Abstract
The establishment of a stable reservoir of latently infected cells allows HIV to persist in the host. Usually, HIV infection of T cells results in integration of the viral genome, with a preference for regions in the human genome containing active genes, viral expression, and production of new viruses. However, in rare cases T cells become latently infected, and this is presumed to be due to a combination of two factors: integrated viruses are not efficiently transcribed and infected T cells revert to a resting memory state. HIV latency has been associated with provirus integration in regions of constitutive heterochromatin, gene deserts, or very highly expressed genes. We have investigated the transcriptional consequences of latent HIV integration into cellular genes and the involvement of chromatin reassembly factors (CRFs) in the transcriptional interference that a host gene exerts on the integrated cryptic HIV promoter. Chimeric transcripts containing sequences from the host gene and HIV can be detected, having been initiated at promoters of either the cell or the virus. Reactivation of HIV downregulates host gene expression. Cryptic promoters might remain inactive due to the repressive chromatin configuration established by CRFs during transcription elongation. Depletion of CRFs such as Spt6, Chd1, and FACT, or the histone chaperones ASF1a and HIRA, promoted HIV reactivation, concomitantly with chromatin relaxation and a decrease in general RNA polymerase activity. Overall, our results indicate that CRFs play a role in maintaining HIV latency by transcriptional interference when the provirus is integrated into an intron of a highly active gene.
In HIV-infected individuals successfully treated with antiretroviral therapy (ART), viremia is controlled to undetectable levels. Nevertheless, viremia appears rapidly after interruption of treatment, consistent with the existence of a latent viral reservoir. This reservoir seems to consist mainly of long-lived latently infected resting memory CD4+ T cells. The absence of viral protein expression shields latently infected cells from the immune system, and latently infected cells can be maintained for years by cellular quiescence (8, 9, 20, 21, 53, 62). Antigen stimulation or cytokine induction can reactivate the latent provirus and lead to viral replication and reinfection. The presence of this reservoir prevents the eradication of the virus and makes the infection a chronic disease. To achieve eradication of HIV from infected patients, it may be necessary to combine ART with drugs able to reactivate dormant viruses (55).
The establishment of HIV latency is a rare event that may occur when infected CD4+ lymphocytes stop proliferating and become resting cells. In addition, to allow cell survival, HIV expression should not occur after integration nor before the cell exits the cell cycle. Latently infected cells contain replication-competent provirus blocked at the transcriptional level by effective and reversible silencing. HIV transcription depends on cellular factors in addition to the viral Tat (transactivator of transcription) protein, and consequently, HIV promoter activity is tightly linked to the level of activation of its host cell. A suboptimal cellular environment for HIV expression at the transcriptional or posttranscriptional level contributes to the maintenance of the latent state (molecular mechanisms of HIV latency are reviewed elsewhere, e.g., in references 3, 11, 12, and 35).
Chromatin plays an essential role in the transcriptional regulation of HIV (48, 50). Key regulatory nucleosomes are positioned around the HIV transcription start site (TSS) when the promoter is inactive and are acetylated or remodeled upon activation (41, 58). Transcription-independent nucleosome rearrangement and histone acetylation are assisted, respectively, by ATP-dependent chromatin remodeling complexes and histone acetyltransferases recruited, at least partially, by Tat to the long terminal repeat (LTR) (43, 54, 56). Tat is an unusual transcription factor in that it binds an RNA element (TAR) at the 5′ end of the viral transcript, not at a DNA binding site. Its main role is to recruit the P-TEFb complex to the LTR to promote efficient elongation (32, 46). Tat is absent from infected cells until inefficient transcription from the HIV promoter depending on host cell factors (Sp1, NF-κB, NFAT, and others) allows some synthesis of this viral protein. After Tat is produced, HIV transcription enters a second, more efficient phase. However, in latently infected cells, HIV transcription is blocked at the initial phase, Tat is absent, and chromatin is a barrier to clearance of the preinitiation complex and polymerase progression. Previous models of HIV latency, cell lines ACH2 and U1, harbored proviruses with mutations in their Tat-TAR transcriptional axis, strengthening the idea that transcription inhibition is key to the establishment and maintenance of HIV latency (18, 19). Thus, a lack of specific host factors, a defective Tat-TAR axis, and a repressive chromatin environment could contribute to postintegration latency.
HIV transcription is influenced by the site of integration in the human genome (29). Survival of the infected cell to allow conversion to the resting state requires integration at a genomic site unfavorable for HIV transcription. HIV integration favors regions with active genes and active epigenetic marks, which most commonly lead to productive infection (6, 51, 59). Rare latent infection has been associated with provirus integration at regions of constitutive heterochromatin such as centromeric alphoid repeats, gene deserts, or introns in very highly expressed genes (24, 28, 39). For the integration into heterochromatin and, probably, gene deserts, local chromatin compaction can easily explain the silencing of the viral promoter. The silenced HIV promoter has been associated with repressive epigenetic marks, histone deacetylases (HDACs) and methyltransferases, heterochromatin proteins, and CpG methylation (4, 17, 26, 33, 60) and also in trans with heterochromatin at the nuclear periphery (16). Transcriptional interference has been proposed as the cause of silencing of HIV integrated at active genes (23, 25, 38, 39). Read-through transcription of the host gene may prevent preinitiation complex formation at the HIV promoter, when the host gene and the provirus have the same or a convergent orientation. Alternatively, it has been proposed that host gene transcription may confer positive effects on HIV expression when the two are in the same orientation (25), but this model is not consistent with the fact that latent HIV does not exhibit a trend toward convergent integration (24, 39). Discrepancies between models may be due to position effects such as differences in the distances between transcriptional start sites and splice sites (25, 38). As active integrations are also common in introns of active genes, various cis and trans determinants may influence whether or not host gene transcription interferes with HIV promoter activity and, ultimately, whether a provirus will become latent.
Using a previously established model of HIV postintegration latency consisting of the infection of Jurkat cells with a green fluorescent protein (GFP)-encoding HIV minigenome and selecting for reversible silent integrations, we investigated the transcriptional consequences of latent HIV integration into introns of cellular genes and the involvement of chromatin reassembly factors (CRFs) in the transcriptional interference that a host gene exerts on the integrated cryptic HIV promoter. Cryptic promoters might remain inactive due to the repressive chromatin configuration established by CRFs during transcription elongation (7, 31, 44). We have previously demonstrated the involvement of CRFs in the silencing of an HIV transcriptional model in Saccharomyces cerevisiae (57). The present paper reports that depletion of human CRFs such as Spt6, Chd1, and FACT, or the histone chaperones ASF1a and HIRA, promotes HIV reactivation, concomitantly with chromatin relaxation and a decrease in general RNA polymerase activity.
MATERIALS AND METHODS
Cell lines, culturing conditions, and cell treatments.
We have used a previously established model of HIV-1 latency based on the infection of Jurkat T cells at a low multiplicity of infection (MOI) with HIV-derived particles containing the minigenome carried on plasmid pEV731 (LTR-Tat-internal ribosome entry site [IRES]-GFP-LTR) (28). Briefly, GFP-positive, productively infected cells were isolated, and clones were obtained, including J-Act C9. GFP-negative cells were treated with tumor necrosis factor alpha (TNF-α), the resulting GFP-positive, latently infected cells were isolated, and clones were generated from single cells. J-Lat H2, A1, and A2 have been previously described (28). Clone J-Lat E27 was newly generated in this study. Additionally, clones J-Lat 6.3 and 8.4 generated similarly with a full-length GFP-encoding HIV-1 vector (HIV-R7/E−/GFP) described elsewhere were also used (28).
Jurkat-derived cells were grown in RPMI 1640 medium (Gibco) supplemented with 10% fetal bovine serum, 2 mM l-glutamine, 100 U/ml penicillin, and 100 mg/ml streptomycin at 37°C under a 5% CO2 atmosphere. When indicated, Jurkat clones were treated with 10 nM phorbol myristate acetate (PMA; Sigma-Aldrich), 400 nM trichostatin A (TSA; Sigma-Aldrich), 10 ng/ml TNF-α (Sigma-Aldrich), and 5 μM 5-aza-2′-deoxycytidine (Sigma-Aldrich) or 10 mM hexamethylene bisacetamide (HMBA; Sigma-Aldrich) (13).
HEK-293T cells were grown under the same conditions as those for Jurkat cells in Dulbecco's modified Eagle's medium (Gibco) supplemented with 10% fetal bovine serum, 100 U/ml penicillin, and 100 mg/ml streptomycin.
Flow cytometry analysis and sorting.
GFP fluorescence was measured in a Cytomics FC500 MPL flow cytometer (Beckman Coulter Inc., Fullerton, CA). A two-parameter analysis was used to distinguish GFP-derived fluorescence from background. Fluorescence was represented in logarithmic scale. The optical alignment was set up using 10-nm fluorescent beads (Flow-Check fluorospheres; Beckman Coulter). Cell sorting was carried out with the Moflo high-speed sorter (Dako-Cytomation, Fort Collins, CO).
Viral production and cell infections.
HEK-293T cells (Clontech) were used to produce viral particles containing the HIV LTR-Tat-IRES-GFP minigenome (pEV731) (29) or the pLKO.1 vector containing different short hairpin RNAs (shRNAs; Sigma Aldrich). 293T cells (2.5 × 106) were transfected with the indicated plasmid of interest (10 μg), pCMVΔR8.91 (15 μg), and pVSVG (5 μg) in 10-cm dishes using calcium phosphate. Medium was collected every 24 h for 2 days and ultracentrifuged for 1 h 30 min at 26,000 rpm and 4°C in a sucrose gradient to concentrate the viral particles. Pellet containing the viruses was dissolved in medium and used for the infection. Cells were infected using the spinoculation system, with centrifugation at 1,200 rpm for 2 h at room temperature. Cells infected with shRNA-expressing pLKO.1 vectors were selected with 2 mg/ml puromycin (Sigma-Aldrich) 24 h after infection.
