Abstract
The switch between the latency and lytic cycles of Kaposi's sarcoma-associated herpesvirus (KSHV) is accompanied by specific alterations of histone codes. Recently, comprehensive analysis of histone modifications of KSHV showed the deposition of H3K27me3 across the KSHV genome with two specific regions occupied by the heterochromatin marker H3K9me3. Here, we show that knockdown of JMJD2A, an H3K9me3 demethylase, attenuates viral titers, whereas its overexpression increases KSHV reactivation. JMJD2A is localized in regions of latent viral chromosomes that are deficient in the H3K9me3 mark, indicating that JMJD2A may be responsible for the low level of this mark on viral chromatin. The presence of JMJD2A on the latent genome maintains H3K9 in unmethylated form and signals the readiness of specific sets of viral genes to be reactivated. The demethylase activity of JMJD2A is important for KSHV reactivation, because a demethylase-deficient mutant cannot restore the JMJD2A knockdown phenotype. Interestingly, we found that the KSHV encoded K-bZIP associated with JMJD2A, resulting in the inhibition of demethylase activity of JMJD2A both in vivo and in vitro. Inhibition of JMJD2A by K-bZIP is likely due to a physical interaction which blocks substrate accessibility. A consequence of such an inhibition is increasing global levels of H3K9me3 and gene silencing. Consistently, K-bZIP overexpression resulted in a repression of ∼80% of the ≥2-fold differentially regulated genes compared to results for the uninduced control cells. The consequences of K-bZIP targeting JMJD2A during viral replication will be discussed. To our knowledge, this is the first description of a viral product shown to be a potent inhibitor of a host cellular histone demethylase.
Kaposi's sarcoma-associated herpesvirus (KSHV), also known as human herpesvirus 8 (HHV-8), is a gammaherpesvirus associated with Kaposi's sarcoma, primary effusion lymphoma (PEL), and multicentric Castleman's disease (MCD) (8). Like all herpesviruses, KSHV has distinct lytic and latent phases, whose interconversion is regulated primarily at the transcriptional level by transactivation/silencing and chromatin remodeling of the viral genome. A long-held view is that during the lytic stage, the herpesvirus genome adopts a state of euchromatin organization, with nearly all viral genes transcribed. In contrast, within latency, viral DNA forms extrachromosomal episomes resembling heterochromatin with only limited actively transcribed regions. Different chromatin conformations are characterized by different histone codes (methylation, acetylation, and phosphorylation of H3 and H4). Using a KSHV viral promoter chip, we previously characterized the changing landscape of acetylated histones during reactivation (9). Two recent reports have provided comprehensive maps of active and repressive histone marks along the entire latent KSHV genome (12, 35). The H3K27me3 is the dominant repressive mark which spreads over the entire genome, whereas the H3K9me3 repressive mark is clustered in two regions corresponding primarily to late genes. Another interesting phenomenon is the coexistence of both active (H3K4me3) and repressive (H3K27me3) histone marks in certain regions. The “bivalent” state of such chromatin is postulated to signify the readiness of target genes to be transcribed upon reactivation. Enzymes catalyzing such histone modifications are histone methylases and demethylases. These enzymes can be further divided among those which target arginine or lysine. As major modifiers of histones, methylases and demethylases are known to regulate chromatin structure and transcriptional programs of the cellular genome and control a variety of biological processes, including stem cell differentiation and oncogenesis (reviewed in references 26 and 29).
The cellular methylases and demethylases are likely to be major regulators of the transcription and replication of chromatinized DNA virus genomes as well. EZH2, an H3K27me2/me3 histone methylase and a member of the Polycomb repressive complex, decorates regions of the KSHV latent genome enriched in H3K27me3 marks, implying a causal relationship (12, 35). SUV39H, an H3K9me3 methylase, is tightly associated with latency-associated nuclear antigen (LANA), a viral latency factor which tethers the viral chromosome to host cell heterochromatin (30), although its occupancy along the viral latent genome was not reported. The involvement of histone demethylases in the modeling of the KSHV latent genome has not been described.
Until recently, histone methylation was thought to be irreversible. With the discovery of LSD1KDM1 (lysine (K)-specific demethylase 1) (32), a dynamic picture of chromatin remodeling via histone methylation/demethylation is emerging. LSD1 demethylates mono- and dimethylated lysine residues of histone H3 via an amine oxidation reaction. More recently, a large family of demethylases containing a Jumonji C (JmjC) domain was identified. Different from LSD1, JmjC domain-containing histone demethylases (JHDMs) catalyze lysine demethylation by an oxidative reaction that can remove all three histone methylation states. Previously, five families of JmjC-containing lysine demethylases (i.e., KDM2 to KDM6) have been identified, each with distinct demethylation specificity, ranging from H3K36me2/me1, H3K9me2/me1, and H3K9me3/me2 to H3K4me3/me2 and H3K27me3/me2 (1, 19). Tsukada et al. recently described a new member, KDM7, with demethylation specificity toward H3K9me2/me1 and H3K27me2/me1 (36), and we have added to this repertoire the newest member, KDM8, which demethylates H3K36me2 (13). Thus, there are now eight families of histone demethylases, consisting of a total of 18 members, each with distinct demethylation target specificity. These enzymes work in concert to effectively “edit” the histone codes, recruiting different sets of transcription factors and thereby regulating transcriptional programs.
JMJD2A, also known as KDM4A, was the first reported trimethyl-specific KDM (20). It contains JmjC and JmjN domains near its N terminus and plant homeodomain (PHD) and Tudor domains near its C terminus (5, 38). An intact JmjC domain is required for the demethylation activity of JMJD2A, because a single amino acid mutation (JMJD2A H188A) in the Fe(II) binding site within the JmjC domain completely abolishes its catalytic activity (20). The C-terminal Tudor domain serves as a chromatin-targeting module which preferentially binds methylated H3K4 and H4K20 (14, 18, 21). Through its JmjC catalytic domain, JMJD2A also binds methylated H3K9 and H3K36 (4). Although JMJD2A exhibits dual specificity toward both tri- and dimethylated forms of H3K9 and H3K36, its primary target is H3K9me3, a repressive mark (5, 6, 20, 38). As such, JMJD2A is generally viewed as a coactivator; for example, this demethylase upregulates androgen receptor responsive genes (33). However, JMJD2A can also function as a repressor, when complexed with retinoblastoma protein, class I histone deacetylase proteins, or N-CoR (11, 40).
In this study, we have characterized the biological effects of modulation of KSHV reactivation and replication by the JMJD2A demethylase. The binding sites of JMJD2A were mapped along the entire latent KSHV genome; these sites were inversely correlated with the presence of the H3K9me3 mark. Interestingly, K-bZIP, a viral transcriptional factor and SUMO (small ubiquitin-like modifier) ligase, was found to be an interacting partner and potent inhibitor of JMJD2A. The significance of these findings in the context of KSHV epigenetic regulation and replication will be described.
MATERIALS AND METHODS
Cells and plasmids.
