Abstract
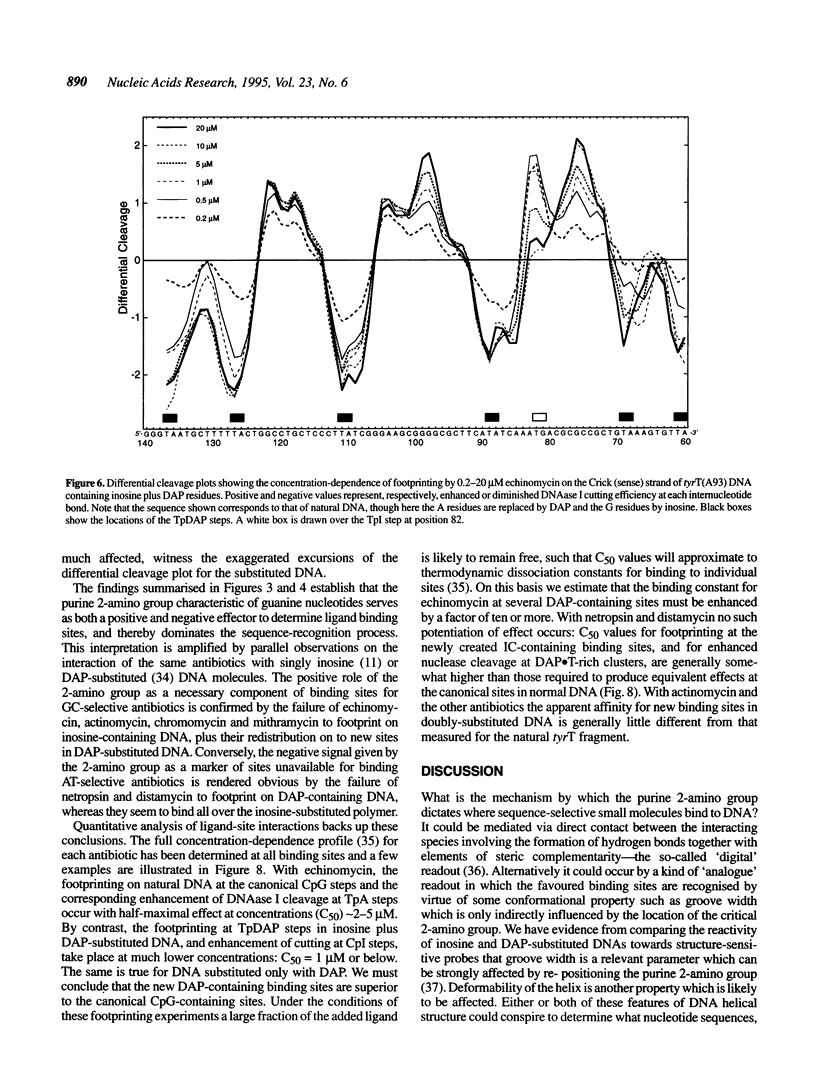
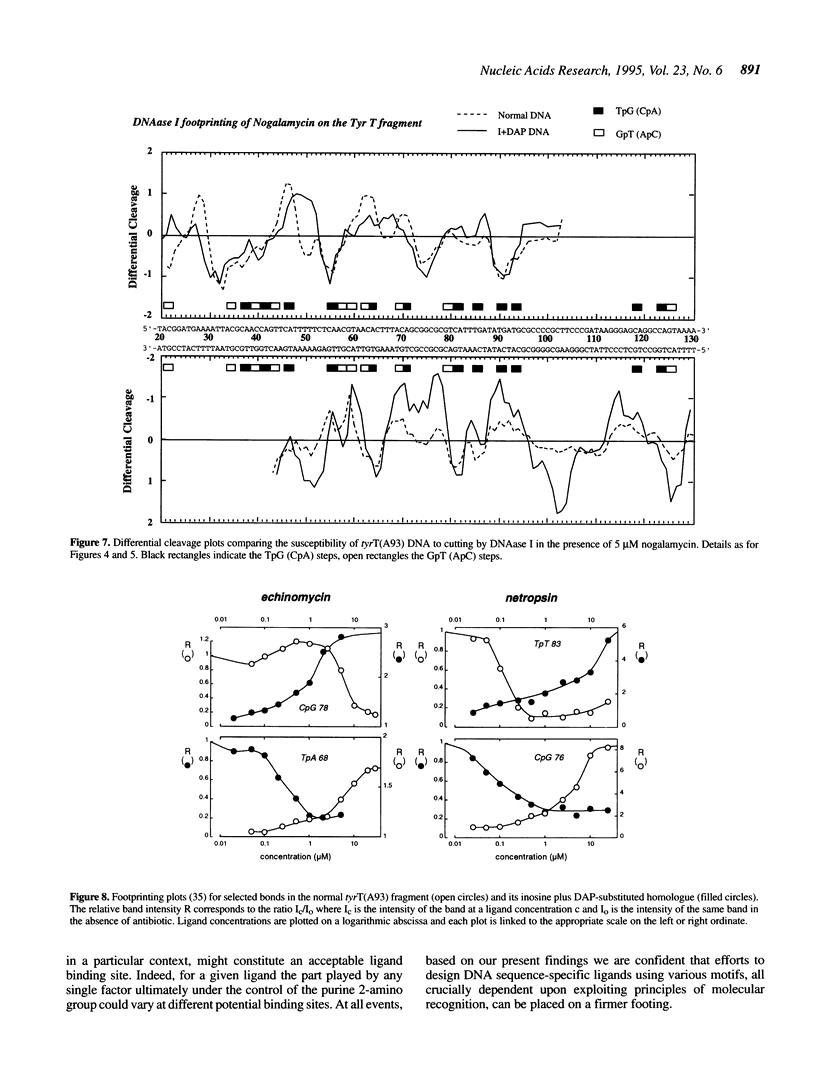
The proposition that the 2-amino group of guanine plays a critical role in determining how antibiotics recognise their binding sites in DNA has been tested by relocating it, using tyrT DNA derivative molecules substituted with inosine plus 2,6-diaminopurine (DAP). Irrespective of their mode of interaction with DNA, such GC-specific antibiotics as actinomycin, echinomycin, mithramycin and chromomycin find new binding sites associated with DAP-containing sequences and are excluded from former canonical sites containing I.C base pairs. The converse is found to be the case for a group of normally AT-selective ligands which bind in the minor groove of the helix, such as netropsin: their preferred sites become shifted to IC-rich clusters. Thus the binding sites of all these antibiotics strictly follow the placement of the purine 2-amino group, which accordingly must serve as both a positive and negative effector. The footprinting profile of the 'threading' intercalator nogalamycin is potentiated in DAP plus inosine-substituted DNA but otherwise remains much the same as seen with natural DNA. The interaction of echinomycin with sites containing the TpDAP step in doubly substituted DNA appears much stronger than its interaction with CpG-containing sites in natural DNA.
Full text
PDF

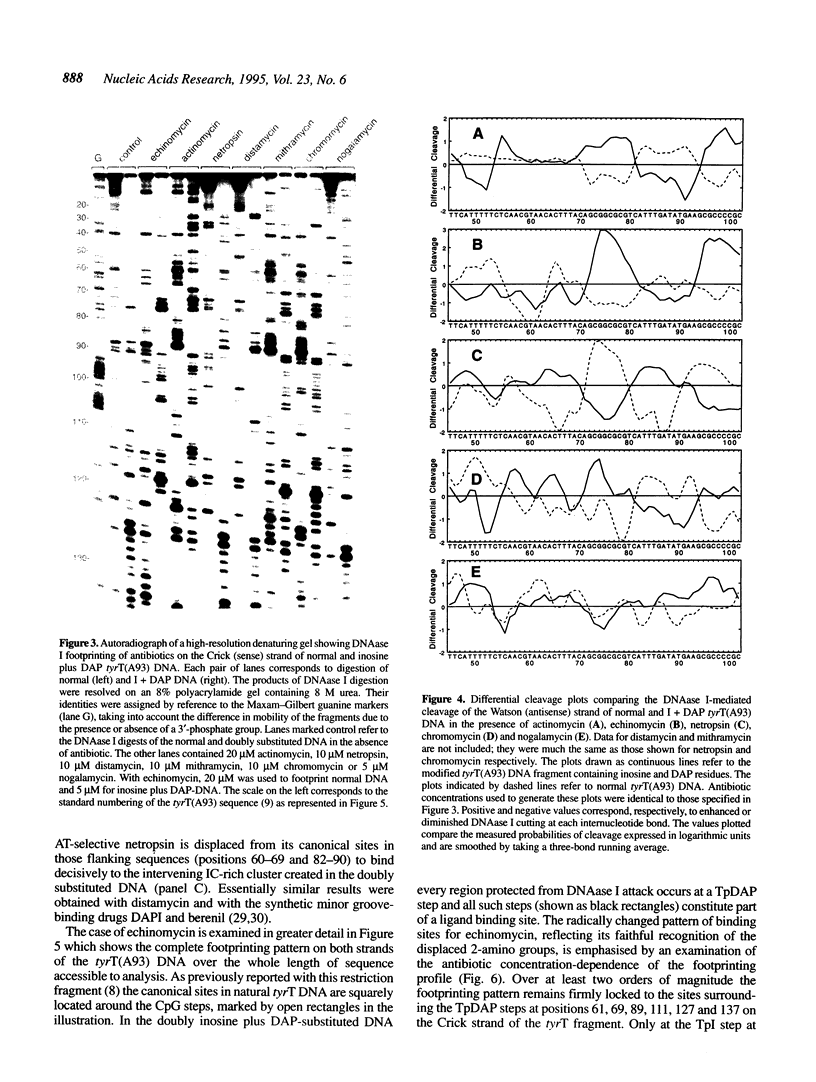
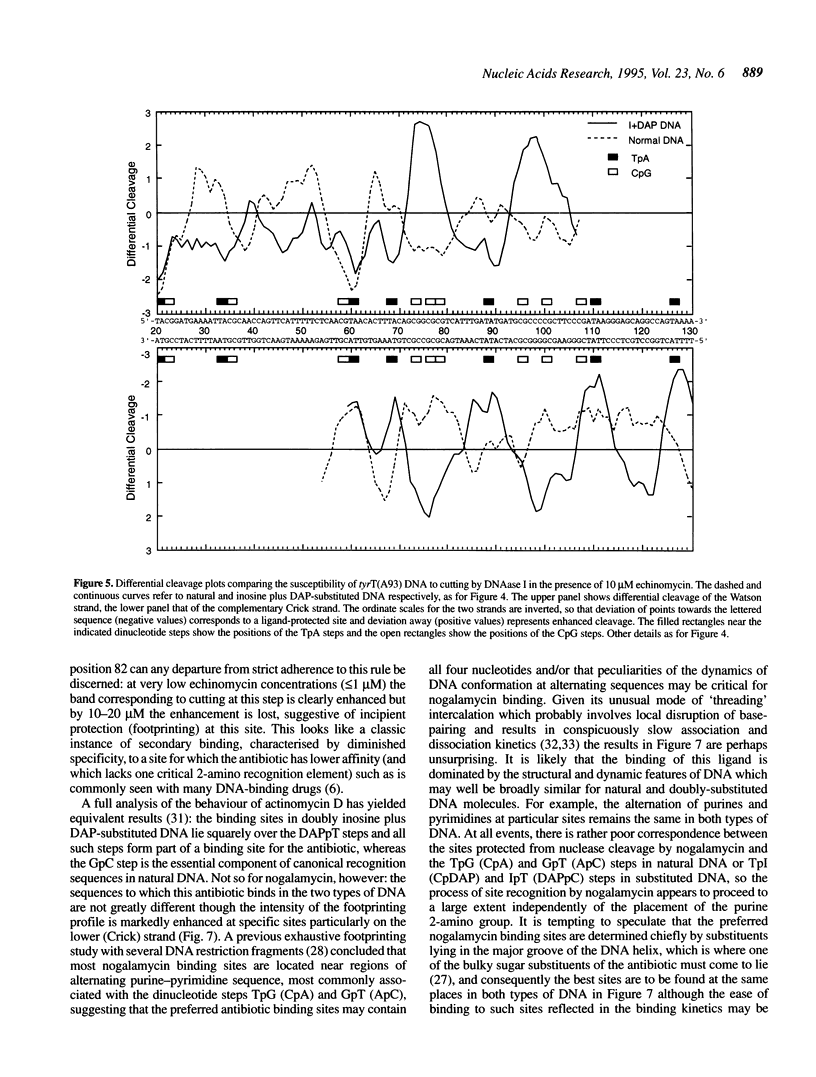

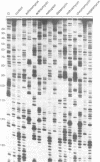
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bloch W. A biochemical perspective of the polymerase chain reaction. Biochemistry. 1991 Mar 19;30(11):2735–2747. doi: 10.1021/bi00225a001. [DOI] [PubMed] [Google Scholar]
