Moerner. 2006. Single molecules of the bacterial actin MreB undergo directed treadmilling motion in Caulobacter crescentus. Proc. Natl. Acad. Sci. U. S. A. 103:10929-10934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30a.Kleinschnitz, E.-M., A. Heichlinger, K. Schirner, J. Winkler, A. Latus, I. Maldener, W. Wohlleben, and G. Muth. 18 January 2011. Proteins encoded by the mre gene cluster in Streptomyces coelicolor A3(2) cooperate in spore wall synthesis. Mol. Micorbiol. doi: 10.1111/j.1365-2958.2010.07529.x. [Epub ahead of print.] [DOI] [PubMed]
- 31.Kruse, T., et al. 2006. Actin homolog MreB and RNA polymerase interact and are both required for chromosome segregation in Escherichia coli. Genes Dev. 20:113-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kruse, T., J. Bork-Jensen, and K. Gerdes. 2005. The morphogenetic MreBCD proteins of Escherichia coli form an essential membrane-bound complex. Mol. Microbiol. 55:78-89. [DOI] [PubMed] [Google Scholar]
- 33.Kruse, T., and K. Gerdes. 2005. Bacterial DNA segregation by the actin-like MreB protein. Trends Cell Biol. 15:343-345. [DOI] [PubMed] [Google Scholar]
- 34.Kruse, T., J. Moller-Jensen, A. Lobner-Olesen, and K. Gerdes. 2003. Dysfunctional MreB inhibits chromosome segregation in Escherichia coli. EMBO J. 22:5283-5292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Letek, M., et al. 2008. DivIVA is required for polar growth in the MreB-lacking rod-shaped actinomycete Corynebacterium glutamicum. J. Bacteriol. 190:3283-3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.MacNeil, D. J., et al. 1992. Analysis of Streptomyces avermitilis genes required for avermectin biosynthesis utilizing a novel integration vector. Gene 111:61-68. [DOI] [PubMed] [Google Scholar]
- 37.Mazza, P., et al. 2006. MreB of Streptomyces coelicolor is not essential for vegetative growth but is required for the integrity of aerial hyphae and spores. Mol. Microbiol. 60:838-852. [DOI] [PubMed] [Google Scholar]
- 38.Nieselt, K., et al. 2010. The dynamic architecture of the metabolic switch in Streptomyces coelicolor. BMC Genomics 11:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Paget, M. S., L. Chamberlin, A. Atrih, S. J. Foster, and M. J. Buttner. 1999. Evidence that the extracytoplasmic function sigma factor sigmaE is required for normal cell wall structure in Streptomyces coelicolor A3(2). J. Bacteriol. 181:204-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pridmore, R. D. 1987. New and versatile cloning vectors with kanamycin-resistance marker. Gene 56:309-312. [DOI] [PubMed] [Google Scholar]
- 41.Redenbach, M., et al. 1996. A set of ordered cosmids and a detailed genetic and physical map for the 8 Mb Streptomyces coelicolor A3(2) chromosome. Mol. Microbiol. 21:77-96. [DOI] [PubMed] [Google Scholar]
- 42.Sali, A., and T. L. Blundell. 1993. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234:779-815. [DOI] [PubMed] [Google Scholar]
- 43.Sambrook, J., and D. W. Russel. 2001. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, New York, NY.
- 44.Schirner, K., and J. Errington. 2009. Influence of heterologous MreB proteins on cell morphology of Bacillus subtilis. Microbiology 155:3611-3621. [DOI] [PubMed] [Google Scholar]
- 45.Söding, J., A. Biegert, and A. N. Lupas. 2005. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 33:W244-W248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tiffert, Y., et al. 2008. The Streptomyces coelicolor GlnR regulon: identification of new GlnR targets and evidence for a central role of GlnR in nitrogen metabolism in actinomycetes. Mol. Microbiol. 67:861-880. [DOI] [PubMed] [Google Scholar]
- 47.Tiyanont, K., et al. 2006. Imaging peptidoglycan biosynthesis in Bacillus subtilis with fluorescent antibiotics. Proc. Natl. Acad. Sci. U. S. A. 103:11033-11038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van den Ent, F., L. A. Amos, and J. Lowe. 2001. Prokaryotic origin of the actin cytoskeleton. Nature 413:39-44. [DOI] [PubMed] [Google Scholar]
- 49.van den Ent, F., C. M. Johnson, L. Persons, P. de Boer, and J. Lowe. 2010. Bacterial actin MreB assembles in complex with cell shape protein RodZ. EMBO J. 29:1081-1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van den Ent, F., J. Moller-Jensen, L. A. Amos, K. Gerdes, and J. Lowe. 2002. F-actin-like filaments formed by plasmid segregation protein ParM. EMBO J. 21:6935-6943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vollmer, W., and S. J. Seligman. 2010. Architecture of peptidoglycan: more data and more models. Trends Microbiol. 18:59-66. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.