Abstract
Catalytically promiscuous enzymes are intermediates in the evolution of new function from an existing pool of protein scaffolds. However, promiscuity will only confer an evolutionary advantage if other useful properties are not compromised, or if there is no ‘negative trade-off’ induced by the mutations that yield promiscuity. Therefore, identification and characterization of negative trade-offs incurred during the emergence of promiscuity is required to further develop the evolutionary models and to optimize in vitro evolution. One potential negative trade-off of catalytic promiscuity is increased susceptibility to inhibition, or inhibitory promiscuity. Here we exploit Cytochrome P450s (CYPs) as a model protein scaffold that spans a vast range of catalytic promiscuity, and apply a quantitative index to determine the relationship between promiscuity of catalysis and promiscuity of inhibition for a series of homologs. The aim of these studies is to begin to identify properties that, in general, correlate with catalytic promiscuity, hypothetically such as inhibitory promiscuity. Interestingly, the data indicate that the potential negative trade-off of inhibitory promiscuity is nearly insignificant because even highly substrate specific CYPs have high inhibitory promiscuity, with little incremental increase in susceptibility to inhibitory interactions as the substrate promiscuity increases across the series of enzymes. In the context of evolution, inhibitory promiscuity is not an obligate negative trade-off of catalytic promiscuity.
Keywords: Enzyme catalysis, evolutionary intermediates, catalytic promiscuity
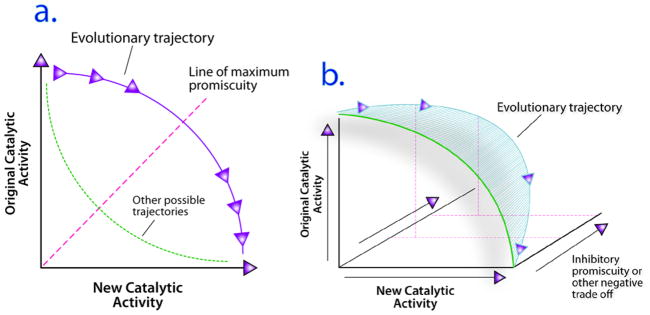
Promiscuity at the molecular level plays a critical role in several cellular processes including signal transduction, immunity, and detoxification (1–9). More broadly, the evolution of new enzymatic activities from a pool of existing protein scaffolds, whether in nature or in vitro directed evolution, may proceed through ‘promiscuous’ intermediates that catalyze multiple reactions (10–19). A common description of this possible evolutionary process and relevant factors is schematized in Figure 1, wherein a specific enzyme undergoes mutation to yield a promiscuous variant that catalyzes a new reaction, without forfeiting its original activity (11, 20). At early stages along the evolutionary trajectory the mutation is neutral with respect to the original function. If this second enzymatic activity provides any survival advantage, then it is retained and, after gene duplication, one copy of the gene encoding the promiscuous enzyme can undergo further refinement to optimize the new function, without loss of the original function. It may be useful to distinguish between ‘promiscuous’ enzymes with catalytic activities unrelated to their presumed biological function, and ‘multifunctional’ enzymes with a clear functional advantage in accepting multiple substrates, as suggested (20). Here we use the term ‘promiscuity’ for both cases, because we aim to understand the physical and chemical properties of enzymes that process multiple substrates, regardless of the functional purpose. Recent studies have suggested that promiscuous intermediates easily evolve into efficient and specific enzymes (11, 20). While this model provides an extremely useful framework to conceptualize evolutionary structure-function relationships, ‘promiscuity’ is not a well-understood property, and it is difficult to quantify, although progress has been made (17, 19).
Figure 1.