Sequencing of flanking genomic regions and PCR products.
The GenomeWalker kit (BD Biosciences) was used to sequence flanking genomic regions according to the manufacturer's instructions. PCR products containing flanking DNA were run in 1% agarose gels with ethidium bromide. Bands obtained were purified from the gel with the QIAquick gel extraction kit (Qiagen) and sequenced.
RNA extraction, reverse transcription-PCR (RT-PCR), and quantitative PCR (qPCR) for expression analysis.
Total RNA was extracted using High Pure RNA isolation (Roche) according to the manufacturer's instructions. cDNA was obtained from 100 ng of total RNA using SuperScript VILO cDNA synthesis (Invitrogen). Indicated gene products were analyzed by PCR with specific oligonucleotides, followed by visualization in an agarose gel. When indicated, quantification of gene products was performed by real-time PCR using LightCycler 480 SYBR green I Master (Roche, Indianapolis, IN). Each value was corrected by human glyceraldehyde-3-phosphate dehydrogenase (GAPDH) or 60S ribosomal protein L31 (RPL31) and expressed as relative units.
Analysis of chromatin accessibility to MNase.
The accessibility of nucleosomal DNA to micrococcal nuclease (MNase) was analyzed as described elsewhere (29), using 0.4 U of MNase for 25 min. Genomic DNA was extracted using the DNeasy blood and tissue kit (Qiagen) according to the manufacturer's instructions. DNA was run in a 1.5% agarose gel and stained with ethidium bromide. The band corresponding to the mononucleosome was cut and purified using the QIAquick gel extraction kit (Qiagen), and DNA was used in a real-time PCR with specific oligonucleotides. Naked genomic DNA was used as a control. DNA was cut with 0.04 U MNase for 5 min, phenol precipitated, run in a 1.5% agarose gel, and stained with ethidium bromide. DNA with a size similar to that of the mononucleosome was cut and purified.
Sequences of shRNAs and PCR primers.
Sequences of oligonucleotides used for conventional or real-time PCR and of short-hairpin interfering RNAs are included in Tables S1 and S2 in the supplemental material.
Yeast experiments.
The wild-type strain BY4741 and the isogenic mutants indicated in Fig. S8 in the supplemental material were transformed with plasmids pTy1-HIV and pTy1-HIVTARless (57) and incubated in yeast extract-peptone-dextrose (YPD) medium until mid-log growth. RNA was extracted, and the levels of HIV-derivative mRNAs were quantified by Northern hybridization as described elsewhere (57).
RESULTS
Characterization of latent HIV integration into introns of active genes.
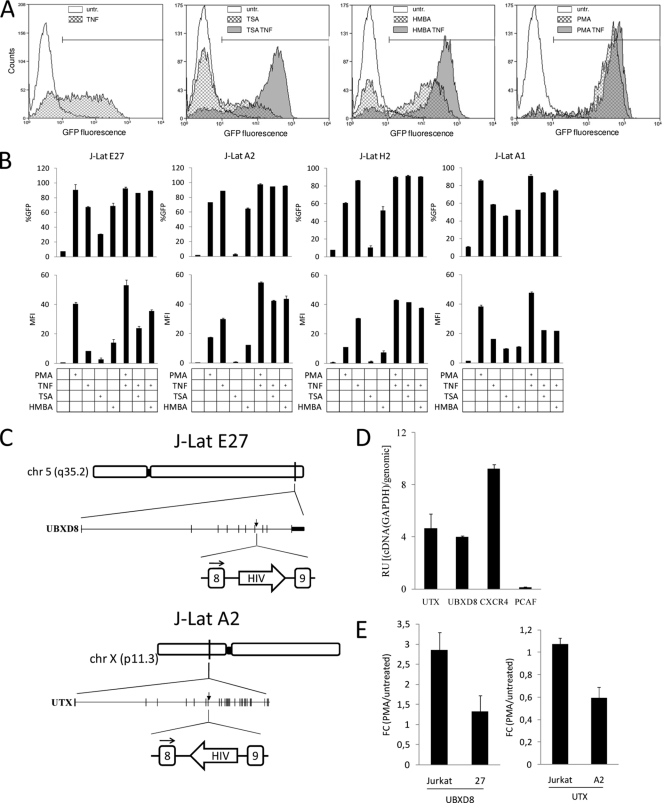
Postintegrative HIV latency has been associated with provirus integration at regions of constitutive heterochromatin such as centromeric alphoid repeats, gene deserts, or introns of very highly expressed genes (39). Four representative Jurkat-derived clones harboring a single latent version of an HIV minigenome expressing Tat and GFP under the control of the viral 5′ LTR (J-Lat) were used to further investigate the causes of HIV promoter repression associated with the site of integration. J-Lat H2 cells contain the HIV-derived provirus at centromeric alphoid repeats (28), while J-Lat A1 cells harbor the HIV minigenome in an intergenic region at position ChXp21.1 (16, 28), and J-Lat A2 cells contain the HIV construct at intron 8 of the UTX gene (ChXp11.3), in a configuration opposite to the transcriptional orientation of this gene (Fig. 1C) (28). Finally, J-Lat E27 cells were prepared for this study with the same HIV vector (pEV731) and procedure as those reported previously (28). Using DNA sequencing, this latent HIV integration was found to occur at intron 8 of the UBXD8 or FAF2 gene (Ch5q35.2), both putatively transcribing in the same direction (Fig. 1C; see also Fig. S1 in the supplemental material). Inducibility of the repressed HIV promoter in these four clones was confirmed by treatment of J-Lat cells with known activators of HIV expression, such as PMA, TNF-α, TSA, and HMBA and measurement of GFP expression by cell cytometry (Fig. 1A and B).
FIG. 1.
Characterization of latent HIV integration into introns of active genes. (A and B) Reactivation of Jurkat clones containing latent HIV minigenome integrations in different genome environments. Cells of clones J-Lat E27, A2, H2, and A1 were incubated with PMA (10 nM), TNF-α (10 ng/ml), TSA (400 nM), or HMBA (10 mM) for 16 h, and HIV-GFP reactivation was followed by fluorescence-activated cell sorting. Data are expressed as percentage of GFP-positive cells (%GFP) or mean fluorescence intensity (MFI). Values represent the mean and range of a representative experiment performed in duplicate. A representative fluorescence-activated cell sorting profile and gating on J-Lat E27 are shown in panel A. (C) Intronic integration of HIV in clones J-Lat E27 and A2. Genome organization of UBXD8 and UTX genes and of the HIV integration in J-Lat E27 and A2 clones. (D) UBXD8 and UTX are active genes in Jurkat cells. RNA was extracted from growing Jurkat cells and used to measure expression of UBXD8, UTX, CXCR4, and PCAF genes by reverse transcription followed by real-time PCR of cDNA (RT-qPCR). In order to compare different amplicons, qPCR was performed in parallel from genomic DNA (gDNA). Data are expressed as relative units (RU) of cDNA amplification/gDNA amplification. Values represent the mean and standard deviation (SD) of a representative experiment performed in triplicate. (E) Effect of PMA treatment on UBXD8 and UTX genes. Jurkat or J-Lat cells were treated or not with PMA (10 nM) for 16 h, and RNA was extracted. UBXD8 and UTX expression was measured by RT-qPCR. GAPDH expression was measured for normalization. Data are expressed as fold change (FC) in expression in PMA-treated cells compared to that in untreated cells.
Transcriptional interference has been proposed to explain HIV promoter repression when integrated into introns of highly expressed genes (3, 38). To investigate the mechanism underlying transcriptional interference and the consequences of HIV integration into human genes and expression, we further characterized the J-Lat clone E27 and, to some extent, the clone A2. First, we confirmed that UBXD8 and UTX genes are basally expressed in Jurkat cells (Fig. 1D). UBXD8 expression has been reported to be increased by mitogenic agents such as PMA (27). We confirmed that UBXD8 was stimulated by PMA in Jurkat cells, but the effect was less marked in J-Lat E27 cells. On the other hand, UTX expression was not responsive to PMA in Jurkat cells but was downregulated in J-Lat A2 cells (Fig. 1E). This suggested that HIV reactivation by PMA could influence expression of the host gene where HIV was integrated (see below).
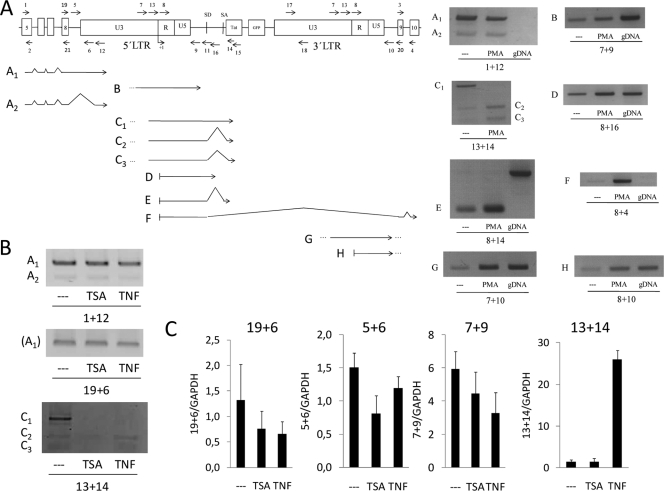
Next, using RT-PCR with oligonucleotides designed to cover several gene and HIV regions, we investigated the effect of the HIV integration into the UBXD8 gene, in terms of transcript species generated (Fig. 2A). J-Lat E27 cells were treated or not with PMA, and RNA was extracted and reverse transcribed with random hexamers prior to PCR amplification. Combining oligonucleotides for UBXD8 exon 5 and U3 of the HIV LTR, we detected transcripts initiated at the UBXD8 promoter retaining intron 8 and extending into the HIV provirus, instead of eliminating them by splicing as expected. In fact, two different products were detected, the second one (A2) due to the presence of a cryptic splicing acceptor in U3 (Fig. 2A; see also Fig. S1 in the supplemental material). These transcripts did not necessarily terminate at the transcription stop site [poly(A)] present in the R region of HIV LTRs, as read-through transcripts were also detected (products B and C1 to C3). Interestingly, splicing of read-through transcripts was enhanced by PMA stimulation. The splicing donor present at the HIV minigenome was recognized, but not the canonical acceptor. Instead, terminal splice sites were generated at two different cryptic sites inside the Tat-encoding sequence (see Fig. S1).
