Abstract
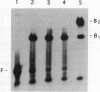
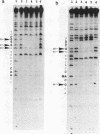
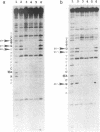
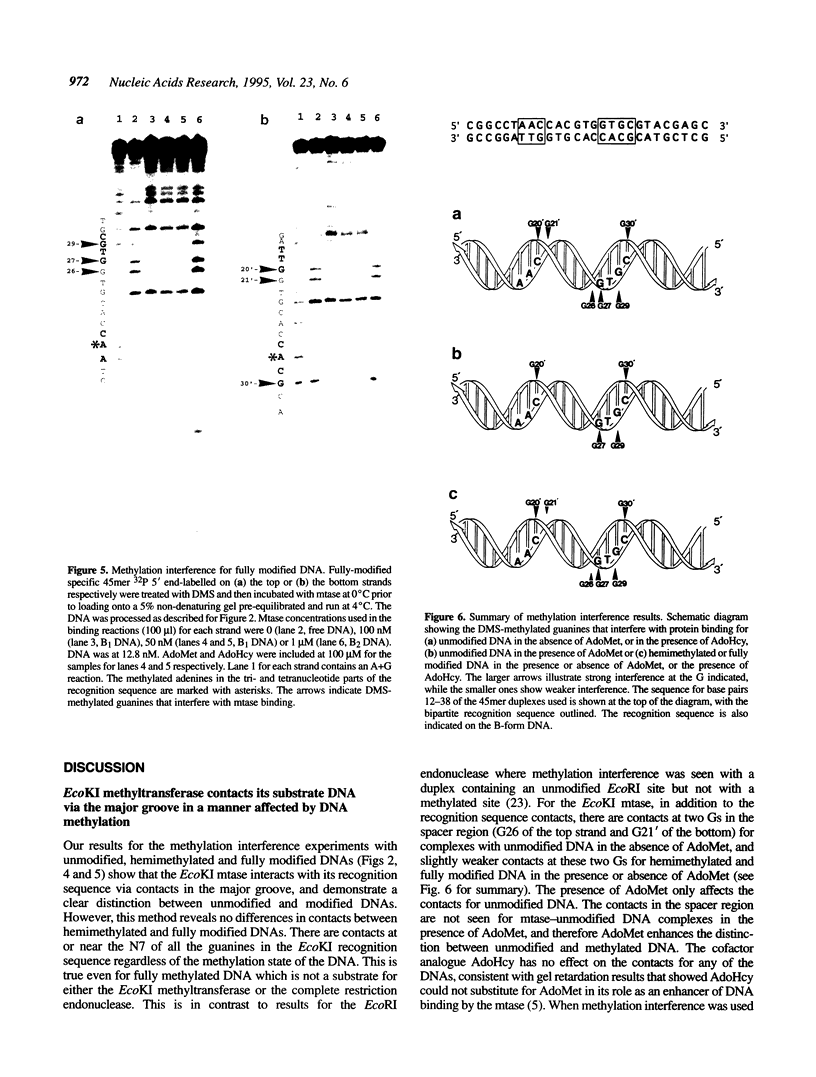
The EcoKI methyltransferase methylates two adenines on opposite strands of its bipartite DNA recognition sequence AAC(N6)GTGC. The enzyme has a strong preference for hemimethylated DNA substrates, but the methylation state of the DNA does not influence its binding affinity. Methylation interference was used to compare the contacts made by the EcoKI methyltransferase with unmodified, hemimethylated or fully modified DNAs. Contacts were seen at or near the N7 position of guanine, in the major groove, for all of the guanines in the EcoKI recognition sequence, and at two guanines on the edge of the intervening spacer sequence. The presence of the cofactor and methyl donor S-adenosyl methionine had a striking effect on the interference pattern for unmodified DNA which could not be mimicked by the presence of the cofactor analogue S-adenosyl homocysteine. In contrast, S-adenosyl methionine had no effect on the interference patterns for either kind of hemimethylated DNA, or for fully modified DNA. Differences between the interference patterns for the unmodified DNA and any of the three forms of methylated DNA provide evidence that methylation of the target sequence influences the conformation of the protein-DNA interface, and illustrate the importance of S-adenosyl methionine in the distinction between unmodified and methylated DNA by the methyltransferase.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abadjieva A., Webb M., Patel J., Zinkevich V., Firman K. Deletions within the DNA recognition subunit of M.EcoR124I that identify a region involved in protein-protein interactions between HsdS and HsdM. J Mol Biol. 1994 Aug 5;241(1):35–43. doi: 10.1006/jmbi.1994.1471. [DOI] [PubMed] [Google Scholar]
- Alves J., Pingoud A., Haupt W., Langowski J., Peters F., Maass G., Wolff C. The influence of sequences adjacent to the recognition site on the cleavage of oligodeoxynucleotides by the EcoRI endonuclease. Eur J Biochem. 1984 Apr 2;140(1):83–92. doi: 10.1111/j.1432-1033.1984.tb08069.x. [DOI] [PubMed] [Google Scholar]
- Bergerat A., Kriebardis A., Guschlbauer W. Preferential site-specific hemimethylation of GATC sites in pBR322 DNA by Dam methyltransferase from Escherichia coli. J Biol Chem. 1989 Mar 5;264(7):4064–4070. [PubMed] [Google Scholar]
- Bickle T. A., Brack C., Yuan R. ATP-induced conformational changes in the restriction endonuclease from Escherichia coli K-12. Proc Natl Acad Sci U S A. 1978 Jul;75(7):3099–3103. doi: 10.1073/pnas.75.7.3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bickle T. A., Krüger D. H. Biology of DNA restriction. Microbiol Rev. 1993 Jun;57(2):434–450. doi: 10.1128/mr.57.2.434-450.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burckhardt J., Weisemann J., Hamilton D. L., Yuan R. Complexes formed between the restriction endonuclease EcoK and heteroduplex DNA. J Mol Biol. 1981 Dec 5;153(2):425–440. doi: 10.1016/0022-2836(81)90287-4. [DOI] [PubMed] [Google Scholar]