Human embryonic kidney epithelial 293T and inducible TREx-F-K-bZIP 293 (3) cells were cultured in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum (FBS) in the presence of 5% CO2. The JMJD2A knockdown TREx-MH-K-Rta BCBL-1 cell line was generated by infection of TREx-MH-K-Rta BCBL-1 cells (27) with lentiviral particles expressing short hairpin RNA (shRNA) targeting JMJD2A and grown in RPMI 1640 containing 15% FBS, 50 μg/ml blasticidin, 100 μg/ml hygromycin (Invitrogen), and 1 μg/ml puromycin. To overexpress wild-type (WT) JMJD2A and its H188A mutant in JMJD2A knockdown TREx-MH-K-Rta BCBL-1 cells, expression vectors containing shRNA-resistant JMJD2A* and JMJD2A*-H188A were transiently transfected into cells using FugeneHD and selected with G418 (150 μg/ml; Cellgro). Expression of each of these constructs was confirmed by Western blot analysis. The Vero-rKSHV.219 cell line was kindly provided by Jeffrey Vieira (University of Washington, Seattle, WA) and cultured as previously described (37). cDNAs encoding K-bZIP, the JMJD2A wild type, and its H188A mutant were cloned into pcDNA3.1-Flag (Invitrogen). To generate recombinant proteins of JMJD2A, WT F-K-bZIP, and its ΔLZ (leucine zipper motif deletion) mutant (16) in the baculovirus protein expression system, the JMJD2A, K-bZIP, and ΔLZ strains were subcloned into pFastBac1.
KSHV reactivation assay using Vero-rKSHV.219 cells.
The Vero-rKSHV.219 cells were infected with lentiviruses carrying shRNAs targeting different regions of JMJD2A. The shRNAs target sequences are shJMJD2A#1 (GCTGCAGTATTGAGATGCTAA); shJMJD2A#2 (CCCGCTTCAAACTGAAATGTA); shJMJD2A#3 (GCCTTGGATCTTTCTGTGAAT), and shJMJD2A#4 (AGATACCGGGAAGATTATATT). Forty-eight hours after transduction, cells were treated with sodium butyrate (1.75 mM) for 48 h. Green fluorescent protein (GFP)- and red fluorescent protein (RFP)-positive cells were visualized by fluorescence microscopy.
Assays of KSHV growth and gene expression in Vero-rKSHV.219 and BCBL-1 cells.
KSHV virions were purified as previously described (2). Briefly, supernatants from 7.5 × 105 control and doxycycline (Dox)-induced (0.2 μg/ml) JMJD2A knockdown TREx-MH-K-Rta BCBL-1 cells were collected at 0, 24, 48, and 72 h after induction. DNA from intact virions was prepared, and the copy number of KSHV DNA purified from viral stocks was estimated by real-time DNA-PCR using primers and TaqMan probes specific for KSHV open reading frame 73 (ORF73).
ChIP assay.
Chromatin immunoprecipitation (ChIP) assays were performed following the protocol described by the Farnham laboratory (provided at http://genomics.ucdavis.edu/farnham). The antibodies used were anti-JMJD2A (Abcam), anti-H3K9me3 (Abcam), and rabbit nonimmune serum IgG (Alpha Diagnostic International). Immunoprecipitated chromatin DNA was analyzed by SYBR green-based quantitative PCR (qPCR) (Bio-Rad).
KSHV tiling array and ChIP-on-chip analysis.
The KSHV tiling array was designed across the KSHV genome sequence (NCBI reference sequence NC_009333.1) and manufactured by Agilent Technologies. The probes were spotted as an 8x15K array format. In addition, probes specific for the promoter region of five human genes were also included in the microarray. This array contains 2,299 probes spanning the entire KSHV genome. Each probe on the array is 60 bp long. Whole ChIP samples and 10 ng of input were linear amplified using a WGA Genomeplex whole genome amplification kit (Sigma). Labeling, hybridization, scanning, and analyzing of the arrays were done at UC Davis Cancer Center Genomics Shared Resource. The enrichment of JMJD2A or H3K9me3 binding to the KSHV genome was calculated by the intensity ratio of immunoprecipitated and input DNA for each array spot, and probe signals were further normalized by the blank subtraction normalization method and Whitehead error model using Genomic Workbench software. Each ChIP-on-chip experiment was performed in triplicate.
Immunocytochemical staining.
293T cells were plated onto 22-mm coverslips at a density of 5 × 104 cells per well in six-well plates. Forty-eight hours after transfection, cells were fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS) for 15 min and then washed twice with 0.1 M glycine-Tris buffer (pH 7.3) and once with PBS. Cells were blocked with 1% bovine serum albumin (BSA) in PBS, followed by incubation with primary antibodies diluted in 1% BSA at 4°C overnight. After being washed three times with PBS, coverslips were incubated with Alexa Fluor 555-conjugated donkey anti-mouse or anti-rabbit IgG and Alexa Fluor 488-conjugated donkey anti-rabbit or anti-mouse IgG (Invitrogen) diluted in 1% BSA at room temperature for 1 h. The cells were washed, mounted in SlowFade Gold antifade reagent with DAPI (4′,6-diamidino-2-phenylindole; Invitrogen), and visualized using a BX61 microscope system equipped with SlideBook 4.1 imaging software (Olympus).
Immunoprecipitation and Western blot analysis.
Transfected 293T cells were collected in modified radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris-HCl [pH 6.7], 1% NP-40, 0.25% sodium deoxycholate, 150 mM NaCl, 1 mM EDTA) supplemented with 1 mM phenylmethylsulfonyl fluoride (PMSF) and 1× protease inhibitor cocktail (Roche). Total cell lysates (TCLs) were incubated with either mouse nonimmune serum IgG, rabbit nonimmune serum IgG, anti-Flag M2 agarose (Sigma), or anti-JMJD2A antibody overnight at 4°C. Immune complexes were captured by protein A and protein G Sepharose beads. Beads were washed, and the bound proteins were analyzed by immunoblotting. Antibodies used for immunoblotting were anti-hemagglutinin (anti-HA) (Santa Cruz), anti-Flag M2 (Sigma), anti-β-actin (Sigma), anti-K-bZIP (Santa Cruz), and anti-H3K9me3 (Abcam).
Demethylation assay and MALDI-time of flight mass spectrometry.
Purified JMJD2A alone or in combination with WT K-bZIP or its ΔLZ mutant were incubated with 10 μM H3K9me3 peptide in demethylation buffer [20 mM Tris-HCl (pH 7.3), 150 mM NaCl, 50 μM (NH4)2Fe(SO4)2+6(H2O), 1 mM α-ketoglutarate, 2 mM ascorbic acid] at 37°C for 4 h. The samples were prepared as previously described (32). Briefly, 4 μl of the sample was desalted with a C18 ZipTip, eluted with 10 mg/ml α-cyano-4-hydroxycinnamic acid matrix-assisted laser desorption ionization (MALDI) matrix in 70% acetonitrile containing 0.1% trifluoroacetic acid (TFA), spotted on a MALDI plate, crystallized, and then analyzed by the University of California, Davis, mass spectrometry facility.
Surface plasmon resonance (SPR) analysis using a Biacore 3000 instrument (Biacore, Pittsburgh, PA).
Biotin-labeled H3K9 or H3K9me3 peptides were immobilized on the streptavidin (SA) sensor chip by using the standard procedure. The Flag, Flag-K-bZIP, and Flag-K-bZIP ΔLZ proteins were individually 2-fold serially diluted from 80 nM to 2.5 nM, and Flag-JMJD2A was 2-fold serially diluted from 20 nM to 0.625 nM in HBS-EP buffer. Samples were injected at 30 μl/min for 3 min, and HBS-EP buffer (Biacore) was pumped into the flow channels at the same flow rate for 5 min to allow the bound Flag fusion protein to dissociate from H3K9me3 peptide. KD (equilibrium dissociation constant) was calculated with biomolecular interaction analysis (BIA) evaluation software by fitting the obtained binding curves with the 1:1 Langmuir binding with drifting baseline model.