FIG. 2.
PCR-mediated identification of transcripts generated at latent HIV integration sites. (A) RNA extracted from J-Lat E27 cells treated or not with PMA (10 nM) for 16 h was used to generate cDNA by reverse transcription with random hexamers. Several oligonucleotide combinations were used on PCR amplification experiments to detect the appearance of different transcripts, shown in the figure below a schematic representation (out of scale) of the HIV minigenome at the UBXD8 intron 8. The HIV integration divides intron 8 into two pieces of 404 bp (upstream) and 1,846 bp (downstream). The positions of PCR primers used are indicated by arrows. Agarose gel electrophoresis of PCR products illustrating each of the cDNA species is shown. When required, DNA sequencing of PCR products was applied to identify the exact nature of a cDNA fragment (see Fig. S1 in the supplemental material). PCR amplification from genomic DNA was performed as a product size control where indicated. (B and C) J-Lat E27 cells were treated or not with TNF-α (10 ng/ml) or TSA (400 nM) for 16 h, and transcripts were characterized by RT-PCR with the oligonucleotide pairs indicated, by conventional PCR (B) or real-time PCR (C). Primer pairs 19 plus 6 and 5 plus 6 detected the transcript named A1, primer pair 7 plus 9 measured transcript B, and primer pair 13 plus 14 measured transcripts C.
Nonetheless, when a combination of primers was used to detect transcripts initiated at the HIV 5′ LTR promoter, unspliced and correctly spliced transcripts were detected (PCR products D and E), and this transcription was enhanced by PMA treatment (Fig. 2A). An unexpected transcription product resulting from a splicing event between the HIV splicing donor and the acceptor at the end of the UBXD8 intron 8 (product F) (see Fig. S1 in the supplemental material) was identified. As this was entirely dependent on the PMA treatment, we speculate that it was initiated at the HIV promoter and could be an example of downstream, truncated host gene products generated by the insertion of lentiviruses. Finally, we also detected the presence of read-through transcripts at the 3′ LTR (product G), as well as transcripts that could be initiated at the 3′ LTR, which may also work as an inducible promoter (product H).
Because PMA stimulates not only HIV activation but also that of UBXD8, we explored the responses to TSA and TNF-α of some of the chimeric UBXD8-HIV RT-PCR products (Fig. 2B and C). Any of these products was enhanced by these HIV inducers, except for the spliced products C2 and C3. This indicated that transcripts containing A1 and A2, B, and C1 products were originated at UBXD8, but C2 and C3 may be originated by an alternative transcription start site downstream of the NF-κB binding sites present at the HIV promoter.
Quantification of transcripts initiated at HIV or host gene promoters.
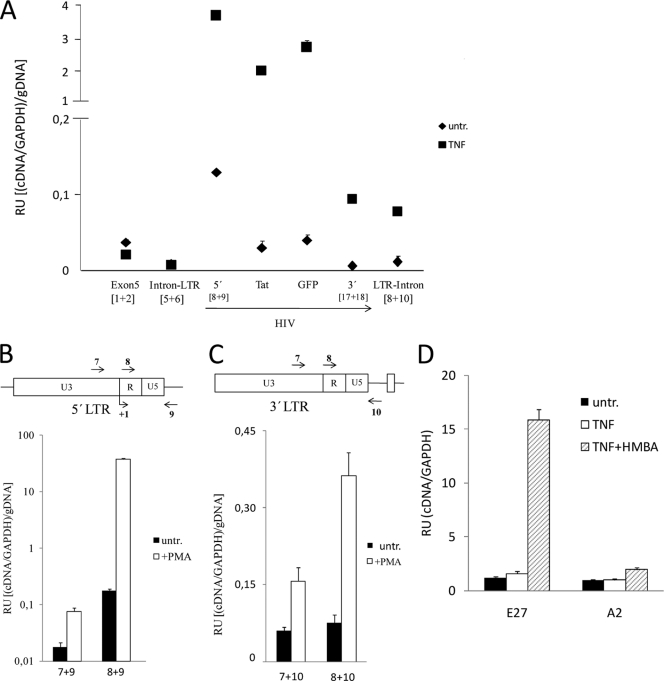
These results suggested that HIV sequences could be included in mature transcripts initiated at the UBXD8 promoter or that UBXD8 downstream exons could be included in transcripts produced due to the reactivation of 5′ or 3′ LTR promoters. In order to quantify the relative frequencies of these events, we performed quantitative PCR on synthesized cDNA generated from J-Lat E27 cells, untreated and treated with TNF-α, which activates the HIV promoter but not UBXD8. In order to compare different amplicons, amplification of cDNA was corrected by amplification of genomic DNA with the same sets of primers (Fig. 3A). This experiment showed that there is much less transcription from UBXD8 into the 5′ LTR (amplicon intron-LTR) than transcription through upstream exons, meaning that most of the UBXD8 transcription would correctly splice intron 8 and the integrated HIV. Similarly, transcripts containing part of intron 8 derived from the HIV genome (amplicon LTR-intron) are not abundant, although their expression significantly increased upon TNF-α treatment.
FIG. 3.
Quantification of transcripts initiated at HIV or host gene promoters. (A) Relative quantification of transcripts generated at the HIV integration site in clone J-Lat E27. J-Lat E27 cells were untreated or treated with TNF-α (10 ng/ml) for 24 h, RNA was extracted, and gene expression was measured by reverse transcription followed by real-time PCR of cDNA (RT-qPCR). Amplicons corresponding to 5′ LTR (R-gag, primers 8 and 9 [Fig. 2A]), Tat, GFP, and 3′ LTR (U3, primers 17 and 18) were used to measure HIV expression. Additionally, amplicons to measure UBXD8 expression at exon 5 (primers 1 and 2) or expression through the boundaries of intron 8 and 5′ LTR (primers 5 and 6) or 3′ LTR and intron 8 (primers 8 and 10) were used. GAPDH expression was measured for normalization. In order to compare different amplicons, qPCR was performed in parallel from genomic DNA (gDNA). Data are expressed as relative units (RU) of (cDNA amplification/GAPDH)/gDNA amplification. Values represent the mean and range of a representative experiment performed in duplicate. (B) HIV transcripts are generated mainly from the viral promoter. J-Lat E27 cells were untreated or treated with PMA (10 nM) for 16 h, RNA was extracted, and transcription through the 5′ LTR was measured by RT-qPCR with the oligonucleotides indicated. GAPDH expression and qPCR from gDNA were used for normalization as in panel A. Data are expressed as in panel A. (C) Downstream transcripts are mainly originated from 3′ LTR. PMA treatment of J-Lat E27 cells was performed as in panel B, and transcription through the 3′ LTR was measured with the qPCR primers indicated. (D) Expression of downstream gene exons is induced upon HIV activation. J-Lat E27 and A2 cells were untreated or treated with TNF-α (10 ng/ml) and HMBA (10 mM) for 24 h, RNA was extracted, and transcription through UBXD8 exon 9 was measured by RT-qPCR. GAPDH expression was measured for normalization. Data are expressed as RU of UBXD8 exon 9/GAPDH expression. In panels B to D, values represent the mean and SD of representative experiments performed in triplicate.
When transcription of HIV regions was analyzed, we unexpectedly observed that transcription from the 5′ LTR promoter in the absence of TNF-α was considerably high, in contrast to the latent phenotype of these cells (GFP negative). Nonetheless, the presence of Tat and GFP-encoding sequences in the transcripts was significantly lower, and the presence of the downstream U3 region was even lower (Fig. 3A). Only correctly terminated transcripts may generate the GFP products that are the basis for the definition of latent or productive infection in this and other cellular models of latency. Similarly, Tat expression was not detected by Western blotting until the HIV promoter was stimulated (data not shown). These results suggest that latency is partially produced by a block in the elongation of HIV transcripts, as previously suggested (1). Upon TNF-α stimulation, all HIV regions were upregulated, but a strong decrease in the presence of region U3 in HIV transcripts was still observed (Fig. 3A). This indicates that latency imposed by a transcription elongation block could be overcome by stimulation of the HIV promoter activity and transcriptional initiation. Similar results concerning a decrease in transcription along the HIV minigenome, under basal and induced conditions, were obtained with J-Lat A2 cells and a heterogeneous population of J-Lat cells (data not shown).
It was ascertained that HIV transcripts mainly initiated at the HIV transcription start site, rather than at upstream UBXD8 regions, even after PMA stimulation, by comparisons with amplification with oligonucleotides 7 plus 9 and oligonucleotides 8 plus 9 (Fig. 3B). Similarly, we showed that transcription downstream of the HIV minigenome, upon stimulation, mainly derived from the 3′ LTR, which can act as a promoter, not from transcripts initiated at the 5′ LTR or UBXD8 promoters unable to terminate at the 3′ LTR poly(A) sequence (Fig. 3C).
Finally, we unequivocally demonstrated that reactivation of HIV when it is integrated cotranscriptionally into a cellular gene may induce the expression of downstream exons. In J-Lat E27, but not in a control clone, expression of UBXD8 exon 9 was increased by TNF-α treatment, most markedly when combined with HMBA, an elongation-enhancing agent (Fig. 3D). Expression of upstream exons was not affected by the combined treatment (data not shown).
HIV induction downregulates host gene expression at the integration site.