- Dervan P. B. Design of sequence-specific DNA-binding molecules. Science. 1986 Apr 25;232(4749):464–471. doi: 10.1126/science.2421408. [DOI] [PubMed] [Google Scholar]
- Drew H. R., Weeks J. R., Travers A. A. Negative supercoiling induces spontaneous unwinding of a bacterial promoter. EMBO J. 1985 Apr;4(4):1025–1032. doi: 10.1002/j.1460-2075.1985.tb03734.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egli M., Williams L. D., Frederick C. A., Rich A. DNA-nogalamycin interactions. Biochemistry. 1991 Feb 5;30(5):1364–1372. doi: 10.1021/bi00219a029. [DOI] [PubMed] [Google Scholar]
- Fox K. R., Brassett C., Waring M. J. Kinetics of dissociation of nogalamycin from DNA: comparison with other anthracycline antibiotics. Biochim Biophys Acta. 1985 Jul 5;840(3):383–392. doi: 10.1016/0304-4165(85)90219-3. [DOI] [PubMed] [Google Scholar]
- Fox K. R., Waring M. J. Evidence of different binding sites for nogalamycin in DNA revealed by association kinetics. Biochim Biophys Acta. 1984 Nov 28;802(2):162–168. doi: 10.1016/0304-4165(84)90157-0. [DOI] [PubMed] [Google Scholar]
- Fox K. R., Waring M. J. Nucleotide sequence binding preferences of nogalamycin investigated by DNase I footprinting. Biochemistry. 1986 Jul 29;25(15):4349–4356. doi: 10.1021/bi00363a026. [DOI] [PubMed] [Google Scholar]
- Gao X. L., Patel D. J. Chromomycin dimer-DNA oligomer complexes. Sequence selectivity and divalent cation specificity. Biochemistry. 1990 Dec 11;29(49):10940–10956. doi: 10.1021/bi00501a012. [DOI] [PubMed] [Google Scholar]
