Abstract
Human immunodeficiency virus type 1 (HIV-1) elite controllers maintain undetectable levels of viral replication in the absence of antiretroviral therapy (ART), but their underlying immunological and virological characteristics may vary. Here, we used a whole-genome transcriptional profiling approach to characterize gene expression signatures of CD4 T cells from an unselected cohort of elite controllers. The transcriptional profiles for the majority of elite controllers were similar to those of ART-treated patients but different from those of HIV-1-negative persons. Yet, a smaller proportion of elite controllers showed an alternative gene expression pattern that was indistinguishable from that of HIV-1-negative persons but different from that of highly active antiretroviral therapy (HAART)-treated individuals. Elite controllers with the latter gene expression signature had significantly higher CD4 T cell counts and lower levels of HIV-1-specific CD8+ T cell responses but did not significantly differ from other elite controllers in terms of HLA class I alleles, HIV-1 viral loads determined by ultrasensitive single-copy PCR assays, or chemokine receptor polymorphisms. Thus, these data identify a specific subgroup of elite controllers whose immunological and gene expression characteristics approximate those of HIV-1-negative persons.
Hope for the development of an effective human immunodeficiency virus type 1 (HIV-1) vaccine or the induction of immune-mediated control of HIV-1 infection is predominantly fueled by the ability of a small proportion of HIV-1-infected individuals to spontaneously control HIV-1 replication (28). A rare subgroup of such persons, termed “elite suppressors” (2) or “elite controllers” (1), maintain levels of HIV-1 replication that are undetectable by standard commercial assays, although residual viral replication can be observed in the majority of these individuals using ultrasensitive assays (24). These patients have moved into the center of current efforts to identify effective host defense mechanisms against HIV-1. Genome-wide association studies (8) and longitudinal investigations of natural-history HIV-1 patient cohorts (6, 13) have shown that expression of specific HLA class I alleles or certain chemokine receptor polymorphisms are significantly associated with the elite controller phenotype. Moreover, previous functional immunology studies demonstrated that HIV-1-specific CD8 T cells in elite controllers have specific cytotoxic (20) and proliferative (16, 19) characteristics and are effective in restricting viral replication (27), while broadly neutralizing antibodies seem to be less involved in immune control in these patients (7).
As the main target of HIV-1, CD4 T cells play a central role in HIV-1 pathogenesis. During progressive infection, these cells continuously decline, most likely as a combined result of immune activation (9), low thymic output, and direct destruction of cells during the HIV-1 replication cycle. Prior work has shown that CD4 T cells from elite controllers are generally capable of supporting HIV-1 infection (3, 14), but specific characteristics of CD4 T cells in elite controllers that distinguish them from alternative patient populations remain unclear. Microarray-based whole-genome transcriptional profiling allows molecular characterization of cellular populations in an unprecedented high-throughput fashion. This strategy has been applied to identify specific gene expression signatures that are associated with distinct clinical outcomes in multiple different disease contexts (32). For instance, specific transcriptional signatures correlate with low levels of immune activation in simian immunodeficiency virus (SIV)-infected sooty mangabeys (4) or with spontaneous allograft tolerance after solid-organ transplantation (22).