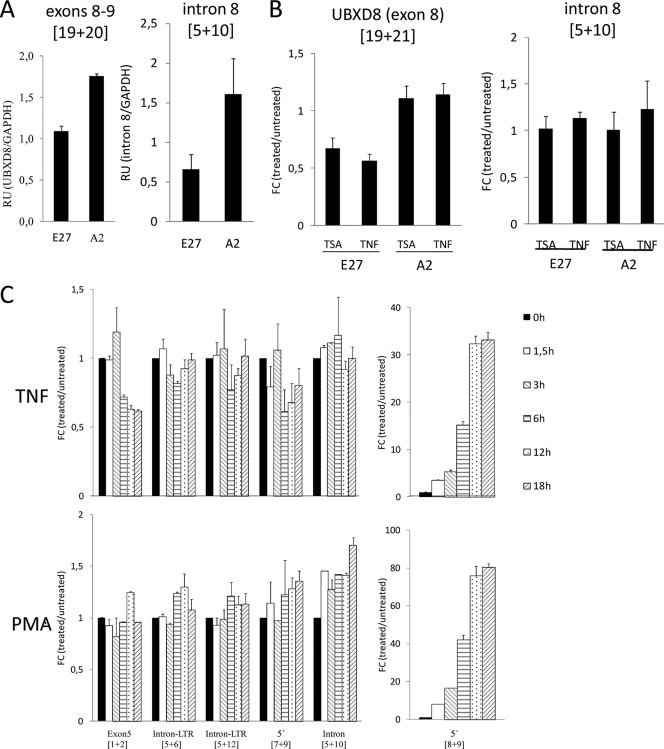
Next, we explored what effects HIV induction might have on the expression of the host gene. Initially, we observed that UBXD8 expression (exon 8-exon 9) was lower in J-Lat E27 than in noninfected control cells (i.e., cells that did not contain HIV in this cellular gene) (Fig. 4A). In Fig. 1E, we have already illustrated the finding that PMA reactivation of latent HIV downregulated UBXD8 in the J-Lat E27 clone and UTX in the A2 clone, compared to parental Jurkat cells. Similarly, TSA or TNF-α induction of HIV downregulated UBXD8 exon 8 in J-Lat E27, but not in a control clone (Fig. 4B), as well as UBXD8-HIV chimeras (Fig. 2C). As a control, we measured expression of the wild-type (unintegrated) intron 8 allele with a primer pair that flanked the integration site. As expected, expression was lower in J-Lat E27 (i.e., 1 intact allele) than in A2 (i.e., 2 alleles) but was not affected by TSA or TNF-α in any case (Fig. 4A and B, right panels).
FIG. 4.
HIV induction downregulates host gene expression at the integration site. (A) UBXD8 expression in J-Lat clones. RNA from J-Lat E27 and A2 cells was extracted, and expression of the wild-type UBXD8 transcript was measured by RT-qPCR with a pair of oligonucleotides in exons 8 and 9 (19 plus 20) flanking the HIV integration site at intron 8 in J-Lat E27. As a control, expression of uninterrupted intron 8 was measured with the primer pair 5 plus 10. GAPDH expression was measured for normalization. (B) UBXD8 expression upon HIV reactivation. J-Lat E27 and A2 cells were untreated or treated with TNF-α (10 ng/ml) or TSA (400 nM) for 24 h, RNA was extracted, and transcription through UBXD8 exon 8 was measured by RT-qPCR with oligonucleotides 19 and 21 or through intact intron 8 with oligonucleotides 5 and 10. GAPDH expression was measured for normalization. Data are expressed as fold change (FC) in UBXD8/GAPDH expression in treated cells compared to that in untreated cells. (C) Time course response to TNF-α and PMA. J-Lat E27 cells were treated with TNF-α (10 ng/ml) or PMA (10 nM) for the time points indicated in the figure, and transcription through different UBXD8-HIV regions was measured by RT-qPCR with the primer pairs indicated. GAPDH expression was measured for normalization. Data are expressed as fold change (FC) in primer pair/GAPDH expression in treated cells compared to that in untreated cells (time zero). Values represent the mean and range of representative experiments performed in duplicate.
Next, we followed changes in expression of the different regions over time after treatment of J-Lat E27 cells with PMA or TNF-α (Fig. 4C). Both treatments induced HIV (5′ LTR). As a consequence, UBXD8 (exon 5) expression, as well as expression of chimeric transcripts entering the 5′ LTR to some extent, was downregulated upon TNF-α treatment. Instead, as PMA also activated UBXD8 (seen in the unintegrated intron 8 PCR), both effects were neutralized.
Preparation of J-Lat clone 6.3 was based on the same methodology but using a full-length HIV clone (HIV-R7/E−/GFP) (28). Reactivation of HIV by TNF-α plus 5-aza-2′-deoxycytidine in this clone, but not in a different clone, promoted downregulation of the host gene PPP5C where HIV is integrated (see Fig. S2 in the supplemental material). In the course of our experiments, we discovered that depletion of serum from the J-Lat cell culture medium promoted reactivation of the HIV promoter and an increase in expression of GFP (see Fig. S3). As a consequence, UBXD8 expression was downregulated, specifically in the J-Lat clone E27, but not in control clones.
Taken together, our data demonstrate that HIV reactivation negatively regulates the expression of upstream exons, presumably by interfering with the activity of the host gene promoter. This indicates that transcriptional interference might be reciprocal, that is, in both directions.
Depletion of chromatin reassembly factors reactivates intron-integrated latent HIV concomitantly with increased chromatin accessibility at the HIV promoter.
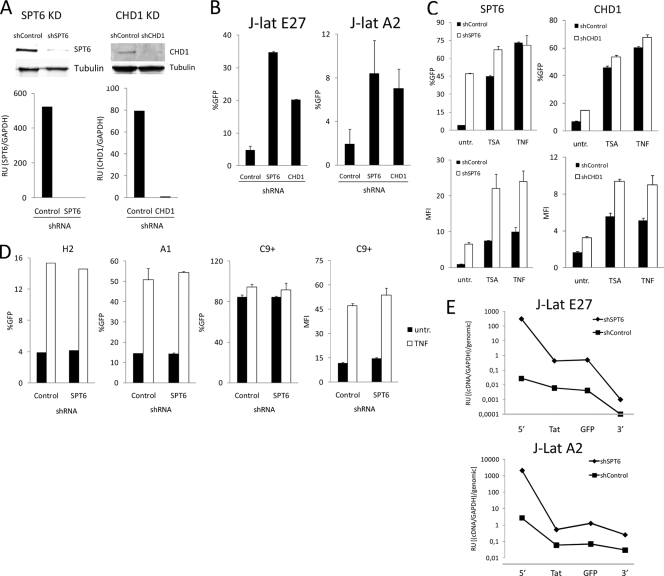
We have previously suggested that depletion of cotranscriptional chromatin reassembly factors (CRFs) could derepress latently integrated HIV proviruses (57). We found that shRNA-mediated depletion of human Spt6 and Chd1 (Fig. 5A) promoted the reactivation of latent HIV in J-Lat clones E27 and A2 harboring the HIV minigenome integrated into introns of active genes (Fig. 5B), but not in clones H2 or A1, in which HIV is not integrated into transcriptional units (Fig. 5D). Moreover, Spt6 and Chd1 depletion enhanced reactivation by drug treatments such as TSA or TNF-α in the J-Lat clone E27 (Fig. 5C). Similarly, Spt6 depletion enhanced reactivation of J-Lat 8.4 cells, containing a latent full-length HIV provirus, by TNF-α plus 5-aza-2′-deoxycytidine (see Fig. S4 in the supplemental material). J-Lat clones H2 and A1 were also reactivated by TNF-α, but Spt6 depletion did not enhance this response (Fig. 5D). J-Act C9 contains a constitutively active version of the HIV minigenome at an intron of the TYK2 gene (Ch19p13.2), integrated contrary to the direction of transcription. In this context, Spt6 depletion did not further enhance basal HIV expression or its response to TNF-α (Fig. 5D).
FIG. 5.
Depletion of chromatin reassembly factors reactivates intron-integrated latent HIV. (A and B) shRNA-mediated depletion of Spt6 and Chd1 in clones J-Lat E27 and A2. Cells of the indicated latently infected clones were infected with control, Spt6, or Chd1 shRNA expression lentiviruses (pLKO.1-Puro), and 7 days after puromycin selection, HIV-GFP reactivation was measured by fluorescence-activated cell sorting and expressed as the percentage of cells that became GFP positive (%GFP). Depletion of these factors was tested by immunoblotting and RT-qPCR with specific antibodies (Spt6 [ab32820; Abcam], Chd1 [H00001105-A01; Abnova]) or primers, respectively, and is shown for clone J-Lat E27 as an example (A). (C) HIV activation by TSA or TNF-α is enhanced in Spt6-depleted J-Lat cells. J-Lat E27 and A2 cells infected with control or Spt6 shRNA expression vectors and puromycin selected for 6 days were treated or not with TSA (400 nM) or TNF-α (10 ng/ml) for 24 h. HIV-GFP expression was measured by fluorescence-activated cell sorting and was expressed as percentage of GFP-positive cells (%GFP) or mean fluorescence intensity (MFI). (D) Spt6 depletion in nonintronic J-Lat clones. Cells of J-Lat clones H2 and A1 and of the constitutively expressing clone J-Act C9 were infected with control or Spt6 shRNA expression lentiviruses and selected in puromycin as in panel A, and HIV-GFP expression was measured by fluorescence-activated cell sorting. A portion of the cells were treated with TNF-α (10 ng/ml) for 24 h prior to fluorescence-activated cell sorting analysis. HIV-GFP expression is expressed as percent GFP-positive cells and as MFI for clone J-Act C9, as percent GFP-positive cells is saturated. (E) Analysis of HIV transcription in J-Lat clones upon Spt6 depletion. Cells of J-Lat E27 and A2 clones depleted of Spt6 as in panel A were RNA extracted, and HIV expression was measured by RT-qPCR with different amplicons covering the HIV genome as in Fig. 3A.