- Goodisman J., Rehfuss R., Ward B., Dabrowiak J. C. Site-specific binding constants for actinomycin D on DNA determined from footprinting studies. Biochemistry. 1992 Feb 4;31(4):1046–1058. doi: 10.1021/bi00119a013. [DOI] [PubMed] [Google Scholar]
- Hurley L. H. DNA and associated targets for drug design. J Med Chem. 1989 Sep;32(9):2027–2033. doi: 10.1021/jm00129a001. [DOI] [PubMed] [Google Scholar]
- Johnston R. F., Pickett S. C., Barker D. L. Autoradiography using storage phosphor technology. Electrophoresis. 1990 May;11(5):355–360. doi: 10.1002/elps.1150110503. [DOI] [PubMed] [Google Scholar]
- Kamitori S., Takusagawa F. Crystal structure of the 2:1 complex between d(GAAGCTTC) and the anticancer drug actinomycin D. J Mol Biol. 1992 May 20;225(2):445–456. doi: 10.1016/0022-2836(92)90931-9. [DOI] [PubMed] [Google Scholar]
- Keniry M. A., Banville D. L., Simmonds P. M., Shafer R. Nuclear magnetic resonance comparison of the binding sites of mithramycin and chromomycin on the self-complementary oligonucleotide d(ACCCGGGT)2. Evidence that the saccharide chains have a role in sequence specificity. J Mol Biol. 1993 Jun 5;231(3):753–767. doi: 10.1006/jmbi.1993.1324. [DOI] [PubMed] [Google Scholar]
- Kopka M. L., Yoon C., Goodsell D., Pjura P., Dickerson R. E. The molecular origin of DNA-drug specificity in netropsin and distamycin. Proc Natl Acad Sci U S A. 1985 Mar;82(5):1376–1380. doi: 10.1073/pnas.82.5.1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen T. A., Kopka M. L., Dickerson R. E. Crystal structure analysis of the B-DNA dodecamer CGTGAATTCACG. Biochemistry. 1991 May 7;30(18):4443–4449. doi: 10.1021/bi00232a010. [DOI] [PubMed] [Google Scholar]