To more closely analyze specific characteristics of CD4 T cells from elite controllers, we conducted a whole-genome transcriptional profiling study of CD4 T cells from an unselected cohort of elite controllers (n = 12) and two background populations of HIV-1-negative persons (n = 9) and HIV-1-infected persons effectively treated with antiretroviral therapy (ART) (n = 14). None of these study persons had detectable HIV-1 viremia by standard commercial assays, and age (P = 0.9) and male/female ratios (P = 0.6) were not statistically different in the study cohorts; moreover, elite controllers did not differ from ART-treated persons in terms of CD4 cell counts (mean of 802/μl versus 789/μl, P = 0.9), but they did have longer known durations of HIV-1 infection (mean of 18 years versus 9 years, P = 0.01). To minimize bias from differential representation of HLA-DR+ CD4 T cells (6% of all CD4 T cells in elite controllers, 8.7% of all CD4 T cells in highly active antiretroviral therapy [HAART]-treated patients, and 4.6% of all CD4 T cells in HIV-1-negative persons) and HLA-DR− CD4 T cells in these study cohorts (11), we focused our investigations on homogenous populations of sorted HLA-DR− CD4 T cell subsets. Briefly, peripheral blood mononuclear cells (PBMC) from study persons were stained with monoclonal antibodies against CD3, CD4, and HLA-DR and subsequently subjected to live sorting at 70 lb/in2 using an Aria cell-sorting device (Becton Dickinson) located in a specifically designated biosafety cabinet. Following mRNA extraction from the sorted cells (RNeasy kit; Qiagen), whole-genome transcriptional profiling was performed using WG-DASL microarrays (Illumina) according to standard protocols. Data retrieved from the Illumina software were background corrected (Illumina beadstudio software) and quantile normalized using the Arraystar normalization function. Analysis was restricted to genes with significant expression (P < 0.01). For quality control, duplicate samples from two HIV-1-negative persons and one ART-treated individual were run on the microarray; the average expression intensity for each transcript was calculated from such replicates and entered in the subsequent data analysis. Differentially expressed genes were detected using the empirical Bayes (eBayes) adjusted t test, as implemented in the Limma package of R (30). Multiple testing was corrected with the Benjamini-Hochberg false discovery rate (FDR). Differentially expressed genes were defined as being at least 2-fold different after log2 transformation and having an FDR P value of <0.05.
Overall, we found that a total of 978 transcripts were expressed differently in HIV-1-negative and ART-treated study persons (see Table S1 in the supplemental material). Of these gene transcripts, 24 were also expressed differently in HIV-1-negative persons and HIV-1 elite controllers, while an additional 4 transcripts were identified that distinguished elite controllers from HIV-1-negative persons without meeting the criteria for differential gene expression in HIV-1-negative persons and ART-treated individuals (see Table S2 in the supplemental material). Interestingly, no differentially expressed genes were detected in elite controllers and ART-treated patients using our defined criteria.
To identify known and predicted interactions of the differentially expressed genes, we used the Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathway Tool (http://www.genome.jp/kegg) (12). These studies showed that transcripts expressed differently in HAART-treated persons and HIV-1-negative individuals included genes encoding chemokines/cytokines and their receptors (see Fig. S1 in the supplemental material) and genes impacting the cell cycle (see Fig. S2 in the supplemental material); these transcripts play important roles in the regulation of immune activation. Gene transcripts known to be involved in intrinsic cellular defense against retroviruses, such as the TRIM (tripartite motif-containing protein) and APOBEC (apolipoprotein B mRNA-editing enzyme catalytic polypeptide) gene families, tetherin/BST2 (bone marrow stromal antigen 2), cyclophilin A, and other genes included in a list of 138 transcripts relevant for HIV-1 pathogenesis based on a literature review in a recent study (26) were not expressed differently in elite controllers and the reference patients, as determined by gene set enrichment analysis (GSEA) (21, 31). Moreover, a similar analysis demonstrated no significant difference in the study cohorts in the expression of a panel of HIV-1 dependency genes identified by a recent small interfering RNA (siRNA)/short hairpin RNA (shRNA) screen (5). However, it is important to recognize that our study was limited to HLA-DR− CD4 T cells, which do not represent the predominant target cells for HIV-1; therefore, additional studies will be necessary to analyze expression signatures of HIV-1 resistance or dependence genes in HLA-DR+ CD4 T cells.