HIV reactivation upon depletion of Spt6 in J-Lat clones E27 and A2 was also analyzed by RT-PCR with oligonucleotides covering different regions of the provirus (Fig. 5E). Spt6 depletion enhanced transcription of the initial region of the provirus by several orders of magnitude, suggesting that it favors promoter activation and initiation, but an elongation block remained. Nonetheless, the overall amount of terminated transcripts was increased upon depletion of Spt6, and this explained the increase in GFP expression. Similarly, depletion of Chd1 enhanced transcription initiation from the HIV promoter (see Fig. S5 in the supplemental material).
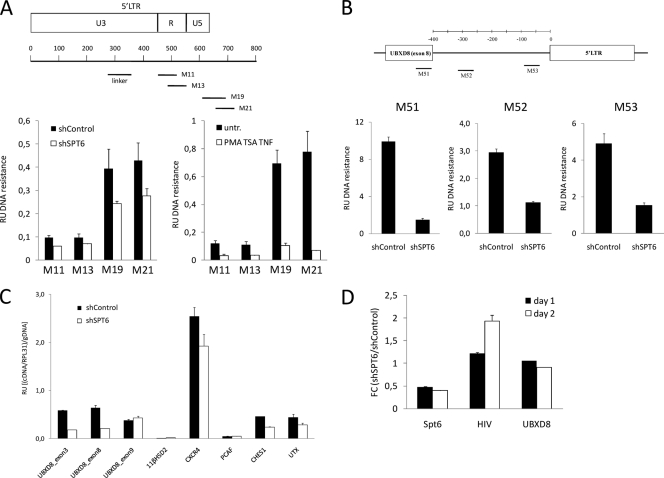
CRFs travel with the elongating RNA polymerase and rebuild nucleosomes after the passage of the transcribing machinery. In the absence of Spt6, we predicted that chromatin would become more relaxed upon transcription of UBXD8 and, consequently, the integrated HIV promoter would be more accessible to transcription factors and preinitiation complex. We investigated chromatin accessibility at the HIV promoter and the UBXD8 gene using a micrococcal nuclease (MNase) sensitivity assay. Upon MNase digestion of chromatin from J-Lat E27 cells expressing Spt6 or control shRNA, the mononucleosomal DNA was purified and the sensitivity of different regions was assessed by quantitative PCR (qPCR) with specific oligonucleotides (Fig. 6A and B). Regions at the HIV promoter, presumably covered by nucleosomes, became more sensitive to MNase upon Spt6 depletion, although not as markedly as when optimal reactivation of HIV was promoted by treatment with PMA, TSA, and TNF-α as a control (Fig. 6A; see also Fig. S6 in the supplemental material). Increased sensitivity to MNase upon Spt6 depletion was not limited to the HIV promoter, but rather it extended to several intron and exon regions of the UBXD8 gene upstream of the site of integration (Fig. 6B), indicating that this was related to the lack of proper chromatin reassembly associated with the movement of the transcriptional machinery along the host gene.
FIG. 6.
Depletion of chromatin reassembly factors reactivates HIV concomitantly with increased chromatin accessibility at the HIV promoter. (A) Increased sensitivity of chromatin to MNase digestion upon Spt6 depletion. J-Lat E27 cells expressing control or Spt6 shRNA were submitted to controlled MNase digestion as indicated in Materials and Methods after nucleus preparation. After purification of mononucleosomal DNA, DNA resistance to MNase was quantified using real-time PCR and oligonucleotides covering HIV regions depicted in the schematic drawing. Signals were normalized to naked DNA purified from the same cells before MNase digestion, PCR amplified with the same set of primers, and to the values of PCR amplification with primers for a region considered to correspond to linker DNA free of nucleosomes. Data are expressed as relative units of DNA resistance to MNase, i.e., (PCR amplification/linker)/gDNA. As a control, MNase resistance of chromatin in J-Lat E27 cells treated with PMA (10 nM), TSA (400 nM), and TNF-α (10 ng/ml) for 4 h, compared to that in untreated cells, was also assayed and is shown in the right panel. (B) UBXD8 chromatin sensitivity to MNase. DNA resistance to MNase at UBXD8 exon 8-intron 8 regions in J-Lat E27 cells expressing control or Spt6 shRNA was determined as described for panel A with three pairs of specific oligonucleotides. (C) Effect of Spt6 depletion on cellular gene expression. RNA was extracted from J-Lat E27 cells expressing control or Spt6 shRNA, and expression of several genes or the indicated exons of the UBXD8 gene was measured by RT-qPCR using specific oligonucleotides. RPL31 expression was measured for normalization. In order to compare different amplicons, qPCR was performed in parallel from genomic DNA (gDNA). Data are expressed as relative units (RU) of (cDNA amplification/RPL31)/gDNA amplification. (D) HIV reactivation occurs earlier than UBXD8 inhibition upon Spt6 depletion. J-Lat E27 cells were infected with control or Spt6 shRNA expression lentiviruses and selected in puromycin. At 1 or 2 days after puromycin addition, RNA was extracted and Spt6, HIV, and UBXD8 gene expression was measured by RT-qPCR. RPL31 expression was measured for normalization. Data are expressed as fold change (FC) of relative units (RU) of cDNA amplification/RPL31 in shSpt6 compared to that in shControl.
According to our data shown above, HIV reactivation could downregulate host gene expression, in particular UBXD8 expression in J-Lat E27 cells. Using RT-qPCR, we investigated whether HIV reactivation upon Spt6 depletion affected UBXD8 expression in this clone (Fig. 6C). Upon depletion of Spt6, expression of UBXD8 was downregulated when oligonucleotides specific for upstream exons were used (exons 3 and 8) but not when downstream exon 9 was measured, due to the herein-reported effect of HIV activation on downstream gene expression. In parallel, we observed that the expression of other active cellular genes (CXCR4, CHES1, and UTX) was also compromised by the depletion of Spt6 (Fig. 6C). Moreover, UBXD8 expression was also affected by Spt6 depletion in a different clone (see Fig. S7 in the supplemental material). Taken together, these findings suggest that UBXD8 downregulation upon Spt6 depletion might be due not only to the reactivation of HIV but also to CRFs being essential for efficient RNA polymerase II (Pol II)-mediated transcription. As UBXD8 downregulation in J-Lat E27 cells could diminish transcriptional interference over the integrated HIV promoter, this by itself could explain why depletion of Spt6 reactivated HIV. In order to clarify whether Spt6 depletion reactivates HIV due to a defect in cotranscriptional chromatin reassembly, before UBXD8 expression is impaired, we performed a time course experiment after infection with the Spt6 shRNA-expressing vector and Spt6, HIV, and UBXD8 expression was monitored by RT-qPCR. Concomitantly with Spt6 inhibition, HIV reactivation was detected, earlier than UBXD8 downregulation (Fig. 6D).
Knockdown of genes required for repression of cryptic promoters reactivates latent HIV.
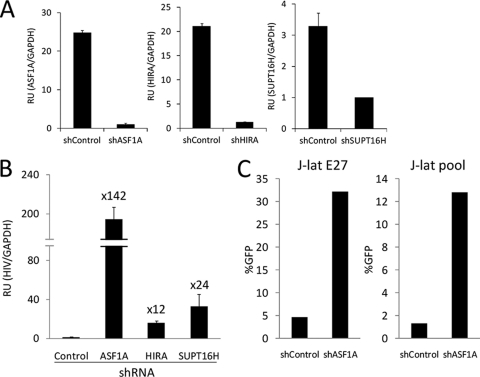
Transcriptional interference has been proposed as an explanation of repression of cryptic promoters from within coding regions. Several chromatin and transcription-related factors have been identified as being involved in this cotranscriptional repression in a yeast genetic screening, including Spt6 and Chd1 (7, 31). Additional factors identified were the histone chaperones HIR1 to -3, HPC2, and ASF1, involved in chromatin assembly, and Spt16, a subunit of the FACT complex, involved in nucleosome disassembly and transcription elongation. Spt6, Chd1, and Spt16 involvement in HIV repression was initially revealed in yeast genetic analysis with a minimal HIV transcriptional system that was blocked due to the assembly of a nucleosome in the initial ca. 200 bp beyond the transcription start site (TAR-encoding region) (57). A TAR-less promoter was derepressed. In addition, the full-length HIV promoter was derepressed in spt6, spt16, and chd1 genetic backgrounds, suggesting an involvement of these factors in the nucleosome-mediated repression of the chimeric yeast-HIV promoter. We have now assessed the involvement of HIR1 to -3, HPC2, and ASF1 in the yeast system and found that the deletion of any of these genes was sufficient to increase the activity of the full-length promoter to the level of the deleted one (see Fig. S8 in the supplemental material).
We tested the participation of the human orthologs of some of these factors in the repression of HIV integrated at active genes by introducing specific shRNA constructs into J-Lat cells. Depletion of HIRA, ASF1a, and Spt16 was measured by RT-qPCR and ranged between 75 and 95% (Fig. 7A). Upon depletion of these three factors, HIV was reactivated in J-Lat E27 cells, most markedly by inhibiting ASF1a expression (Fig. 7B). Comparable results were obtained by repeating this experiment in the J-Lat A2 clone (data not shown) and in the J-Lat heterogeneous population (Fig. 7C). As occurred upon Spt6 depletion, and concomitantly with HIV reactivation by depletion of HIRA, ASF1a, and Spt16 in the J-Lat E27 cells, UBXD8 expression was downregulated. In a different clone, only ASF1a depletion downregulated UBXD8 and other genes; HIRA or Spt16 inhibition did not (see Fig. S9 in the supplemental material), supporting the idea that reactivation of the provirus is due not merely to downregulation of the host genes in the knockdown.
FIG. 7.