- Low C. M., Drew H. R., Waring M. J. Sequence-specific binding of echinomycin to DNA: evidence for conformational changes affecting flanking sequences. Nucleic Acids Res. 1984 Jun 25;12(12):4865–4879. doi: 10.1093/nar/12.12.4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchand C., Bailly C., McLean M. J., Moroney S. E., Waring M. J. The 2-amino group of guanine is absolutely required for specific binding of the anti-cancer antibiotic echinomycin to DNA. Nucleic Acids Res. 1992 Nov 11;20(21):5601–5606. doi: 10.1093/nar/20.21.5601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portugal J., Waring M. J. Assignment of DNA binding sites for 4',6-diamidine-2-phenylindole and bisbenzimide (Hoechst 33258). A comparative footprinting study. Biochim Biophys Acta. 1988 Feb 28;949(2):158–168. doi: 10.1016/0167-4781(88)90079-6. [DOI] [PubMed] [Google Scholar]
- Portugal J., Waring M. J. Comparison of binding sites in DNA for berenil, netropsin and distamycin. A footprinting study. Eur J Biochem. 1987 Sep 1;167(2):281–289. doi: 10.1111/j.1432-1033.1987.tb13334.x. [DOI] [PubMed] [Google Scholar]
- Sastry M., Patel D. J. Solution structure of the mithramycin dimer-DNA complex. Biochemistry. 1993 Jul 6;32(26):6588–6604. doi: 10.1021/bi00077a012. [DOI] [PubMed] [Google Scholar]
- Sayers E. W., Waring M. J. Footprinting titration studies on the binding of echinomycin to DNA incapable of forming Hoogsteen base pairs. Biochemistry. 1993 Sep 7;32(35):9094–9107. doi: 10.1021/bi00086a014. [DOI] [PubMed] [Google Scholar]
- Smith C. K., Davies G. J., Dodson E. J., Moore M. H. DNA-nogalamycin interactions: the crystal structure of d(TGATCA) complexed with nogalamycin. Biochemistry. 1995 Jan 17;34(2):415–425. doi: 10.1021/bi00002a005. [DOI] [PubMed] [Google Scholar]