RESULTS
Modulation of JMJD2A expression affects KSHV reactivation in the Vero-rKHSV.219 model.
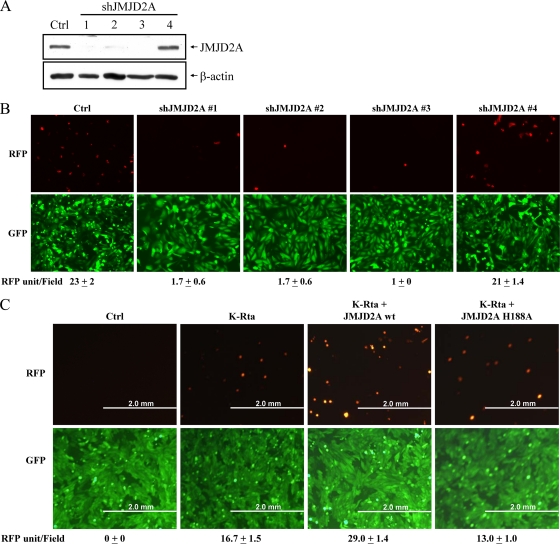
Transcriptional reprogramming, accompanied by chromatin remodeling of the viral genome, is thought to be the principal mechanism underlying herpesvirus reactivation. Posttranslational modification of histones, in particular methylation, is viewed as a driver of chromatin remodeling. We have been interested in the role of Jumonji C-containing histone demethylases in KSHV reactivation. Based on a preliminary screening, we found that shRNAs targeting JMJD2A severely impacted viral reactivation in the Vero-rKSHV.219 model (37). The Vero-rKHSV.219 cell line harbors viral genomes with a constitutively (EF-1α promoter) expressed GFP gene and a reactivation-induced (PAN promoter) RFP gene. This cell line was infected with lentiviruses carrying shRNAs targeting different regions of JMJD2A, followed by treatment with sodium butyrate for 48 h. As shown in Fig. 1A and B, three different JMJD2A shRNAs, which effectively knocked down the expression of JMJD2A, significantly reduced viral reactivation compared to that of the vector control. An shRNA which was ineffective in knocking down JMJD2A expression did not have such an effect (Fig. 1B). To extend these observations, we overexpressed JMJD2A in Vero-rKSHV.219 cells by cotransfection of JMJD2A with K-Rta (as an inducer for reactivation); viral reactivation increased about 1.8-fold (Fig. 1C). These data suggest that JMJD2A and, by extension, H3K9me3 demethylation is a critical step in KSHV reactivation or replication. To study whether the demethylase activity is important, we took advantage of the JMJD2A H188A mutant, which is catalytically inactive. In cotransfection with K-Rta, this mutant failed to increase the reactivation titer, suggesting that the demethylase activity is relevant for JMJD2A function as a modulator of KSHV replication (Fig. 1C).
FIG. 1.
Effect of JMJD2A modulation on K-Rta-mediated KSHV reactivation. (A) Total cell lysates (TCLs) from Vero-rKSHV.219 cells infected with control lentivirus or lentiviruses expressing different shRNA (1, 2, 3, or 4) directly targeting different regions of JMJD2A were collected 48 h after infection. Knockdown of JMJD2A was visualized by immunoblotting using an antibody specific for JMJD2A. (B) Vero-rKSHV.219 cells were infected as described for panel A and were reactivated with sodium butyrate. Two days after reactivation, the cells were visualized by fluorescence microscopy. Top, RFP fluorescence (lytic-infected cells); bottom, GFP fluorescence (latent-infected cells). The mean number of RFP-positive cells per field is listed. (C) Vero-rKSHV.219 cells transfected with control, K-Rta alone, K-Rta plus JMJD2A, or K-Rta plus the JMJD2A H188A mutant were visualized by fluorescent microscope. The mean number of RFP positive cells per field is listed.
Modulation of JMJD2A expression affects KSHV replication in the BCBL-1 model of latency.
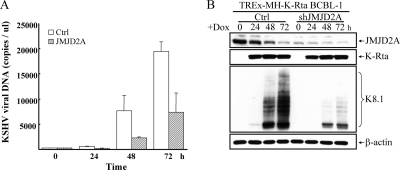
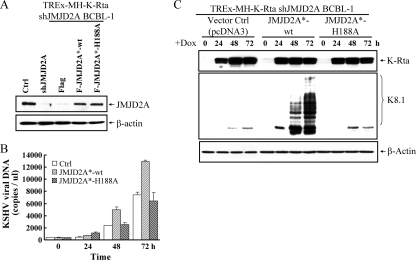
As an independent confirmation of the role of JMJD2A in KSHV replication, we employed the TREx-MH-K-Rta BCBL-1 model, a pleural effusion B-lymphoma cell line harboring a latent KSHV genome and a doxycycline-inducible K-Rta gene (27). The knockdown of JMJD2A was achieved by infection of TREx-MH-K-Rta BCBL-1 cells with lentivirus vector containing shJMJD2A. A pool of infected clones was collected for analysis. Reactivation was carried out by treatment with Dox to induce K-Rta. Supernatants containing KSHV virions were collected before and after K-Rta induction in the TREx-MH-K-Rta BCBL-1 shJMJD2A cell line and the vector-infected parental cell line. Virion-associated DNA copy number was assayed by real-time quantitative PCR (qPCR). JMJD2A knockdown significantly inhibited viral reactivation and reduced viral production by ∼2- to 3-fold compared to that of the vector-infected control at 48 and 72 h after induction (Fig. 2A). In agreement with the inhibition of virus production, protein expression of a late gene, K8.1, was reduced in JMJD2A knockdown cells (Fig. 2B). Effective knockdown of JMJD2A during viral reactivation was confirmed by immunoblotting (Fig. 2B, top). With the stable JMJD2A knockdown cells, we attempted rescue experiments using wild-type JMJD2A or the JMJD2A H188A mutant. To this end, we developed shRNA-resistant JMJD2A*-wt and JMJD2A*-H188A mutants which were individually transfected into the TREx-MH-K-Rta BCBL-1 shJMJD2A cell line. The immunoblotting experiment confirmed the shRNA-resistant nature of these constructs and indicated similar expression levels of JMJD2A*-wt and its demethylase-dead JMJD2A*-H188A in the respective cell lines. In addition, their expression levels are similar to the endogenous JMJD2A level of the parental cells (Fig. 3 A). Introduction of JMJD2A*-wt significantly increased lytic replication of KSHV in JMJD2A knockdown BCBL-1 cells, as measured by supernatant virion DNA (Fig. 3B) and by the expression of K8.1 (Fig. 3C). In contrast, the shRNA-resistant JMJD2A*-H188A mutant was unable to restore KSHV lytic replication (Fig. 3B and C). These data suggest that JMJD2A plays an important role in KSHV reactivation and replication in both Vero and BCBL-1 cells and that the demethylation activity of JMJD2A is an integral part of its function.
FIG. 2.
JMJD2A plays an essential role in KSHV reactivation. (A) Supernatants from control and JMJD2A knockdown TREx-MH-K-Rta BCBL-1 cells (TREx-MH-K-Rta shRNA BCBL-1) were collected at days 0, 1, 2, and 3 after doxycycline (Dox; 0.1 μg/ml) treatment, and the levels of virion-associated DNA were determined by real-time qPCR. Input DNA was normalized to 1. Mean ± SD. (B) TCLs from the cells described for panel A were immunoblotted with antibodies as indicated.