Knockdown of genes required for repression of cryptic promoters reactivates latent HIV. (A and B) J-Lat E27 cells were infected with control, ASF1a, HIRA, or SUPT16H shRNA expression lentiviruses and selected in puromycin as described for Fig. 5A, and HIV-GFP expression was measured by RT-qPCR with oligonucleotides 8 and 9. GAPDH expression was used for normalization. The fold change of HIV induction in cellular factor-depleted cells compared to that in control cells is indicated. Depletion of these factors was tested by RT-qPCR with specific primers and is shown in panel A. Values represent the mean and range of a representative experiment performed in duplicate. (C) J-Lat E27 and J-Lat pool cells were infected with control or ASF1a shRNA as described for panels A and B, and HIV-GFP expression was measured by fluorescence-activated cell sorting. Data are expressed as percentages of GFP-positive cells (%GFP).
Our results show that in certain genomic environments, chromatin has a repressive effect on the initiation of HIV transcription. When integrated within coding regions, CRFs participate in the maintenance of this repressive chromatin organization at the HIV promoter, and this could be the origin of the transcriptional interference that a transcribing unit exerts on an internal promoter.
DISCUSSION
HIV transcription depends on host cellular transcription factors and consequently on the activation state of the infected cell, on the viral Tat protein, and on the chromatin state of the viral promoter at the integration site. When HIV infects a homogeneous cell population, viral expression is heterogeneous among individual integration events (clones) (29). This heterogeneity can be attributed to the integration site and, consequently, to the chromatin state of the HIV promoter that influences the accessibility of the transcription factors and the loading of the preinitiation complex. It cannot be ruled out that chromatin compaction may also influence progression of the elongating polymerase, termination, or reinitiation. HIV is a highly effective virus that is expressed in the majority of infected cells, allowing viral replication, presumably by integration into open chromatin represented by active genes (51, 59). Occasionally, the HIV provirus is not basally expressed but can be reactivated by disturbing the equilibrium between activation and repression with agents that interfere with chromatin, such as histone deacetylase inhibitors, or inducers of cell signaling that activate transcription factors. This behavior resembles viral latency, which in vivo certainly depends on additional components (11, 35). Analysis of the integration sites that are associated with a latent phenotype has shown that it is produced by integration into regions of obviously compacted chromatin or, more importantly, into introns of highly expressed genes (24, 28, 39). It is not obvious how the chromatin environment could be a determinant for repression when a provirus is embedded in a transcribing gene. Instead, it has been proposed that read-through transcription from an upstream promoter may interfere with HIV transcription by disturbing assembly of the preinitiation complex, irrespective of the relative orientation between the host gene and the provirus (23, 38). Nonetheless, there have also been described situations in which upstream transcription could indeed enhance transcription from HIV-1 proviruses that are in the same orientation as the host gene (25). Position effects may explain the discrepancy between the different models used. The mechanism by which transcriptional interference is exerted has not previously been unraveled. Here, we propose that the chromatin reassembly machinery and associated factors traveling with the elongating form of RNA polymerase II (Pol II) through the HIV promoter actively maintain a chromatin environment refractory to HIV promoter activation.
In this paper, we have addressed the question of how HIV latency is established upon integration into a highly expressed gene, in particular, what transcriptional consequences the provirus integration has for the host genes and for the virus itself and what host factors are involved in maintaining HIV repression in this context. For this, we have used a minimal model system proven to be useful for studying the genomic determinants of postintegration latency. Two J-Lat clones harboring HIV integrations into introns of highly active genes, in the same direction as or opposite to the transcription of the host gene, have been analyzed and compared to clones where the HIV provirus is in other genomic environments.
We observed that HIV integration into an intron of a gene does not abolish expression of its normally spliced transcript, although expression is diminished to some extent. Analyzing the J-Lat clone E27, we were not able to determine the proportion in which normally spliced UBXD8 transcripts derived from the intact or the HIV-containing allele, although we have measured expression of the unintegrated allele (intron 8) and it was normal, proportional to the number of copies in the genome for different clones. Host gene expression was further decreased when HIV was reactivated, presumably at the provirus-containing allele, as the unintegrated allele was not affected by treatments that induce HIV (Fig. 4). Moreover, in clone A2, as HIV is in the single-allele UTX gene (chromosome X), we were able to ascertain that normally spliced UTX transcripts are being produced despite the provirus integration but that this expression is reduced upon HIV reactivation. The reason for this is not clear, and possible mechanisms have not yet been investigated, but it could be due to transcription factor or polymerase trapping by the strong initiating activity of the HIV promoter, a putative form of transcriptional interference. In this line, we have measured by chromatin immunoprecipitation (ChIP) a strong recruitment of Sp1 to the HIV LTR upon TNF-α treatment (see Fig. S10 in the supplemental material). It has recently been proposed that enhancer-blockers and insulators could resemble specialized promoters that sequester the transcriptional machinery (49). The activated HIV promoter could be acting as one of these enhancer-blockers.
A relative comparison of transcripts representing host gene exons or the stimulated initiating HIV indicated that HIV transcription is comparable to or higher than host gene transcription (Fig. 3). Notably, host gene downregulation is much lower than the activation of HIV observed upon stimulation. A limited decrease in host gene expression upon latent HIV reactivation has been also observed in a study (38) that analyzed J-Lat clones 9.2 and 15.4 generated with a full-length, GFP-expressing HIV genome (28). We also observed this with the related J-Lat clone 6.3 (see Fig. S2 in the supplemental material). In a different report, downregulation of the host gene upon latent HIV reactivation by Tat expression was not attributed to transcriptional interference, but rather to a Tat effect, as it affected both alleles (15).
Lenasi et al. (38) reported that host-viral chimeric transcripts at the infected allele in J-Lat 9.2 and 15.4 cells terminate at the poly(A) contained in the 5′ LTR (R-U5 junction). In that case, chimeric transcripts were due to the appearance of cryptic splicing acceptor sites at the intron preceding the 5′ LTR. We have also characterized the existence of several species of chimeric transcripts in J-Lat E27 cells extending into the 5′ LTR and transcripts that cross the 5′ LTR without terminating at the poly(A), with different splicing variants (Fig. 2). Similarly, we have identified transcription through the 3′ LTR. This indicates that poly(A) function is leaky; otherwise, all transcription of the infected allele would terminate at the HIV LTRs and no correctly spliced gene transcripts would be detected. The existence of mature, chimeric transcripts containing the U3 region of the 5′ LTR has been used to argue that transcriptional interference may occur. Moreover, transcription through the 5′ LTR, presumably initiated at an upstream host gene and terminating at the 5′ LTR poly(A), can be detected in heterogeneous populations of HIV-infected primary T cells (38). Nonetheless, transcription interference would still take place even if chimeric transcripts were not detected, because the provirus-containing intron could be efficiently removed from mature transcripts and be present only in short-lived pre-mRNAs. Relative quantification of transcripts shows that chimeric transcripts are significantly less abundant than are transcripts containing upstream exons (chimeric plus wild type).
Interestingly, the alternative splicing varies as a function of PMA treatment: the intron 8/5′ LTR containing the A1 band is decreased compared to spliced A2, while the unspliced C1 product was substituted for the splicing variants C2 and C3. Apparently, PMA stimulation of the host gene promoter favors splicing events, although in all three cases splicing was accepted at cryptic sites, at either the 5′ LTR or the Tat sequences. The reciprocal relationship between transcription elongation and splicing has been extensively reported: elongation rates control alternative splicing and splicing factors can, in turn, modulate Pol II elongation (34, 45, 52). It has to be noted that upon TNF-α treatment, HIV transcripts that initiated shortly upstream of the canonical transcription start site are detected.
Upon stimulation, transcripts initiated at the HIV 5′ LTR are produced, with several splicing variants. Unexpectedly, a transcript that uses the HIV splicing donor and UBXD8 exon 9 acceptor is highly induced. This could explain in part why the relative quantification of HIV transcripts covering different regions shows that Tat, GFP, and 3′ LTR are underrepresented compared to the 5′ LTR region (Fig. 3). Nonetheless, an elongation block or incomplete read-through elongation after HIV transcription initiation also seems to exist, as has been reported previously (1, 36, 47). In agreement with this, exogenously added Tat can reactivate latent HIV (40, 42). Another consequence of our study is that simple measurement of a reporter gene such as GFP is an oversimplification of the transcriptional events taking place around the provirus.
Transcription from the 3′ LTR is also induced upon cell stimulation, being responsible for the expression of downstream UBXD8 exons, most strongly if elongation is stimulated with HMBA in addition to TNF-α treatment (Fig. 3). Despite the 3′ LTR being identical to the 5′ LTR, its promoter activity is always lower, basally and after induction, due to interference of the upstream HIV promoter in the downstream LTR (14, 23). Expression of downstream host gene exons from either or both LTRs could lead to the synthesis of truncated proteins, this being one of the bases of the mutagenic capacity of retroviruses. However, this was not the case in clone E27 according to the predictions made from the sequencing data on amplified cDNAs.
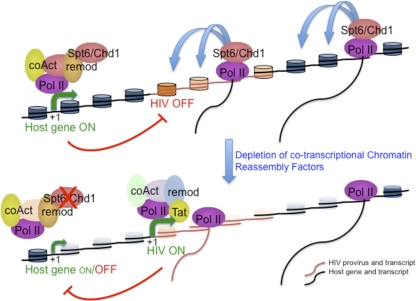
Transcriptional interference has been proposed to explain how a highly transcribing gene may maintain as silent a downstream HIV promoter leading to HIV latency. We report that components of the machinery which reassemble chromatin concomitantly with transit of the transcribing RNA polymerase are necessary for the repression of the cryptic HIV promoter in this context. Upon depletion of these factors, HIV reactivation occurs when integrated into introns of active genes, but not for other genome sites, concomitantly with increased chromatin accessibility at the HIV promoter and host gene. This effect produces synergies with other treatments that induce HIV promoter activation and could be explored as therapeutic interventions against the latent reservoir of the virus, although the general effects of knocking down these factors on gene expression would in principle discourage this.