- Van Dyke M. W., Dervan P. B. Chromomycin, mithramycin, and olivomycin binding sites on heterogeneous deoxyribonucleic acid. Footprinting with (methidiumpropyl-EDTA)iron(II). Biochemistry. 1983 May 10;22(10):2373–2377. doi: 10.1021/bi00279a011. [DOI] [PubMed] [Google Scholar]
- Van Dyke M. W., Hertzberg R. P., Dervan P. B. Map of distamycin, netropsin, and actinomycin binding sites on heterogeneous DNA: DNA cleavage-inhibition patterns with methidiumpropyl-EDTA.Fe(II). Proc Natl Acad Sci U S A. 1982 Sep;79(18):5470–5474. doi: 10.1073/pnas.79.18.5470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward B., Rehfuss R., Goodisman J., Dabrowiak J. C. Determination of netropsin-DNA binding constants from footprinting data. Biochemistry. 1988 Feb 23;27(4):1198–1205. doi: 10.1021/bi00404a020. [DOI] [PubMed] [Google Scholar]
- Waring M. J., Bailly C. The purine 2-amino group as a critical recognition element for binding of small molecules to DNA. Gene. 1994 Nov 4;149(1):69–79. doi: 10.1016/0378-1119(94)90414-6. [DOI] [PubMed] [Google Scholar]
- Waring M. J. Drugs which affect the structure and function of DNA. Nature. 1968 Sep 28;219(5161):1320–1325. doi: 10.1038/2191320a0. [DOI] [PubMed] [Google Scholar]
- Waring M. J., Wakelin L. P. Echinomycin: a bifunctional intercalating antibiotic. Nature. 1974 Dec 20;252(5485):653–657. doi: 10.1038/252653a0. [DOI] [PubMed] [Google Scholar]
- van Houte L. P., van Garderen C. J., Patel D. J. The antitumor drug nogalamycin forms two different intercalation complexes with d(GCGT).d(ACGC). Biochemistry. 1993 Feb 16;32(6):1667–1674. doi: 10.1021/bi00057a034. [DOI] [PubMed] [Google Scholar]