A. Promiscuous Intermediate Model for Evolution of New Enzymatic Function. Enzymes with high catalytic specificity for one reaction (original catalytic activity) move along the indicated trajectory. At early stages they become promiscuous, gaining new catalytic activity without forfeiting the original activity. The dotted line indicates that other evolutionary trajectories are possible. B. Three-dimensional evolutionary trajectory relating catalytic function toward original and new substrates with susceptibility to inhibition. Susceptibility to inhibition could be a significant negative trade-off that increases the evolutionary distance from any intermediate to the desired evolutionary endpoint on the new activity axis. Each incremental increase in catalytic promiscuity has an associated cost of potential new inhibitory interaction with other ligands; the negative trade-off is revealed as an increase in the distance between the green two-dimensional curve and the blue trajectory as the catalytic promiscuity increases. The greater the increase in distance between the blue and green curves, the greater the negative trade-off. The detailed shape of the new trajectory is not known for any enzyme, but it ultimately restricts the possible evolutionary trajectories by defining the extent of negative trade-off.
For example, natural evolution of new function does not happen in isolation. Rather, each evolutionary intermediate must function in the context of the existing niche that includes other enzymes and a ‘background’ of potential ligands. Within a niche, a loss due to mutation in any useful trait is referred to as a ‘negative trade-off’(20). The evolutionary trajectory of an enzyme is unlikely to be a two dimensional process as depicted in the model in Figure 1a, but it will trace a multidimensional surface that includes the relationships between promiscuity and other properties that comprise the overall fitness of the enzyme. That is, there will be a third collective coordinate for negative trade-offs, as in Figure 1b. In this case, ‘promiscuity’ per se is not an obvious advantage in the evolutionary process unless it is achieved without significant negative trade-offs (20). Whereas a highly promiscuous enzyme would provide a very efficient template from which to evolve many new functions from a single protein sequence, such an intermediate would unlikely be neutral with respect to all of the original properties. If the magnitude of any negative trade-off is coupled exactly to the magnitude of promiscuity, then the mutational neutrality will not be maintained in all dimensions of fitness space and the path towards new function will have a different shape than if the putative negative trade-off is not correlated with the promiscuity in any dimension. Ultimately, the relationship between any potential negative trade-off and catalytic promiscuity will determine whether evolution will follow that path. These concepts underscore the need to define the relationship between catalytic promiscuity and other protein properties.
Speculatively, for example, a new promiscuous enzyme would have increased susceptibility to inhibition by ligands that do not inhibit the ‘original’ enzyme and this would be a negative trade-off. This is based on the observation that structurally diverse drugs can bind to and inhibit a single enzyme, without yielding products (consider antibiotics, statins, etc.). In contrast catalysis requires proper alignment of multiple reactive functional groups. Each incremental step towards promiscuity, i.e. towards the diagonal of Figure 1a, will only be advantageous if the new catalytic activity is not accompanied by an equal or greater increase in susceptibility to ‘new’ inhibitory interactions that, at ambient ligand concentrations, limit the original enzyme activity. For the example here, inhibitory promiscuity is a potential negative trade-off that adds a new dimension to yield the scheme in Figure 1b, and can be substituted by any parameter hypothesized to be a negative trade-off.
We emphasize that the current studies are not intended to establish specific evolutionary mechanisms, which may best be accomplished by successive rounds of random mutagenesis and relevant selection protocols. Rather we aim to understand more generally the relationships between catalytic promiscuity and other properties of enzymes that could limit or define evolutionary pathways. An understanding of these relationships is necessary to understand the role of catalytic promiscuity in evolution and to advance the incomplete model of Figure 1. The Cytochrome P450s provide an excellent test case to further understand these relationships because, as a family with highly conserved scaffold, they span a very wide range of catalytic promiscuity. Human hepatic CYPs dominate drug metabolism and have very broad substrate selectivities (19, 21). Here we use these multifunctional CYPs as a model for physico-chemical traits of enzymes near the limits of catalytic promiscuity. Other CYPs, including bacterial and human isoforms used here, contribute to specific biosynthetic pathways and are very substrate specific (22, 23). Collectively, the CYP superfamily provides insight into the relationship between catalytic and inhibitory promiscuity. The results indicate a surprising level of inhibitory promiscuity even for substrate-specific CYPs, and they further suggest that because inhibitory promiscuity is already present in highly specific enzymes, there is little cost associated with evolutionary steps toward catalytic promiscuity.