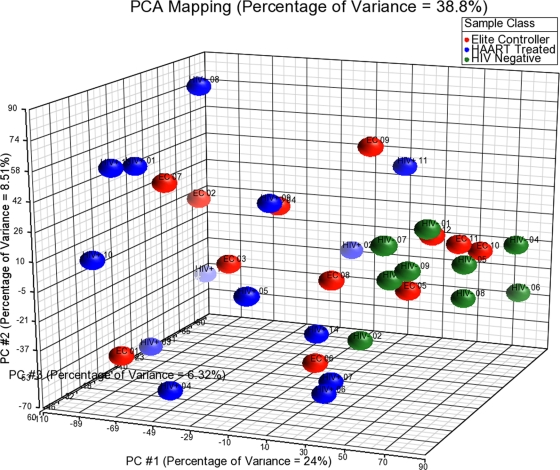
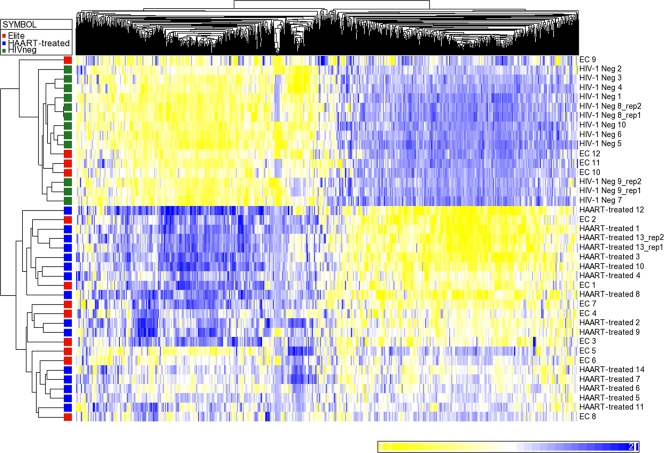
To more closely compare gene expression signatures for individual persons in the three patient groups, we entered expression data for all transcripts of the microarray into a principal component analysis (PCA), using Partek software (Fig. 1). This computational dimension reduction method uses orthogonal transformation of correlated variables to generate a new set of decorrelated variables (called principal components) that can be plotted in a new coordinate system in which axes follow the direction of greatest variance in the data set (15). Principal component analysis of gene expression characteristics using our comprehensive, unfiltered data set demonstrated that six elite controllers (EC 5, 8, 9, 10, 11, and 12) grouped more closely with HIV-1-negative persons than the remaining six elite controllers (EC 1, 2, 3, 4, 6, and 7) whose gene expression patterns resembled HAART-treated persons. To further investigate this, we analyzed the gene expression characteristics of elite controllers with regard to the 978 transcripts that, per our previous analysis, significantly distinguished HIV-1-negative persons from HAART-treated individuals. For this purpose, z-score standardized data from all three patient groups were entered into an unsupervised hierarchical linkage clustering analysis with the Euclidean metric (Fig. 2). As expected, this analysis revealed clearly distinguishable patterns of gene expression in HIV-1-negative persons and ART-treated HIV-1-infected persons. Interestingly, gene expression networks in elite controllers were split into two distinct subgroups: in the majority of these patients (n = 8), the transcriptional profiles of elite controllers clustered with those of HAART-treated persons; however, a group of 4 elite controllers (EC 9, 10, 11, and 12) was observed whose gene expression patterns clustered closely with those of HIV-1-negative persons. When directly comparing the gene expression patterns of the two subgroups of elite controllers, no differentially expressed transcripts were identified using our previously established criteria, most likely due to the small patient subgroups. However, after the criteria for differential gene expression were relaxed to a fold change of 1.5 and an FDR-adjusted P value of 0.11, 159 transcripts that distinguished the two study groups were identified (see Table S3 in the supplemental material). These genes included genes encoding components of the mitogen-activated protein kinase (MAPK)/focal adhesion kinase (FAK) signaling pathway and transcripts involved in cell cycle regulation and other currently unidentified biological functions.
FIG. 1.
Principal component analysis of transcriptional profiles of CD4 T cells from elite controllers, HIV-1-negative persons, and ART-treated individuals, using unfiltered data from all transcripts included in the microarray.
FIG. 2.