FIG. 3.
Demethylase activity of JMJD2A is essential for KSHV reactivation. (A) Immunoblotting of JMJD2A expression in control, JMJD2A knockdown, and shRNA-resistant Flag-tag-expressing control vector (Flag), JMJD2A*-wt, and JMJD2A*-H188A mutant-transfected TREx-MH-K-Rta shJMJD2A BCBL-1 cells. (B) Virion-associated DNA from shRNA-resistant Flag, JMJD2A*-wt, and the JMJD2A*-H188A mutant-transfected TREx-MH-K-Rta shJMJD2A BCBL-1 cells was collected at days 0, 1, 2, and 3 after Dox (0.1 μg/μl) induction. Viral titer was determined and normalized as described for Fig. 1A. (C) TCLs from the cells described for panel B were immunoblotted with antibodies as indicated.
Occupancy of JMJD2A on latent KSHV chromatin inversely correlates with H3K9me3 repressive marks.
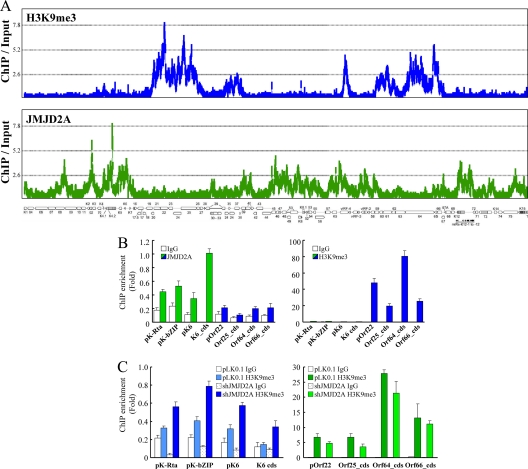
To determine whether JMJD2A is directly involved in regulating the histone landscape of KSHV chromatin, we performed a ChIP-on-chip experiment by hybridizing DNA from JMJD2A-associated chromatin with a KSHV genome tiling array designed by our laboratory. Input (nonenriched) DNA and DNA from JMJD2A immunoprecipitates were labeled with Cy3 and Cy5, respectively, and cohybridized on one tiling array. In parallel, we also analyzed the occupancy of H3K9me3, the substrate of JMJD2A, along the viral latent genome. In agreement with previous reports that had investigated H3K9me3 patterns on the KSHV genome, we found that H3K9me3 was located mainly in two distinct regions of the KSHV genome (Fig. 4 A, top). Interestingly, JMJD2A was associated with most of the viral latent genome except for the two regions marked by H3K9me3 (Fig. 4A, bottom); this finding is consistent with our hypothesis that JMJD2A functions to maintain H3K9 in an unmethylated form, thereby allowing this residue to be acetylated upon the recruitment of an acetylase. The ChIP-on-chip data were confirmed by gene-specific real-time qPCR analysis using four locations representing H3K9me3-enriched regions (Fig. 4A, marked in blue; promoter of Orf22 and coding regions of Orf25, Orf64, and Orf66) and regions devoid of H3K9me3 (Fig. 4A, marked in green; promoter regions of Orf50/K-Rta, K-bZIP, and K6 and the coding region of K6). The binding of JMJD2A on K-Rta, K-bZIP, and K6 promoters and the K6 coding region was significantly higher than that on the H3K9me3-enriched region (Fig. 4B, left). Conversely, the H3K9me3 mark is enriched in the promoter of Orf22 and coding regions of Orf25, Orf64, and Orf66 to a much higher degree than for the JMJD2A binding regions (Fig. 4B, right). Knockdown of JMJD2A increased the H3K9me3 level in JMJD2A binding regions but not in the H3K9me3-enriched regions (Fig. 4C). These data, taken together, suggest that the presence of JMJD2A on the latent genome is to prevent H3K9 residues from being methylated.
FIG. 4.
ChIP-on-chip analysis of JMJD2A binding and H3K9me3 occupancy across the KSHV genome during latency. (A) Chromatin immunoprecipitation (ChIP) assay was performed on noninduced TREx-MH-K-Rta BCBL-1 cells using anti-JMJD2A antibody, anti-H3K9me3 antibody, or rabbit IgG. ChIP-on-chip analysis and peak detection were performed as described in Materials and Methods. JMJD2A and H3K9me3 binding patterns along the latent KSHV genome are displayed, and the genomic locations of the KSHV ORFs are depicted below the histograms. (B) ChIP-on-chip data were verified by real-time qPCR using primers specific for KSHV K-Rta, K-bZIP, K6, and Orf22 promoters (pK-Rta, pK-bZIP, pK6, and pOrf22; green bars) and coding sequences (cds) for K6, Orf25, Orf64, and Orf66 (K6_cds, Orf25_cds, Orf64_cds, and Orf66_cds; blue bars). (C) ChIP assays were done on control and TREx-MH-K-Rta shJMJD2A BCBL-1 cells using anti-H3K9me3 antibody and rabbit IgG. ChIP DNA levels were determined using primers as described for panel B.
K-bZIP interacts with JMJD2A and inhibits its demethylase activity.