Materials and Methods
Materials
Recombinant CYP Supersomes™ were purchased from Becton Dickinson (San Jose, CA). NADPH was obtained from EMD Chemicals (Gibbstown, NJ). HPLC solvents were from J. T. Baker (Phillipsburg, NJ). All other chemicals were purchased from Sigma-Aldrich (St. Louis, MO) and were of the highest grade available.
Determination of Catalytic and Inhibitory Parameters
Kinetic constants for CYP-catalyzed turnover were determined using a substrate depletion approach (25, 26). Briefly, incubations consisted of 1 pmol recombinant CYP enzyme, 3 mM magnesium chloride and substrate (0.001 – 25 μM, final concentration) in 100 mM potassium phosphate buffer (pH 7.4) in 250 μL. Reactions (performed in duplicate) were initiated with the addition of 1 mM NADPH (final concentration) and allowed to proceed for 0, 5, 10 or 20 minutes before being quenched with 2 volumes of acetonitrile containing 0.1 μM tolbutamide as an internal standard. To determine IC50 values, a similar incubation procedure was used. Probe substrates were incubated at their previously determined Km values (data not shown) and inhibitor concentrations ranged from 0 – 25 μM. Incubations (n = 3) were quenched after 10 minutes as described above. Samples were vortex-mixed and centrifuged prior to LC-MS/MS analysis.
Liquid Chromatography/Tandem Mass Spectral Analysis
All samples were analyzed on an Applied Biosystems API4000 Q-trap mass spectrometer (Applied Biosystems, Foster City, CA) that was operated in triple-quadrupole mode. The LC-MS/MS system was comprised of an electrospray ionization source coupled to a binary pump with in-line solvent degasser (Shimadzu, Columbia, MD) and a LEAP CTC HTS PAL autosampler (CTC Analytics, Carrboro, NC). Chromatographic separation was achieved using a mobile phase system consisting of 0.1% formic acid in acetonitrile and 0.1% formic acid in 5 mM ammonium formate with a Gemini C18 2.0 × 30 mm 5 μm column (Phenomenex, Torrance, CA). MS/MS conditions were optimized for each compound prior to analysis.
Statistical Analysis
Substrate depletion rate constants were determined by fitting the percentage of substrate remaining versus time to first-order decay functions, as [Substrate] vs. ln time. Data points which resulted in the substrate depletion plot deviating from linearity were not included in determining the depletion rate constants. Michaelis constants (Km) were obtained by plotting the substrate depletion rate constants as a function of the substrate concentration using the following equation (where kdep is the substrate depletion rate constant, Km is the Michaelis constant, [S] is the substrate concentration and kdep:[S]=0represents the maximal theoretical rate of substrate depletion):
IC50 values were determined by fitting inhibition data to the following equation, using the algorithms contained in GraphPad Prism 7.0 (Graphpad Software Inc., San Diego, CA):
where % activity refers to the percent activity remaining relative to a control with no inhibitor present, max and min are the maximum and minimum percent activities remaining in the presence of inhibitor and [I] is the inhibitor concentration.