The situation described here resembles the mechanism reported in Saccharomyces cerevisiae to suppress cryptic transcription from within coding regions. In a first report, the transcription elongation factor Spt6 was identified as a repressor of transcription initiation from cryptic promoters by maintaining the normal chromatin structure during transcription elongation (31). A yeast spt6 mutant permitted aberrant initiation from within coding regions, and transcribed chromatin became hypersensitive to micrococcal nuclease. Concomitantly with activation of the cryptic promoter, the level of transcription of the wild-type host gene decreased. More recently, a comprehensive analysis of cryptic transcription identified at least 50 factors, many involved in chromatin structure and transcription, required to repress cryptic promoters throughout the yeast genome (7). These factors would maintain the global integrity of gene expression during normal growth but may also allow the expression of alternative genetic information under altered genetic or physiological conditions. These factors include histone-encoding genes; histone regulators, such as Hir1 to -3 or Hpc2; chromatin assembly and remodeling factors, such as Spt6, Chd1, Asf1, and Spt16; histone deacetylases and accompanying factors; components of the mediator complex; and transcription elongation factors, such as Spt4 and Spt5 (orthologs of subunits of the DSIF complex, a known inhibitor of HIV elongation) (22, 46), among others.
By using a chimeric yeast-HIV promoter system, we initially found that spt6, chd1, and spt16 mutants derepressed the viral promoter (57). We have now added Hir1 to -3, Hpc2, and Asf1A to the list of factors that mediate HIV repression in this model system. RNA interference-mediated depletion of all these factors in the human HIV latency J-Lat system shows their involvement in the repression of HIV when integrated at certain coding regions. Further, all these factors can be related to the process of chromatin reassembly associated with transcription elongation, either as histone chaperones or as chromatin remodeling or disassembly/reassembly factors. Accordingly, we propose a model in which CRFs are involved in the repression of the HIV promoter by transcriptional interference when integrated at highly active transcription units (Fig. 8). The transcriptional machinery elongating through a highly expressed cellular gene containing an integrated HIV provirus includes CRFs that maintain a repressive chromatin configuration. In the absence of any chromatin reassembly factor, nucleosomes are poorly rebuilt and the chromatin configuration allows transcription factors and the transcriptional machinery to access the HIV promoter. Concomitantly, host gene expression decreases due to the global effect of CRF depletion, contributing to a reduction in transcriptional interference, and HIV activation further downregulates the upstream promoter.
FIG. 8.
A model for the involvement of chromatin reassembly factors in the repression of integrated HIV promoter by transcriptional interference.
The histone chaperone Spt6, in addition to depositing histones during reassembly, is known to interact with the RNA Pol II C-terminal domain (CTD) and Set2, a histone methyltransferase that methylates H3K36 through transcription elongation (5, 30, 64, 65). H3K36 methylation is a mark for histone deacetylation (37). Set2, histone deacetylases, and the histone modifications that they determine provide restoration of normal chromatin in the wake of elongating RNA Pol II and prevent inappropriate initiation within coding regions masked by chromatin. This would include initiation at the HIV promoter in certain chromatin environments. In agreement with this model, histone deacetylase (HDAC) inhibitors are known inducers of HIV transcription (56, 63), and this is also true in the context of the latent integrations at active coding regions analyzed here.
CRFs participate in the mechanism that controls the equilibrium between activation and repression of HIV when integrated in the human genome, which depends greatly on the chromatin environment at the integration site. Disturbance of this equilibrium, by depleting CRFs, for example, makes some of the latent integrations become activated without the need for further activating stimuli. Similarly, it has been reported that transcriptional activators are dispensable for transcription in the absence of Spt6-mediated chromatin reassembly of particular yeast promoter regions (2). Many of the factors involved in preventing internal cryptic promoter initiation in yeast, including Spt2, Spt4, Spt5, Spt6, and Spt16 (7), were originally identified in genetic screening based on the yeast Ty1 retroelement (10, 61). It is, therefore, plausible that the repressive function of CRFs to maintain the global integrity of gene expression is conserved from yeast to humans and may have been evolutionarily shaped by the selective pressure of endogenous retroelements. It would be interesting to investigate what the global consequences of Spt6 inhibition on the expression and mobility of these elements are and whether the observed downregulation of host genes is due to Spt6 being essential for transcription or to side effects produced by the induction of overlapping cryptic promoters.
Infection of T cells with an HIV vector leads to viral expression in the vast majority of integration events. Latency is rare, as in the in vivo situation. Integration into heterochromatin or gene deserts favors the establishment of latency, but it can also occur with the integration into introns of active genes (3, 39). We have found that depletion of Spt6 reactivates latent HIV when inserted into an active gene (J-Lat clones E27 and A2) but does not further activate HIV in an active clone (J-Act C9) where the host gene (TYK2) is expressed. Since we have observed TYK2 downregulation upon Spt6 depletion (data not shown), the ability of CRFs to repress HIV must be context dependent. The easiest explanation is that chromatin reassembly exerts its repressive effect when the host gene is highly expressed and polymerases are thoroughly reading the host gene. Accordingly, in active host genes, latency seems to be favored when they are highly expressed (3, 39). Nonetheless, local determinants such as position, orientation, and distance to other regulatory elements (enhancers, insulators, and splice sites) may also be of consideration. After repression is set, the situation is maintained by CRFs and histone deacetylation. Perturbing the equilibrium of promoter chromatin leads to HIV reactivation, but once the promoter becomes active, these perturbations have a minor effect.
Further work is required to clarify whether, in addition to CRFs, other cis or trans determinants may participate in transcriptional interference of the viral promoter.
Supplementary Material
Acknowledgments
This work was supported by grants from the Fundación para la Investigación y Prevencion del SIDA en España (FIPSE, 36602/06), Instituto de Salud Carlos III-Fondo de Investigación Sanitaria (PI05/1831), Ministerio de Ciencia e Innovación (MICINN) and FEDER (BFU2008-00359/BMC), and the Generalitat de Catalunya (2009-SGR-1222) to A.J. and from the MICINN (BFU2007-67575-C03-02/BMC) and the Andalusian Government (P07-CVI-02623) to S.C. E.G. was the recipient of a fellowship from the Generalitat de Catalunya and also supported by intramural resources from the Centre de Regulació Genòmica and Institut de Biologia Molecular de Barcelona-CSIC. J.-M.T. was the recipient of a fellowship from the Fondation pour la Recherche Medicale and a JAE-Doc contract from CSIC-MICINN.
Footnotes
Published ahead of print on 26 January 2011.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Adams, M., et al. 1994. Cellular latency in human immunodeficiency virus-infected individuals with high CD4 levels can be detected by the presence of promoter-proximal transcripts. Proc. Natl. Acad. Sci. U. S. A. 91:3862-3866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adkins, M. W., and J. K. Tyler. 2006. Transcriptional activators are dispensable for transcription in the absence of Spt6-mediated chromatin reassembly of promoter regions. Mol. Cell 21:405-416. [DOI] [PubMed] [Google Scholar]
- 3.Bisgrove, D., M. Lewinski, F. Bushman, and E. Verdin. 2005. Molecular mechanisms of HIV-1 proviral latency. Expert Rev. Anti Infect. Ther. 3:805-814. [DOI] [PubMed] [Google Scholar]
- 4.Blazkova, J., et al. 2009. CpG methylation controls reactivation of HIV from latency. PLoS Pathog. 5:e1000554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bortvin, A., and F. Winston. 1996. Evidence that Spt6p controls chromatin structure by a direct interaction with histones. Science 272:1473-1476. [DOI] [PubMed] [Google Scholar]
- 6.Brady, T., et al. 2009. HIV integration site distributions in resting and activated CD4+ T cells infected in culture. AIDS 23:1461-1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheung, V., et al. 2008. Chromatin- and transcription-related factors repress transcription from within coding regions throughout the Saccharomyces cerevisiae genome. PLoS Biol. 6:e277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chun, T. W., and A. S. Fauci. 1999. Latent reservoirs of HIV: obstacles to the eradication of virus. Proc. Natl. Acad. Sci. U. S. A. 96:10958-10961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chun, T. W., et al. 1997. Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proc. Natl. Acad. Sci. U. S. A. 94:13193-13197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clark-Adams, C. D., D. Norris, M. A. Osley, J. S. Fassler, and F. Winston. 1988. Changes in histone gene dosage alter transcription in yeast. Genes Dev. 2:150-159. [DOI] [PubMed] [Google Scholar]
- 11.Coiras, M., M. R. Lopez-Huertas, M. Perez-Olmeda, and J. Alcami. 2009. Understanding HIV-1 latency provides clues for the eradication of long-term reservoirs. Nat. Rev. Microbiol. 7:798-812. [DOI] [PubMed] [Google Scholar]
- 12.Colin, L., and C. Van Lint. 2009. Molecular control of HIV-1 postintegration latency: implications for the development of new therapeutic strategies. Retrovirology 6:111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Contreras, X., M. Barboric, T. Lenasi, and B. M. Peterlin. 2007. HMBA releases P-TEFb from HEXIM1 and 7SK snRNA via PI3K/Akt and activates HIV transcription. PLoS Pathog. 3:1459-1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cullen, B. R., P. T. Lomedico, and G. Ju. 1984. Transcriptional interference in avian retroviruses—implications for the promoter insertion model of leukaemogenesis. Nature 307:241-245. [DOI] [PubMed] [Google Scholar]