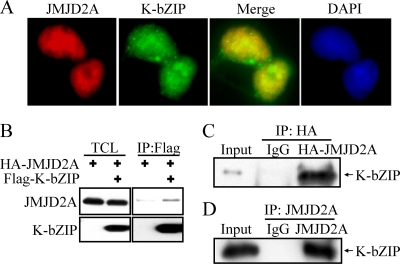
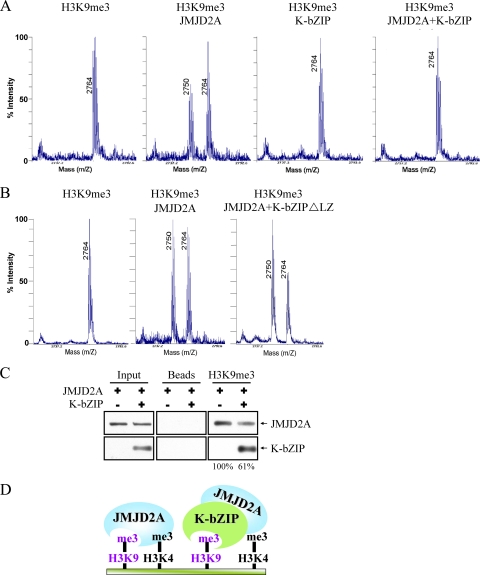
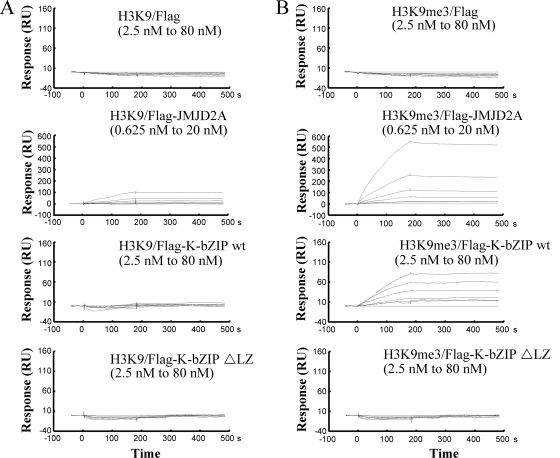
If JMJD2A is important for KSHV replication, we surmised that the virus has evolved a means to modulate expression or activity of this demethylase. To first address this, we analyzed viral proteins which interact with JMJD2A. After a survey of several KSHV early gene products, K-bZIP was found to be a strong interacting partner of JMJD2A. Immunostaining results showed that JMJD2A colocalized with K-bZIP, a KSHV viral transcriptional repressor, in the cell nucleus (Fig. 5 A). This finding suggests that JMJD2A may be a cellular target of KSHV K-bZIP. To confirm this interaction, 293T cells were cotransfected with HA-tagged JMJD2A and Flag-tagged K-bZIP. Anti-Flag antibody was used to precipitate K-bZIP and blotted with anti-HA antibody to detect HA-JMJD2A. The data demonstrated positive interaction between these two gene products (Fig. 5B). The reciprocal experiments confirmed such an interaction (Fig. 5C). Importantly, this interaction was also shown in naturally KSHV-infected BCBL-1 cells after K-Rta induction (Fig. 5D). Because K-bZIP directly targets JMJD2A, we wanted to determine if K-bZIP binding affects the demethylase activity of JMJD2A. In vitro demethylase assays were performed in demethylation buffer containing either the H3K9me3 substrate peptide alone or in combination with K-bZIP, JMJD2A, and K-bZIP plus JMJD2A. As expected, mass spectrometric analysis demonstrated a shift in the mass of the H3K9me3 peptide by 14 Da in the H3K9me3 peptide plus JMJD2A assay, indicating that JMJD2A demethylates H3K9me3 to H3K9me2 (Fig. 6 A, left two panels). As expected, K-bZIP alone did not have demethylation activity, but its presence completely inhibits the demethylation activity of JMJD2A (Fig. 6A, third and fourth panels). In addition, the leucine zipper motif deletion mutant of K-bZIP (K-bZIP ΔLZ) that disrupts the ability of K-bZIP to homodimerize no longer inhibited the demethylation activity of JMJD2A (Fig. 6B). This result indicated that the inhibition of JMJD2A is specific for wild-type K-bZIP and that the leucine zipper motif of K-bZIP is important for this inhibition. In addition, Biacore analysis (Fig. 7 A and B, second panels) confirmed the direct interaction of JMJD2A with H3K9me3 peptide but not the unmethylated counterpart. Interestingly, K-bZIP, but not K-bZIP ΔLZ, also binds H3K9me3 peptide with a high affinity (Fig. 7B, third and fourth panels). In a competition assay, the presence of K-bZIP decreased the binding of JMJD2A to H3K9me3-conjugated beads (Fig. 6C). These data suggest that by binding both JMJD2A and H3K9me3 substrate, K-bZIP directly blocks substrate accessibility to JMJD2A (Fig. 6D). To our knowledge, this is the first viral protein shown to directly inhibit host histone demethylase activity.
FIG. 5.
K-bZIP interacts with JMJD2A. (A) TREx-F-K-bZIP 293 cells were treated with Dox (1 μg/μl) for 48 h. Cells were fixed, stained with antibody specific for Flag and JMJD2A, and mounted in SlowFade Gold antifade reagent with DAPI. (B) 293T cells were cotransfected with pcDNA3-HA-JMJD2A and pcDNA3-Flag-KbZIP. Forty-eight hours after transfection, TCLs were immunoprecipitated with anti-Flag antibody. Immunoprecipitates were analyzed by immunoblotting using anti-JMJD2A antibody. (C) 293T cells were cotransfected with pcDNA3-Flag-JMJD2A and pcDNA3-HA-KbZIP. Forty-eight hours after transfection, TCLs were immunoprecipitated with mouse IgG or anti-HA antibody. Immunoprecipitates were analyzed by immunoblotting using anti-K-bZIP antibody. (D) TREx-MH-K-Rta BCBL-1 cells were treated with Dox (1 μg/μl) for 48 h. TCLs were immunoprecipitated with rabbit IgG or anti-JMJD2A antibody. Immunoblotting was performed using anti-K-bZIP antibody.
FIG. 6.
K-bZIP inhibits the demethylase activity of JMJD2A. (A) In vitro demethylation assays were performed under the conditions of H3K9me3 peptide alone, H3K9me3 peptide plus JMJD2A, H3K9me3 peptide plus K-bZIP, or H3K9me3 peptide plus JMJD2A and K-bZIP. Samples were analyzed using mass spectrophotometry. (B) In vitro demethylase assays were performed under the conditions of H3K9me3 peptide alone, H3K9me3 peptide plus JMJD2A, or H3K9me3 peptide plus JMJD2A and K-bZIP ΔLZ. Samples were analyzed as described for panel A. (C) H3K9me3-conjugated beads were used to pull down purified JMJD2A protein (0.5 μm). Purified K-bZIP protein (0.5 μM) was added to compete for the JMJD2A binding to the H3K9me3 peptide. Unconjugated beads were used as the negative control. (D) A model depicting K-bZIP inhibition of JMJD2A demethylase activity. K-bZIP can directly interact with JMJD2A and inhibit JMJD2A catalytic function by blocking its substrate accessibility.
FIG. 7.
Direct binding of JMJD2A and K-bZIP to H3K9me3. Biacore analysis of the interactions between H3K9 (A) or H3K9me3 (B) and JMJD2A, K-bZIP, or the K-bZIP ΔLZ mutant. H3K9 or H3K9me3 peptide was immobilized to an SA sensor chip separately. Flag, Flag-K-bZIP wt, and Flag-K-bZIP ΔLZ were 2-fold serially diluted from 80 nM to 2.5 nM. Flag-JMJD2A was serially diluted from 20 nM to 0.625 nM. Solution of Flag, Flag-JMJD2A, Flag-K-bZIP, or the Flag-K-bZIP ΔLZ mutant was passed over the chip. Flag-JMJD2A and K-bZIP bound to H3K9me3 with apparent KDs of ∼319 nM and ∼10.6 nM, respectively. The KD was nonanalyzable for all proteins in H3K9 peptide and Flag and Flag-K-bZIP ΔLZ protein in H3K9me3 peptide.
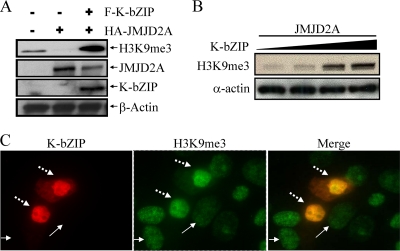
K-bZIP inhibits JMJD2A activity in vivo and increases the level of global H3K9me3 mark.
To demonstrate that the inhibition of JMJD2A by K-bZIP also occurs in vivo, 293T cells were transfected with JMJD2A alone or in combination with K-bZIP. Consistent with the in vitro result, overexpression of JMJD2A diminished the endogenous H3K9me3 level; this was completely restored in the presence of K-bZIP (Fig. 8 A). Transient cotransfection of various amounts of K-bZIP with JMJD2A showed that the increase of global H3K9me3 is K-bZIP dose dependent (Fig. 8B). Immunocytochemical staining confirmed the higher H3K9me3 intensity in Flag-K-bZIP-overexpressing cells (dashed arrows) than in nontransfected cells that do not express K-bZIP (solid arrows) (Fig. 8C). These results strongly suggest that K-bZIP is a potent JMJD2A inhibitor and an inducer of H3K9me3.
FIG. 8.