Clustering analysis of the differentially expressed genes in CD4 T cells from elite controllers, HIV-1-negative persons, and ART-treated individuals. This analysis was performed using all transcripts expressed differently in HIV-1-negative persons and ART-treated individuals. Replicate samples were included into this figure for the sole purpose of highlighting the high reproducibility of microarray data; calculations of differentially expressed genes were performed using the average of two replicate samples (rep1 and rep2). Elite controllers EC 1 to EC 12 were studied. Neg, negative.
We subsequently attempted to determine whether these different transcriptional gene profiles in elite controllers were correlated with specific clinical, immunological, or immunogenetic characteristics of the respective patient subgroups. We observed that CD4 T cell counts in elite controllers clustering with HIV-1-negative persons were significantly higher than the counts in the alternative group of elite controllers (mean [median] absolute counts, 1,148/μl [1,172/μl] versus 629/μl [800/μl], P = 0.01; mean [median] relative counts, 50.7% [51.5%] versus 39.2% [45.75%], P = 0.05). Elite controllers whose transcriptional CD4 T cell profiles clustered with those of HIV-1-negative persons had a smaller magnitude of HIV-1-specific CD8 T cell responses than the remaining elite controllers (mean [median] of 3,820 [3,850] spot-forming cells versus 8,892 [9,177] spot-forming cells, P = 0.03), as determined by comprehensive screening using gamma interferon enzyme-linked immunospot (ELISPOT) assays and a library of overlapping HIV-1 peptides spanning the entire HIV-1 proteome using a method described in our prior work (17). Finally, we were able to analyze the plasma viremia of 10 elite controllers (7 patients whose transcriptional profiles clustered with those of HAART-treated patients and 3 patients whose profiles clustered with those of HIV-1-negative persons) for whom sufficient material was available, using an ultrasensitive single-digit PCR protocol described elsewhere (23). This analysis demonstrated lower HIV-1 copy numbers in the plasma of elite controllers whose gene expression profiles clustered with those of HIV-1-negative persons compared to the remaining elite controllers (mean [median] of 1.16 [0.3] copies/ml versus 9.69 [3.38] copies/ml), although differences were not significant in this small patient population (P = 0.24). There were also no differences between the two subgroups of elite controllers in terms of allele frequencies for HLA-B57 (P = 0.4) or HLA-B27 (P = 0.42), duration of known HIV-1 infection (P = 0.5), or the frequency of CCR5 Δ32 alleles (P = 0.6).
Elite controllers are defined as a group of HIV-1-infected patients with undetectable viremia in the absence of antiretroviral therapy, but it is now well recognized that their clinical and immunological properties may be quite heterogeneous. For instance, in some of these patients, a progressive decline of CD4 T cells, sometimes to a level of <200/μl, or other AIDS-defining clinical events have been observed (11, 24), requiring initiation of antiretroviral therapy (29) despite undetectable viral loads. Here, we have analyzed an unselected cohort of elite controllers who all had stable CD4 T cell counts within normal limits for >10 years and who did not show any clinical signs of HIV-1 disease progression. For these patients, who all phenotypically and clinically resemble healthy individuals, the transcriptional profiling approach showed that at the level of CD4 T cell gene expression signatures, most elite controllers behave similar to ART-treated patients, specifically with regard to expression of gene transcripts involved in immune activation. It will be important to determine whether these elite controllers whose transcriptional profiles resemble those of ART-treated patients are prone to experience typical complications of chronic treated HIV-1 infection, such as an increased risk for cardiovascular comorbidities (10) and an overall life expectancy that is reduced in comparison to HIV-1-negative persons (18).
Interestingly, the transcriptional profiling approach also identified the presence of a small subgroup of HIV-1 elite controllers who in addition to undetectable viral loads maintain high-normal relative and absolute CD4 cell counts and whose transcriptional gene expression profiles in CD4 T cells are indistinguishable from those of HIV-1-negative persons, while being distinct from those of ART-treated patients. Although our observations were made in a relatively small group of patients and have to be confirmed using larger patient populations, they indicate that a subgroup of extremely rare elite controllers exists who appear to be as close as possible to a true “functional cure” of HIV-1 infection and who may be of specific interest for recent efforts in designing strategies leading to a cure of HIV-1 infection (25, 33). The closer investigation of this extremely rare subgroup of untreated HIV-1 patients with spontaneous viral control and a transcriptional gene profile that is indistinguishable from that of HIV-1-negative persons may increase our understanding of how long-term drug-free remissions of HIV-1 infection may be induced in a broader population of HIV-1-infected persons.