- Burckhardt J., Weisemann J., Yuan R. Characterization of the DNA methylase activity of the restriction enzyme from Escherichia coli K. J Biol Chem. 1981 Apr 25;256(8):4024–4032. [PubMed] [Google Scholar]
- Clark L., Nicholson J., Hay R. T. Enhancer binding protein (EBP1) makes base and backbone contacts over one complete turn of the DNA double helix. J Mol Biol. 1989 Apr 20;206(4):615–626. doi: 10.1016/0022-2836(89)90570-6. [DOI] [PubMed] [Google Scholar]
- Cooper L. P., Dryden D. T. The domains of a type I DNA methyltransferase. Interactions and role in recognition of DNA methylation. J Mol Biol. 1994 Mar 4;236(4):1011–1021. doi: 10.1016/0022-2836(94)90008-6. [DOI] [PubMed] [Google Scholar]
- Cowan G. M., Gann A. A., Murray N. E. Conservation of complex DNA recognition domains between families of restriction enzymes. Cell. 1989 Jan 13;56(1):103–109. doi: 10.1016/0092-8674(89)90988-4. [DOI] [PubMed] [Google Scholar]
- Derbyshire K. M., Grindley N. D. Binding of the IS903 transposase to its inverted repeat in vitro. EMBO J. 1992 Sep;11(9):3449–3455. doi: 10.1002/j.1460-2075.1992.tb05424.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dryden D. T., Cooper L. P., Murray N. E. Purification and characterization of the methyltransferase from the type 1 restriction and modification system of Escherichia coli K12. J Biol Chem. 1993 Jun 25;268(18):13228–13236. [PubMed] [Google Scholar]
- Gann A. A., Campbell A. J., Collins J. F., Coulson A. F., Murray N. E. Reassortment of DNA recognition domains and the evolution of new specificities. Mol Microbiol. 1987 Jul;1(1):13–22. doi: 10.1111/j.1365-2958.1987.tb00521.x. [DOI] [PubMed] [Google Scholar]
- Hadi S. M., Bickle T. A., Yuan R. The role of S-adenosylmethionine in the cleavage of deoxyribonucleic acid by the restriction endonuclease from Escherichia coli K. J Biol Chem. 1975 Jun 10;250(11):4159–4164. [PubMed] [Google Scholar]
- Hendrickson W., Schleif R. A dimer of AraC protein contacts three adjacent major groove regions of the araI DNA site. Proc Natl Acad Sci U S A. 1985 May;82(10):3129–3133. doi: 10.1073/pnas.82.10.3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannan P., Cowan G. M., Daniel A. S., Gann A. A., Murray N. E. Conservation of organization in the specificity polypeptides of two families of type I restriction enzymes. J Mol Biol. 1989 Oct 5;209(3):335–344. doi: 10.1016/0022-2836(89)90001-6. [DOI] [PubMed] [Google Scholar]
- Klimasauskas S., Kumar S., Roberts R. J., Cheng X. HhaI methyltransferase flips its target base out of the DNA helix. Cell. 1994 Jan 28;76(2):357–369. doi: 10.1016/0092-8674(94)90342-5. [DOI] [PubMed] [Google Scholar]
- Kneale G. G. A symmetrical model for the domain structure of type I DNA methyltransferases. J Mol Biol. 1994 Oct 14;243(1):1–5. doi: 10.1006/jmbi.1994.1624. [DOI] [PubMed] [Google Scholar]
- Labahn J., Granzin J., Schluckebier G., Robinson D. P., Jack W. E., Schildkraut I., Saenger W. Three-dimensional structure of the adenine-specific DNA methyltransferase M.Taq I in complex with the cofactor S-adenosylmethionine. Proc Natl Acad Sci U S A. 1994 Nov 8;91(23):10957–10961. doi: 10.1073/pnas.91.23.10957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu A. L., Jack W. E., Modrich P. DNA determinants important in sequence recognition by Eco RI endonuclease. J Biol Chem. 1981 Dec 25;256(24):13200–13206. [PubMed] [Google Scholar]