In vivo inhibition of JMJD2A demethylase activity by K-bZIP. (A) 293T cells were transiently transfected with Flag-K-bZIP and HA-JMJD2A. Forty-eight hours posttransfection, TCLs were collected, and immunoblotting was performed using antibodies specific for H3K9me3, HA-tag, and Flag-tag. β-Actin was used as a loading control. (B) Dose-dependent inhibition of JMJD2A demethylase activity by K-bZIP was analyzed in 293T cells. 293T cells were cotransfected with HA-JMJD2A and different amounts of Flag-K-bZIP. Immunoblotting was done as described for panel A. (C) 293 cells were transiently transfected with pcDNA3-Flag-K-bZIP for 48 h. Cells were fixed with 4% paraformaldehyde and stained with antibodies specific for K-bZIP and H3K9me3. K-bZIP and H3K9me3 were revealed by Alexa Fluor 555-conjugated anti-mouse (red) and Alexa Fluor 488-conjugated anti-rabbit (green) secondary antibodies, respectively. Dashed arrows and solid arrows designate K-bZIP-overexpressing or nonexpressing (nontransfected) cells (left) and H3K9me3 high- or low-level expression (middle), respectively. The merged image is shown in the right panel.
The potential role of JMJD2A inhibition by K-bZIP in KSHV replication.
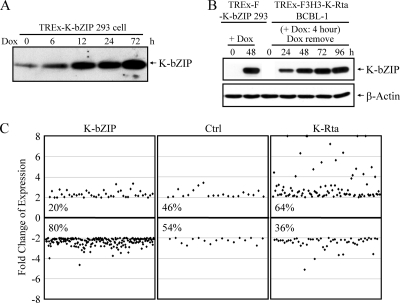
With the finding that JMJD2A directly associates with the KSHV genome, it is conceivable that K-bZIP's modulation of JMJD2A activity can directly affect viral replication and latency. We are, however, severely handicapped by the lack of knowledge of the role of K-bZIP in KSHV replication, let alone its inhibition of JMJD2A (see Discussion). Several reports showed that K-bZIP represses host gene activities and functions, including those involved in immune responses (15, 23-25, 28, 34). By elevating the H3K9me3 mark, K-bZIP may help shut off host genes, a general characteristic of herpesvirus infection (7, 10). We therefore tested the possibility that K-bZIP generally represses host gene expression. Using an inducible TREx-F-K-bZIP 293 cell line which expresses K-bZIP at a level comparable to that in BCBL-1 after K-Rta induction for 48 h (Fig. 9 B), we analyzed the expression pattern of host genes using Affymetrix HG-U133Q GeneChip arrays. Using a 2-fold cutoff, 250 genes were differentially expressed upon K-bZIP overexpression. Among them, about 80% were repressed, and only 20% were activated (Fig. 9C, left). Profiling of a K-Rta-inducible 293 cell line (TREx-F-K-Rta-293) yielded an expression pattern very different from that obtained with K-bZIP in that 60% of the differentially expressed genes were activated, while only 40% were repressed (Fig. 9C, right). This result is consistent with the notion that K-bZIP is a general transcriptional repressor, and it functions to shut off a subset of host genes to provide a favorable environment for the early phase of KSHV infection.
FIG. 9.
Global repression of host gene expression by K-bZIP. (A) TREx-F-K-bZIP 293 cells were treated with Dox (1 μg/μl) for 0, 6, 12, 24, and 72 h. Cells were harvested and TCLs were analyzed by immunoblotting using anti-K-bZIP antibody. β-Actin was used as a loading control. (B) TREx-F-K-bZIP 293 cells were treated with Dox (1 μg/μl) for 0 and 48 h. TREx-F3H3-K-Rta BCBL-1 cells were treated with Dox (1 μg/μl) for 4 h, and cells were harvested at 24, 48, 72, and 96 h after removal of Dox. TCLs were analyzed as described for panel A. (C) Control (mock-transfected TREx 293 cells), TREx-F-K-bZIP, and TREx-F-K-Rta 293 cells were treated with Dox for 72 h. Total RNA was prepared, and comprehensive gene expression profiling was performed using Affymetrix HG-U133A microarray. Genes differentially regulated by K-bZIP and K-Rta were identified by comparison analysis using a 2-fold cutoff for expression changes between groups.
DISCUSSION
It has long been postulated that epigenetic regulation, by which DNA or histone modification alters the chromatin structure, plays an important role in the transcriptional regulation of mammalian gene expression and transcriptional reactivation of herpesvirus genomes. Histone deacetylase inhibitors, such as butyrate and trichostatin A, are well-recognized activators of viral reactivation from the latent viral state. By the same token, histone methylases and demethylases are also likely to be major regulators of viral reactivation, although relatively few studies have been devoted to this subject. Ours is among the first to examine the direct relationship between histone demethylases and KSHV reactivation. Two links are reported here. First, JMJD2A targets specific regions of the viral latent genome for binding and plays a role in maintaining the local chromatin conformation and transcriptional potential of the adjacent genes. In turn, JMJD2A is targeted by the viral protein K-bZIP for inhibition via direct blockade of the demethylase catalytic activity toward the H3K9me3 substrate. These findings illustrate how herpesviruses have evolved mechanisms to exploit and subvert the host histone methylation/demethylation machinery to regulate their replication. These findings are likely to be generalized for regulation of other viruses as well.
Our studies were prompted by shRNA-based genetic screening of known histone demethylases involved in the reactivation of the latent KSHV genome. The Vero-rKSHV.219 cell line, which displays RFP signals when reactivated, provides an effective way of screening. This assay identified JMJD2A as a significant inducer of KSHV replication and reactivation. Our findings were further validated by multiple shRNAs targeting JMJD2A in Vero-rKSHV.219 cells and by an independent reactivation assay using the TREx-MH-K-Rta BCBL-1 cell line. In addition, overexpression of JMJD2A increases virus titers after K-Rta-mediated reactivation of latent genomes in Vero-rKSHV.219 cells. This occurred only in the presence of K-Rta, indicating that JMJD2A overexpression alone was not sufficient to induce the reactivation of the latent genome, a result consistent with the report of Toth et al. (35). Thus, JMJD2A functions to provide a favorable chromatin environment, allowing K-Rta to be recruited and to execute its transcriptional reprogramming. The requirement for catalytic activity of JMJD2A was shown by the finding that the JMJD2A H188A mutant cannot rescue the deficient phenotype caused by shRNA knockdown of JMJD2A. The most striking result of this study is the finding that JMJD2A decorates a large portion of the latent KSHV genome, excluding those regions enriched in H3K9me3 mark (mostly late genes). The inverse relationship of occupancy by JMJD2A and H3K9me3 suggests that JMJD2A protects H3K9 from being methylated and ensures its readiness to be acetylated. Indeed, the regions of the KSHV genome occupied by JMJD2A are enriched in H3K9Ac marks during the latent state and further enhanced upon reactivation (35). These regions are also enriched for H3K4me3, an activation mark, as well as H3K27me3, a repressive mark. Such a “bivalent” state often indicates the potential of a gene to be turned “on” or “off,” pending the presence of an activator or a repressor. In addition to the function of JMJD2A in maintaining the H3K9 landscape of the viral chromatin, JMJD2A is also the demethylase for H3K36me3, although with much less potency. The importance of the H3K36me3 histone code in KSHV latency and reactivation remains to be explored. There are also other potential functions of JMJD2A in viral replication. For instance, we found in the reporter assay that JMJD2A is a strong coactivator of K-Rta (data not show), suggesting that perhaps JMJD2A also plays a role in host gene activation by K-Rta.