Microarray data accession number.
All data were deposited in a public repository (GEO) and can be accessed at http://www.ncbi.nlm.nih.gov/geo (accession no. GSE23879).
Supplementary Material
Acknowledgments
This study was supported by the U.S. National Institutes of Health (grant AI078799 to X.G.Y.) and by a Feasibility Grant (to M.L.) of the Harvard University Center for AIDS Research. X.G.Y. and M.L. are both recipients of the Doris Duke Clinical Scientist Development Award. F.V. is supported by a Canadian Institutes of Health Research and a Ragon Institute Fellowship. Work in the laboratory of J.M.-P. was supported by the Spanish AIDS network (RD06/0006) and the Catalan HIV Vaccine Development Program (HIVACAT). A.T. is supported by the Swiss National Science Foundation. Recruitment of study patients was supported by the Bill and Melinda Gates Foundation and the Mark and Lisa Schwartz Foundation.
The authors are grateful to the International HIV Controllers Study for providing patient samples. QP1 and RCASBP(A)gfp plasmids to perform the ultrasensitive HIV-1 viremia assay were kindly provided by M. Kearney from the National Cancer Institute.
Footnotes
Published ahead of print on 22 December 2010.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Baker, B. M., B. L. Block, A. C. Rothchild, and B. D. Walker. 2009. Elite control of HIV infection: implications for vaccine design. Expert Opin. Biol. Ther. 9:55-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blankson, J. N. 2010. Control of HIV-1 replication in elite suppressors. Discov. Med. 9:261-266. [PubMed] [Google Scholar]
- 3.Blankson, J. N., et al. 2007. Isolation and characterization of replication-competent human immunodeficiency virus type 1 from a subset of elite suppressors. J. Virol. 81:2508-2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bosinger, S. E., et al. 2009. Global genomic analysis reveals rapid control of a robust innate response in SIV-infected sooty mangabeys. J. Clin. Invest. 119:3556-3572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brass, A. L., et al. 2008. Identification of host proteins required for HIV infection through a functional genomic screen. Science 319:921-926. [DOI] [PubMed] [Google Scholar]
- 6.Dean, M., et al. 1996. Genetic restriction of HIV-1 infection and progression to AIDS by a deletion allele of the CKR5 structural gene. Hemophilia Growth and Development Study, Multicenter AIDS Cohort Study, Multicenter Hemophilia Cohort Study, San Francisco City Cohort, ALIVE Study. Science 273:1856-1862. [DOI] [PubMed] [Google Scholar]
- 7.Doria-Rose, N. A., et al. 2010. Breadth of human immunodeficiency virus-specific neutralizing activity in sera: clustering analysis and association with clinical variables. J. Virol. 84:1631-1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fellay, J., et al. 2007. A whole-genome association study of major determinants for host control of HIV-1. Science 317:944-947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hazenberg, M. D., et al. 2000. T-cell division in human immunodeficiency virus (HIV)-1 infection is mainly due to immune activation: a longitudinal analysis in patients before and during highly active antiretroviral therapy (HAART). Blood 95:249-255. [PubMed] [Google Scholar]
- 10.Hsue, P. Y., et al. 2009. Role of viral replication, antiretroviral therapy, and immunodeficiency in HIV-associated atherosclerosis. AIDS 23:1059-1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hunt, P. W., et al. 2008. Relationship between T cell activation and CD4+ T cell count in HIV-seropositive individuals with undetectable plasma HIV RNA levels in the absence of therapy. J. Infect. Dis. 197:126-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kanehisa, M., S. Goto, M. Furumichi, M. Tanabe, and M. Hirakawa. 2010. KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res. 38:D355-D360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaslow, R. A., et al. 1996. Influence of combinations of human major histocompatibility complex genes on the course of HIV-1 infection. Nat. Med. 2:405-411. [DOI] [PubMed] [Google Scholar]