- Meister J., MacWilliams M., Hübner P., Jütte H., Skrzypek E., Piekarowicz A., Bickle T. A. Macroevolution by transposition: drastic modification of DNA recognition by a type I restriction enzyme following Tn5 transposition. EMBO J. 1993 Dec;12(12):4585–4591. doi: 10.1002/j.1460-2075.1993.tb06147.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papavassiliou A. G. 1,10-Phenanthroline-copper ion nuclease footprinting of DNA-protein complexes in situ following mobility-shift electrophoresis assays. Methods Mol Biol. 1994;30:43–78. doi: 10.1385/0-89603-256-6:43. [DOI] [PubMed] [Google Scholar]
- Papp P. P., Chattoraj D. K. Missing-base and ethylation interference footprinting of P1 plasmid replication initiator. Nucleic Acids Res. 1994 Jan 25;22(2):152–157. doi: 10.1093/nar/22.2.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papp P. P., Chattoraj D. K., Schneider T. D. Information analysis of sequences that bind the replication initiator RepA. J Mol Biol. 1993 Sep 20;233(2):219–230. doi: 10.1006/jmbi.1993.1501. [DOI] [PubMed] [Google Scholar]
- Powell L. M., Dryden D. T., Willcock D. F., Pain R. H., Murray N. E. DNA recognition by the EcoK methyltransferase. The influence of DNA methylation and the cofactor S-adenosyl-L-methionine. J Mol Biol. 1993 Nov 5;234(1):60–71. doi: 10.1006/jmbi.1993.1563. [DOI] [PubMed] [Google Scholar]
- Sain B., Murray N. E. The hsd (host specificity) genes of E. coli K 12. Mol Gen Genet. 1980;180(1):35–46. doi: 10.1007/BF00267350. [DOI] [PubMed] [Google Scholar]
- Siebenlist U., Gilbert W. Contacts between Escherichia coli RNA polymerase and an early promoter of phage T7. Proc Natl Acad Sci U S A. 1980 Jan;77(1):122–126. doi: 10.1073/pnas.77.1.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suri B., Nagaraja V., Bickle T. A. Bacterial DNA modification. Curr Top Microbiol Immunol. 1984;108:1–9. doi: 10.1007/978-3-642-69370-0_1. [DOI] [PubMed] [Google Scholar]
- Taylor I. A., Davis K. G., Watts D., Kneale G. G. DNA-binding induces a major structural transition in a type I methyltransferase. EMBO J. 1994 Dec 1;13(23):5772–5778. doi: 10.1002/j.1460-2075.1994.tb06915.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor J. D., Halford S. E. The activity of the EcoRV restriction endonuclease is influenced by flanking DNA sequences both inside and outside the DNA-protein complex. Biochemistry. 1992 Jan 14;31(1):90–97. doi: 10.1021/bi00116a014. [DOI] [PubMed] [Google Scholar]
- Willcock D. F., Dryden D. T., Murray N. E. A mutational analysis of the two motifs common to adenine methyltransferases. EMBO J. 1994 Aug 15;13(16):3902–3908. doi: 10.1002/j.1460-2075.1994.tb06701.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson G. G., Murray N. E. Restriction and modification systems. Annu Rev Genet. 1991;25:585–627. doi: 10.1146/annurev.ge.25.120191.003101. [DOI] [PubMed] [Google Scholar]
- van de Wetering M., Clevers H. Sequence-specific interaction of the HMG box proteins TCF-1 and SRY occurs within the minor groove of a Watson-Crick double helix. EMBO J. 1992 Aug;11(8):3039–3044. doi: 10.1002/j.1460-2075.1992.tb05374.x. [DOI] [PMC free article] [PubMed] [Google Scholar]