Among the KSHV-encoded viral proteins studied, K-bZIP was found to be an interacting partner of JMJD2A. The interaction can be detected at the endogenous level and confirmed by colocalization studies. Significantly, K-bZIP was found to be a potent inhibitor of JMJD2A, based on the results from the in vitro H3K9me3 demethylation assay as well as the in vivo restoration of the H3K9me3 mark by K-bZIP in cells overexpressing JMJD2A. There are several nonmutually exclusive mechanisms which can account for this inhibition. First, K-bZIP binding may directly alter the conformation of the JmjC domain and interfere with its cofactor (e.g., alpha-ketoglutarate and ferric ion) binding. We have no evidence either for or against this, and more detailed biochemical or structural analysis is required to resolve this issue. Second, K-bZIP may block the accessibility of substrate to JMJD2A. We found that K-bZIP also recognizes the H3K9me3 moiety and thus likely interferes with JMJD2A binding to this substrate. Third, K-bZIP, a SUMO ligase, is able to sumoylate JMJD2A, which in turn inhibits the catalytic ability of JMJD2A. Although we found that K-bZIP is capable of sumoylating JMJD2A, this seems to be an unlikely scenario, because the in vitro demethylation assay does not have the components necessary for sumoylation, yet K-bZIP still inhibits JMJD2A. However, sumoylation of JMJD2A may have other yet-to-be-discovered effects inside the cell. Based on the above considerations, we propose that K-bZIP inhibits JMJD2A by blocking the substrate binding in situ, and it is designed to affect those genes targeted by JMJD2A (Fig. 6D). Although this is the first time a viral gene product has been shown to block histone demethylase by direct interaction, there is a precedent for a cellular transcription factor involved in a similar type of action. C-myc was shown to bind and inhibit histone demethylase, thereby increasing the level of H3K4me3 with consequent activation of the myc target genes (31).
What is the biological consequence of K-bZIP's inhibition of JMJD2A in the context of KSHV replication or latency? At present, we do not have a definitive answer. The role of K-bZIP in KSHV replication remains elusive. Some reports show that it is required for replication (17, 22), but the requirement can be bypassed if K-Rta is overexpressed (17). Others showed that K-bZIP is not absolutely required for lytic replication, but the copy number of the latent genome in the culture is decreased (Y. Yuan, personal communication). It thus appears that the principal function of K-bZIP is to serve a modulatory role, whose requirement depends on the particular cellular context. We and others showed that K-bZIP modulates the activity of K-Rta, p53, cyclin-dependent kinase (CDK), CCAAT/enhancer binding protein (C/EBP), and p21 (9, 15, 16, 28, 39), most of which rely on the repressive function of K-bZIP. Thus, K-bZIP may not be absolutely required for KSHV replication in vitro, but it serves to facilitate the process by modulating viral transcription, latency entry, and host gene expression, including those involved in cell cycle progression and immune response. In so doing, it provides a more favorable environment for viral replication. The present study provides a mechanistic basis as to how K-bZIP represses gene transcription in general.
A predicted consequence of K-bZIP inhibition of JMJD2A is the increase of the global H3K9me3 level, which we have detected in 293T cells overexpressing K-bZIP, either in transiently transfected cells or an inducible stable cell line. The increase of H3K9me3 leads to heterochromatinization of the host genome and general silencing of host genes. Supporting this notion, K-bZIP was reported to suppress the expression of host immune-related genes (23). Although we have carefully adjusted the conditions so that the K-bZIP expression level is similar to that in naturally infected cells, this is nevertheless an artificial condition, because K-bZIP does not exist in isolated form without other viral proteins. Nevertheless, our experiments demonstrate the potential for K-bZIP to shut off host genes; this may provide a host environment in favor of viral replication. However, it is noteworthy that not all cell types respond to K-bZIP overexpression in this way. It is possible that for those cells in which K-bZIP overexpression does not increase the global level of H3K9me3, there exist other H3K9me3 demethylases which K-bZIP cannot effectively inhibit. It is curious that the H3K9me3 marks associated with these two regions of latent genome do not significantly change during reactivation and, in fact, slightly increase at 12 h after reactivation (35). While we do not understand why these repressive marks are not removed during lytic replication, this increase is consistent with the elevated level of K-bZIP at that time point.
In summary, this study presents several new findings: (i) histone demethylase JMJD2A is involved in KSHV replication, (ii) JMJD2A decorates KSHV latent viral chromatin to maintain H3K9 in the unmethylated state in specific regions, (iii) K-bZIP associates with and directly inhibits JMJD2A activity, and (iv) as a consequence, K-bZIP increases the global level of H3K9me3, which in part contributes to its general transcriptional repressive activity. These findings provide new insights into epigenetic regulation of viral replication as well as viral regulation of host epigenetic machinery.
Acknowledgments
This work was supported by NIH grants (R01 CA114575, CA150197, and DE019085). The UC Davis Cancer Center Genomics Shared Resource is supported by Cancer Center Support Grant P30 CA93373-01 (Ralph W. deVere White) from NCI.
We thank Jeffrey Vieira (University of Washington) for providing the Vero-rKSHV.219 cells and Ryan R. Davis (UCDCC Genomics Shared Resource) for his expert technical assistance for the cDNA microarray and tiling array experiments. We thank Yoshihiro Izumiya for supporting the tiling array experiments (grant R01 CA147791A1).
We have no potential conflicts of interest.
Footnotes
Published ahead of print on 12 January 2011.