Calculation of catalytic promiscuity, Jcat, and inhibitory promiscuity, Jinh, was based on the previously described method that quantifies the probability that any enzyme will metabolize a specific substrate from a set of substrates, wherein the probability is normalized to account for chemical dissimilarity among substrates in the set (19, 24). Substrates that are chemically very different from others in the set are weighted more. For each of the 65 compounds studied here, 161 chemical properties were assigned a score of ‘1’ if the compound contained that property or ‘0’ if it did not. For each of the 161 compounds, its’ chemical similarity to others in the set was calculated as described below. Briefly, the Jcat is defined, as in reference 19, by:
where ei is the kcat/KM for the ith substrate in a basis set with N substrates, is the average distance in chemical space of the ith substrate from the other members of the set and δset is a measure of the chemical space covered by the set, with a theoretical maximum of δset = 1. More specifically, for a pair of chemicals A and B, where a is the number of features present only in A, b is the number of features present only in B and c is the number of features present in both A and B, the Tanimoto distance is . For substrates in a set, we can also define as the mean Tanimoto distance from a member i to all the other members in the set. For the calculations here, there are 161 chemical properties used to determine the 〈δ〉i for each of the 65 substrates. The overall set dissimilarity δset serves as an upper bound for : if k is the number of features present in at least one but not all of k the members of the set, and lis the number of features present in all members of the set, then . Finally, yields the normalized mean distance for each substrate i. With this measure of distance in chemical space between substrates, the substrates that are furthest from others in the chemical space are weighted more heavily in the calculation of promiscuity. If many structurally similar substrates are metabolized with the same efficiency then the resulting Jcat value will not be as great as when the substrates are far from each other in chemical space.
For inhibition, the Jinh value is the susceptibility to inhibition and is
where xi is the inhibitory potency of an inhibitor from a set containing M inhibitors and 〈δ〉i is the average distance in chemical space from other inhibitors. As with the catalytic parameters, inhibitors that are chemically very different from others in the set are weighted more heavily in the determination of inhibitory promiscuity.
Results
Promiscuity parameters
Eight hepatic human CYP isoforms, typically considered to be detoxification enzymes, and two CYPs with high catalytic activity toward defined endogenous substrates were compared directly for their ability to metabolize 65 substrates. These substrates collectively span a wide range of the 166-dimensional chemical space, with a δset = 0.99, as defined in the Methods. This value of δset, which has a theoretical maximum value of 1.0, confirms that the substrates and inhibitors used here are extrememly diverse with respect to chemical space. The catalytic parameters kcat, KM, and kcat/KM for each CYP isoform and its probe substrate are summarized in Supplemental Information. The promiscuity values (J-values) obtained for each CYP, calculated as described in Methods, are summarized in Table 1. Briefly, the J-value is a normalized distribution of kcat/KM values for the substrates in the basis set. A wide distribution implies a promiscuous enzyme and a narrow distribution reflects high specificity (see methods). The scale ranges from 1, the upper limit of promiscuity, to 0, which describes a completely substrate specific enzyme. Promiscuity values (Jcat) for some of these isoforms were previously reported based on a smaller basis set of substrates, and the values reported here with a significantly expanded basis set further validate and extend our previous approach (19). J-values obtained from basis sets that sample more chemical space will be more predictive than J-values obtained from a basis set of chemically similar substrates. Notably, the hepatic CYP enzymes not previously studied, CYP2E1, CYP3A5, CYP2C8, and CYP1A2, are highly promiscuous, as expected. The promiscuity parameters for the biosynthetic CYP26A1 and the fatty acid metabolizing CYP4F12, which are usually considered to be substrate specific, are much lower. The hepatic drug metabolizing CYPs, with J-values > 0.7 clearly segregate from the biosynthetic CYPs, with J-values ≤ 0.30.
Table 1.