- 15.De Marco, A., et al. 2008. Intragenic transcriptional cis-activation of the human immunodeficiency virus 1 does not result in allele-specific inhibition of the endogenous gene. Retrovirology 5:98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dieudonne, M., et al. 2009. Transcriptional competence of the integrated HIV-1 provirus at the nuclear periphery. EMBO J. 28:2231-2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.du Chene, I., et al. 2007. Suv39H1 and HP1gamma are responsible for chromatin-mediated HIV-1 transcriptional silencing and post-integration latency. EMBO J. 26:424-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Emiliani, S., et al. 1998. Mutations in the tat gene are responsible for human immunodeficiency virus type 1 postintegration latency in the U1 cell line. J. Virol. 72:1666-1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Emiliani, S., et al. 1996. A point mutation in the HIV-1 Tat responsive element is associated with postintegration latency. Proc. Natl. Acad. Sci. U. S. A. 93:6377-6381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Finzi, D., et al. 1999. Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nat. Med. 5:512-517. [DOI] [PubMed] [Google Scholar]
- 21.Finzi, D., et al. 1997. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science 278:1295-1300. [DOI] [PubMed] [Google Scholar]
- 22.Garber, M. E., and K. A. Jones. 1999. HIV-1 Tat: coping with negative elongation factors. Curr. Opin. Immunol. 11:460-465. [DOI] [PubMed] [Google Scholar]
- 23.Greger, I. H., F. Demarchi, M. Giacca, and N. J. Proudfoot. 1998. Transcriptional interference perturbs the binding of Sp1 to the HIV-1 promoter. Nucleic Acids Res. 26:1294-1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Han, Y., et al. 2004. Resting CD4+ T cells from human immunodeficiency virus type 1 (HIV-1)-infected individuals carry integrated HIV-1 genomes within actively transcribed host genes. J. Virol. 78:6122-6133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Han, Y., et al. 2008. Orientation-dependent regulation of integrated HIV-1 expression by host gene transcriptional readthrough. Cell Host Microbe 4:134-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Imai, K., H. Togami, and T. Okamoto. 2010. Involvement of histone H3 lysine 9 (H3K9) methyltransferase G9a in the maintenance of HIV-1 latency and its reactivation by BIX01294. J. Biol. Chem. 285:16538-16545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Imai, Y., et al. 2002. Cloning and characterization of the highly expressed ETEA gene from blood cells of atopic dermatitis patients. Biochem. Biophys. Res. Commun. 297:1282-1290. [DOI] [PubMed] [Google Scholar]
- 28.Jordan, A., D. Bisgrove, and E. Verdin. 2003. HIV reproducibly establishes a latent infection after acute infection of T cells in vitro. EMBO J. 22:1868-1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jordan, A., P. Defechereux, and E. Verdin. 2001. The site of HIV-1 integration in the human genome determines basal transcriptional activity and response to Tat transactivation. EMBO J. 20:1726-1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Joshi, A. A., and K. Struhl. 2005. Eaf3 chromodomain interaction with methylated H3-K36 links histone deacetylation to Pol II elongation. Mol. Cell 20:971-978. [DOI] [PubMed] [Google Scholar]
- 31.Kaplan, C. D., L. Laprade, and F. Winston. 2003. Transcription elongation factors repress transcription initiation from cryptic sites. Science 301:1096-1099. [DOI] [PubMed] [Google Scholar]
- 32.Karn, J. 1999. Tackling Tat. J. Mol. Biol. 293:235-254. [DOI] [PubMed] [Google Scholar]
- 33.Kauder, S. E., A. Bosque, A. Lindqvist, V. Planelles, and E. Verdin. 2009. Epigenetic regulation of HIV-1 latency by cytosine methylation. PLoS Pathog. 5:e1000495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kornblihtt, A. R. 2007. Coupling transcription and alternative splicing. Adv. Exp. Med. Biol. 623:175-189. [DOI] [PubMed] [Google Scholar]
- 35.Lassen, K., Y. Han, Y. Zhou, J. Siliciano, and R. F. Siliciano. 2004. The multifactorial nature of HIV-1 latency. Trends Mol. Med. 10:525-531. [DOI] [PubMed] [Google Scholar]
- 36.Lassen, K. G., J. R. Bailey, and R. F. Siliciano. 2004. Analysis of human immunodeficiency virus type 1 transcriptional elongation in resting CD4+ T cells in vivo. J. Virol. 78:9105-9114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee, J. S., and A. Shilatifard. 2007. A site to remember: H3K36 methylation a mark for histone deacetylation. Mutat. Res. 618:130-134. [DOI] [PubMed] [Google Scholar]
- 38.Lenasi, T., X. Contreras, and B. M. Peterlin. 2008. Transcriptional interference antagonizes proviral gene expression to promote HIV latency. Cell Host Microbe 4:123-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lewinski, M. K., et al. 2005. Genome-wide analysis of chromosomal features repressing human immunodeficiency virus transcription. J. Virol. 79:6610-6619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lin, X., et al. 2003. Transcriptional profiles of latent human immunodeficiency virus in infected individuals: effects of Tat on the host and reservoir. J. Virol. 77:8227-8236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lusic, M., A. Marcello, A. Cereseto, and M. Giacca. 2003. Regulation of HIV-1 gene expression by histone acetylation and factor recruitment at the LTR promoter. EMBO J. 22:6550-6561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Macias, D., R. Oya, L. Saniger, F. Martin, and F. Luque. 2009. A lentiviral vector that activates latent human immunodeficiency virus-1 proviruses by the overexpression of tat and that kills the infected cells. Hum. Gene Ther. 20:1259-1268. [DOI] [PubMed] [Google Scholar]
- 43.Mahmoudi, T., et al. 2006. The SWI/SNF chromatin-remodeling complex is a cofactor for Tat transactivation of the HIV promoter. J. Biol. Chem. 281:19960-19968. [DOI] [PubMed] [Google Scholar]
- 44.Mason, P. B., and K. Struhl. 2003. The FACT complex travels with elongating RNA polymerase II and is important for the fidelity of transcriptional initiation in vivo. Mol. Cell. Biol. 23:8323-8333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pandit, S., D. Wang, and X. D. Fu. 2008. Functional integration of transcriptional and RNA processing machineries. Curr. Opin. Cell Biol. 20:260-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Peterlin, B. M., and D. H. Price. 2006. Controlling the elongation phase of transcription with P-TEFb. Mol. Cell 23:297-305. [DOI] [PubMed] [Google Scholar]
- 47.Pomerantz, R. J., D. Trono, M. B. Feinberg, and D. Baltimore. 1990. Cells nonproductively infected with HIV-1 exhibit an aberrant pattern of viral RNA expression: a molecular model for latency. Cell 61:1271-1276. [DOI] [PubMed] [Google Scholar]
- 48.Quivy, V., S. De Walque, and C. Van Lint. 2007. Chromatin-associated regulation of HIV-1 transcription: implications for the development of therapeutic strategies. Subcell. Biochem. 41:371-396. [PubMed] [Google Scholar]
- 49.Raab, J. R., and R. T. Kamakaka. 2010. Insulators and promoters: closer than we think. Nat. Rev. Genet. 11:439-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sadowski, I., P. Lourenco, and T. Malcolm. 2008. Factors controlling chromatin organization and nucleosome positioning for establishment and maintenance of HIV latency. Curr. HIV Res. 6:286-295. [DOI] [PubMed] [Google Scholar]
- 51.Schroder, A. R., et al. 2002. HIV-1 integration in the human genome favors active genes and local hotspots. Cell 110:521-529. [DOI] [PubMed] [Google Scholar]
- 52.Schwartz, S., and G. Ast. 2010. Chromatin density and splicing destiny: on the cross-talk between chromatin structure and splicing. EMBO J. 29:1629-1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Siliciano, J. D., et al. 2003. Long-term follow-up studies confirm the stability of the latent reservoir for HIV-1 in resting CD4+ T cells. Nat. Med. 9:727-728. [DOI] [PubMed] [Google Scholar]
- 54.Treand, C., et al. 2006. Requirement for SWI/SNF chromatin-remodeling complex in Tat-mediated activation of the HIV-1 promoter. EMBO J. 25:1690-1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Trono, D., et al. 2010. HIV persistence and the prospect of long-term drug-free remissions for HIV-infected individuals. Science 329:174-180. [DOI] [PubMed] [Google Scholar]
- 56.Van Lint, C., S. Emiliani, M. Ott, and E. Verdin. 1996. Transcriptional activation and chromatin remodeling of the HIV-1 promoter in response to histone acetylation. EMBO J. 15:1112-1120. [PMC free article] [PubMed] [Google Scholar]
- 57.Vanti, M., et al. 2009. Yeast genetic analysis reveals the involvement of chromatin reassembly factors in repressing HIV-1 basal transcription. PLoS Genet. 5:e1000339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Verdin, E., P. Paras, Jr., and C. Van Lint. 1993. Chromatin disruption in the promoter of human immunodeficiency virus type 1 during transcriptional activation. EMBO J. 12:3249-3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang, G. P., A. Ciuffi, J. Leipzig, C. C. Berry, and F. D. Bushman. 2007. HIV integration site selection: analysis by massively parallel pyrosequencing reveals association with epigenetic modifications. Genome Res. 17:1186-1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Williams, S. A., et al. 2006. NF-kappaB p50 promotes HIV latency through HDAC recruitment and repression of transcriptional initiation. EMBO J. 25:139-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Winston, F., D. T. Chaleff, B. Valent, and G. R. Fink. 1984. Mutations affecting Ty-mediated expression of the HIS4 gene of Saccharomyces cerevisiae. Genetics 107:179-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wong, J. K., et al. 1997. Recovery of replication-competent HIV despite prolonged suppression of plasma viremia. Science 278:1291-1295. [DOI] [PubMed] [Google Scholar]
- 63.Ylisastigui, L., N. M. Archin, G. Lehrman, R. J. Bosch, and D. M. Margolis. 2004. Coaxing HIV-1 from resting CD4 T cells: histone deacetylase inhibition allows latent viral expression. AIDS 18:1101-1108. [DOI] [PubMed] [Google Scholar]
- 64.Yoh, S. M., J. S. Lucas, and K. A. Jones. 2008. The Iws1:Spt6:CTD complex controls cotranscriptional mRNA biosynthesis and HYPB/Setd2-mediated histone H3K36 methylation. Genes Dev. 22:3422-3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Youdell, M. L., et al. 2008. Roles for Ctk1 and Spt6 in regulating the different methylation states of histone H3 lysine 36. Mol. Cell. Biol. 28:4915-4926. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.