REFERENCES
- 1.Agger, K., J. Christensen, P. A. Cloos, and K. Helin. 2008. The emerging functions of histone demethylases. Curr. Opin. Genet. Dev. 18:159-168. [DOI] [PubMed] [Google Scholar]
- 2.Chang, P. C., et al. 2009. Kruppel-associated box domain-associated protein-1 as a latency regulator for Kaposi's sarcoma-associated herpesvirus and its modulation by the viral protein kinase. Cancer Res. 69:5681-5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chang, P. C., et al. 2010. Kaposi's sarcoma-associated herpesvirus (KSHV) encodes a SUMO E3 ligase that is SIM-dependent and SUMO-2/3-specific. J. Biol. Chem. 285:5266-5273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen, Z., et al. 2007. Structural basis of the recognition of a methylated histone tail by JMJD2A. Proc. Natl. Acad. Sci. U. S. A. 104:10818-10823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, Z., et al. 2006. Structural insights into histone demethylation by JMJD2 family members. Cell 125:691-702. [DOI] [PubMed] [Google Scholar]
- 6.Couture, J. F., E. Collazo, P. A. Ortiz-Tello, J. S. Brunzelle, and R. C. Trievel. 2007. Specificity and mechanism of JMJD2A, a trimethyllysine-specific histone demethylase. Nat. Struct. Mol. Biol. 14:689-695. [DOI] [PubMed] [Google Scholar]
- 7.Covarrubias, S., J. M. Richner, K. Clyde, Y. J. Lee, and B. A. Glaunsinger. 2009. Host shutoff is a conserved phenotype of gammaherpesvirus infection and is orchestrated exclusively from the cytoplasm. J. Virol. 83:9554-9566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dupin, N., et al. 1999. Distribution of human herpesvirus-8 latently infected cells in Kaposi's sarcoma, multicentric Castleman's disease, and primary effusion lymphoma. Proc. Natl. Acad. Sci. U. S. A. 96:4546-4551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ellison, T. J., Y. Izumiya, C. Izumiya, P. A. Luciw, and H. J. Kung. 2009. A comprehensive analysis of recruitment and transactivation potential of K-Rta and K-bZIP during reactivation of Kaposi's sarcoma-associated herpesvirus. Virology 387:76-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Glaunsinger, B. A., and D. E. Ganem. 2006. Messenger RNA turnover and its regulation in herpesviral infection. Adv. Virus Res. 66:337-394. [DOI] [PubMed] [Google Scholar]
- 11.Gray, S. G., et al. 2005. Functional characterization of JMJD2A, a histone deacetylase- and retinoblastoma-binding protein. J. Biol. Chem. 280:28507-28518. [DOI] [PubMed] [Google Scholar]
- 12.Gunther, T., and A. Grundhoff. 2010. The epigenetic landscape of latent Kaposi sarcoma-associated herpesvirus genomes. PLoS Pathog. 6:e1000935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hsia, D. A., et al. 2010. KDM8, a H3K36me2 histone demethylase that acts in the cyclin A1 coding region to regulate cancer cell proliferation. Proc. Natl. Acad. Sci. U. S. A. 107:9671-9676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang, Y., J. Fang, M. T. Bedford, Y. Zhang, and R. M. Xu. 2006. Recognition of histone H3 lysine-4 methylation by the double tudor domain of JMJD2A. Science 312:748-751. [DOI] [PubMed] [Google Scholar]
- 15.Izumiya, Y., et al. 2003. Kaposi's sarcoma-associated herpesvirus K-bZIP is a coregulator of K-Rta: physical association and promoter-dependent transcriptional repression. J. Virol. 77:1441-1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Izumiya, Y., et al. 2003. Cell cycle regulation by Kaposi's sarcoma-associated herpesvirus K-bZIP: direct interaction with cyclin-CDK2 and induction of G1 growth arrest. J. Virol. 77:9652-9661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kato-Noah, T., et al. 2007. Overexpression of the Kaposi's sarcoma-associated herpesvirus transactivator K-Rta can complement a K-bZIP deletion BACmid and yields an enhanced growth phenotype. J. Virol. 81:13519-13532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim, J., et al. 2006. Tudor, MBT and chromo domains gauge the degree of lysine methylation. EMBO Rep. 7:397-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klose, R. J., E. M. Kallin, and Y. Zhang. 2006. JmjC-domain-containing proteins and histone demethylation. Nat. Rev. Genet. 7:715-727. [DOI] [PubMed] [Google Scholar]
- 20.Klose, R. J., et al. 2006. The transcriptional repressor JHDM3A demethylates trimethyl histone H3 lysine 9 and lysine 36. Nature 442:312-316. [DOI] [PubMed] [Google Scholar]
- 21.Lee, J., J. R. Thompson, M. V. Botuyan, and G. Mer. 2008. Distinct binding modes specify the recognition of methylated histones H3K4 and H4K20 by JMJD2A-tudor. Nat. Struct. Mol. Biol. 15:109-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lefort, S., and L. Flamand. 2009. Kaposi's sarcoma-associated herpesvirus K-bZIP protein is necessary for lytic viral gene expression, DNA replication, and virion production in primary effusion lymphoma cell lines. J. Virol. 83:5869-5880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lefort, S., A. Gravel, and L. Flamand. 2010. Repression of interferon-alpha stimulated genes expression by Kaposi's sarcoma-associated herpesvirus K-bZIP protein. Virology 408:14-30. [DOI] [PubMed] [Google Scholar]
- 24.Lefort, S., A. Soucy-Faulkner, N. Grandvaux, and L. Flamand. 2007. Binding of Kaposi's sarcoma-associated herpesvirus K-bZIP to interferon-responsive factor 3 elements modulates antiviral gene expression. J. Virol. 81:10950-10960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liao, W., Y. Tang, S. F. Lin, H. J. Kung, and C. Z. Giam. 2003. K-bZIP of Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8 (KSHV/HHV-8) binds KSHV/HHV-8 Rta and represses Rta-mediated transactivation. J. Virol. 77:3809-3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lim, S., E. Metzger, R. Schule, J. Kirfel, and R. Buettner. 2010. Epigenetic regulation of cancer growth by histone demethylases. Int. J. Cancer 127:1991-1998. [DOI] [PubMed] [Google Scholar]
- 27.Nakamura, H., et al. 2003. Global changes in Kaposi's sarcoma-associated virus gene expression patterns following expression of a tetracycline-inducible Rta transactivator. J. Virol. 77:4205-4220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park, J., et al. 2000. The K-bZIP protein from Kaposi's sarcoma-associated herpesvirus interacts with p53 and represses its transcriptional activity. J. Virol. 74:11977-11982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pasini, D., et al. 2008. Regulation of stem cell differentiation by histone methyltransferases and demethylases. Cold Spring Harb. Symp. Quant. Biol. 73:253-263. [DOI] [PubMed] [Google Scholar]
- 30.Sakakibara, S., et al. 2004. Accumulation of heterochromatin components on the terminal repeat sequence of Kaposi's sarcoma-associated herpesvirus mediated by the latency-associated nuclear antigen. J. Virol. 78:7299-7310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Secombe, J., L. Li, L. Carlos, and R. N. Eisenman. 2007. The Trithorax group protein Lid is a trimethyl histone H3K4 demethylase required for dMyc-induced cell growth. Genes Dev. 21:537-551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shi, Y., et al. 2004. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 119:941-953. [DOI] [PubMed] [Google Scholar]
- 33.Shin, S., and R. Janknecht. 2007. Activation of androgen receptor by histone demethylases JMJD2A and JMJD2D. Biochem. Biophys. Res. Commun. 359:742-746. [DOI] [PubMed] [Google Scholar]
- 34.Tomita, M., J. Choe, T. Tsukazaki, and N. Mori. 2004. The Kaposi's sarcoma-associated herpesvirus K-bZIP protein represses transforming growth factor beta signaling through interaction with CREB-binding protein. Oncogene 23:8272-8281. [DOI] [PubMed] [Google Scholar]
- 35.Toth, Z., et al. 2010. Epigenetic analysis of KSHV latent and lytic genomes. PLoS Pathog. 6:e1001013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tsukada, Y., T. Ishitani, and K. I. Nakayama. 2010. KDM7 is a dual demethylase for histone H3 Lys 9 and Lys 27 and functions in brain development. Genes Dev. 24:432-437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vieira, J., and P. M. O'Hearn. 2004. Use of the red fluorescent protein as a marker of Kaposi's sarcoma-associated herpesvirus lytic gene expression. Virology 325:225-240. [DOI] [PubMed] [Google Scholar]
- 38.Whetstine, J. R., et al. 2006. Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases. Cell 125:467-481. [DOI] [PubMed] [Google Scholar]
- 39.Wu, F. Y., et al. 2002. Lytic replication-associated protein (RAP) encoded by Kaposi sarcoma-associated herpesvirus causes p21CIP-1-mediated G1 cell cycle arrest through CCAAT/enhancer-binding protein-alpha. Proc. Natl. Acad. Sci. U. S. A. 99:10683-10688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang, D., H. G. Yoon, and J. Wong. 2005. JMJD2A is a novel N-CoR-interacting protein and is involved in repression of the human transcription factor achaete scute-like homologue 2 (ASCL2/Hash2). Mol. Cell. Biol. 25:6404-6414. [DOI] [PMC free article] [PubMed] [Google Scholar]