- 14.Lamine, A., et al. 2007. Replication-competent HIV strains infect HIV controllers despite undetectable viremia (ANRS EP36 study). AIDS 21:1043-1045. [DOI] [PubMed] [Google Scholar]
- 15.Li, L. 2010. Dimension reduction for high-dimensional data. Methods Mol. Biol. 620:417-434. [DOI] [PubMed] [Google Scholar]
- 16.Lichterfeld, M., et al. 2004. Loss of HIV-1-specific CD8+ T cell proliferation after acute HIV-1 infection and restoration by vaccine-induced HIV-1-specific CD4+ T cells. J. Exp. Med. 200:701-712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lichterfeld, M., et al. 2007. A viral CTL escape mutation leading to immunoglobulin-like transcript 4-mediated functional inhibition of myelomonocytic cells. J. Exp. Med. 204:2813-2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lohse, N., et al. 2007. Survival of persons with and without HIV infection in Denmark, 1995-2005. Ann. Intern. Med. 146:87-95. [DOI] [PubMed] [Google Scholar]
- 19.Migueles, S. A., et al. 2002. HIV-specific CD8+ T cell proliferation is coupled to perforin expression and is maintained in nonprogressors. Nat. Immunol. 3:1061-1068. [DOI] [PubMed] [Google Scholar]
- 20.Migueles, S. A., et al. 2008. Lytic granule loading of CD8+ T cells is required for HIV-infected cell elimination associated with immune control. Immunity 29:1009-1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mootha, V. K., et al. 2003. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 34:267-273. [DOI] [PubMed] [Google Scholar]
- 22.Newell, K. A., et al. 2010. Identification of a B cell signature associated with renal transplant tolerance in humans. J. Clin. Invest. 120:1836-1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Palmer, S., et al. 2008. Low-level viremia persists for at least 7 years in patients on suppressive antiretroviral therapy. Proc. Natl. Acad. Sci. U. S. A. 105:3879-3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pereyra, F., et al. 2009. Persistent low-level viremia in HIV-1 elite controllers and relationship to immunologic parameters. J. Infect. Dis. 200:984-990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Richman, D. D., et al. 2009. The challenge of finding a cure for HIV infection. Science 323:1304-1307. [DOI] [PubMed] [Google Scholar]
- 26.Rotger, M., et al. 2010. Genome-wide mRNA expression correlates of viral control in CD4+ T-cells from HIV-1-infected individuals. PLoS Pathog. 6:e1000781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saez-Cirion, A., et al. 2007. HIV controllers exhibit potent CD8 T cell capacity to suppress HIV infection ex vivo and peculiar cytotoxic T lymphocyte activation phenotype. Proc. Natl. Acad. Sci. U. S. A. 104:6776-6781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saez-Cirion, A., G. Pancino, M. Sinet, A. Venet, and O. Lambotte. 2007. HIV controllers: how do they tame the virus? Trends Immunol. 28:532-540. [DOI] [PubMed] [Google Scholar]
- 29.Sedaghat, A. R., et al. 2009. T cell dynamics and the response to HAART in a cohort of HIV-1-infected elite suppressors. Clin. Infect. Dis. 49:1763-1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smyth, G. K. 2005. Limma: linear models for microarray data, p. 397-420. In R. Gentleman, V. Carey, S. Dudoit, R. Irizarry, and W. Huber (ed.), Bioinformatics and computational biology solutions using R and Bioconductor. Springer, New York, NY.
- 31.Subramanian, A., et al. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U. S. A. 102:15545-15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Telenti, A., and D. B. Goldstein. 2006. Genomics meets HIV-1. Nat. Rev. Microbiol. 4:865-873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Trono, D., et al. 2010. HIV persistence and the prospect of long-term drug-free remissions for HIV-infected individuals. Science 329:174-180. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.