Promiscuity Parameters
| CYP isoform | Jcat | Jinh |
|---|---|---|
| Detoxification CYPs | ||
| CYP3A4 | 0.83 | 0.78 |
| CYP3A5 | 0.82 | |
| CYP2D6 | 0.81 | 0.82 |
| CYP2E1 | 0.77 | |
| CYP2C8 | 0.69 | 0.85 |
| CYP2C9 | 0.65 | 0.80 |
| CYP2C19 | 0.70 | 0.84 |
| CYP1A2 | 0.81 | 0.79 |
| Substrate specific CYPs | ||
| CYP26A1 | 0.30 | 0.58 |
| CYP4F12 | 0.16 | 0.65 |
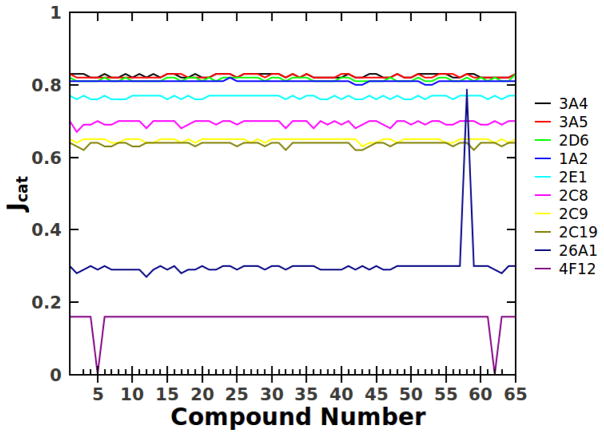
In order to validate the approach and this basis set of substrates, statistical ‘jacknife’ methods were used; for each data set, each substrate was removed from the basis set and the Jcat value was recalculated. If the basis set is adequately unbiased, then removal of any individual substrate has no effect of the recovered J-value, if the enzyme is ‘promiscuous.’ In contrast, for substrate specific enzymes, removal of the ‘cognate substrate’ from the calculation results in a large increase or decrease in the J-value and removal of other substrates has negligible effect. The results of these jackknife calculations are summarized in Figure 2. Clearly the detoxification CYPs are highly promiscuous, whereas CYPs 26A1 and 4F12 are highly specific.
Figure 2.
Jacknife analysis of Jcat values for all CYPS in Table 1. Each compound is removed from the basis set and the Jcat recalculated. The analysis validates that the basis set of substrates does not bias the calculation of Jcat and further differentiates promiscuous enzymes from specific enzymes. See text for further explanation.
Inhibitory promiscuity
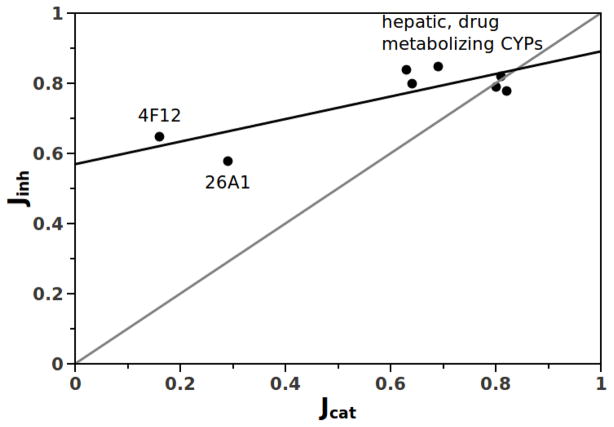
The calculation of inhibitory promiscuity has been done previously for CYPs 2D6, 1A2, 3A4, 2C9, 2C8, and 2C19 using a basis set of 65 inhibitors (24). Here we have calculated the inhibitory Jinh values for CYP26A1, and CYP4F12, using the basis set of compounds listed in Supplemental Information, in each isoform-specific probe reaction. These inhibitors span a wide range of structural space, as did the substrates. A plot of Jinh vs. Jcat is shown in Figure 3. The diagonal line in Figure 3 represents a line of slope 1, and therefore is the expected line along which the data would lie if catalytic promiscuity and inhibitory promiscuity were exactly correlated. This line represents the negative trade-off of inhibitory promiscuity that evolving organisms would ‘pay’ if each incremental increase in catalytic promiscuity were accompanied by an equal incremental increase in inhibitory promiscuity. The relationship would not, a priori, be linear, but if it is the slope could in principle be 1, less than 1, or greater than 1. A slope much greater than 1 would, for example, constitute a significant negative trade-off, and a slope much less than 1 would constitute a minimal negative trade-off. As can be seen, the data for the hepatic CYPs are clustered near this diagonal at the top right, suggesting that these enzymes are near the functional limit of both catalytic and inhibitory promiscuity. In contrast, the CYPs, 26A1 and CYP4F12 lie near the lower limits of catalytic promiscuity but are clearly above the diagonal. Although the Jinh values of CYP26A1 and CYP4F12, are relatively low compared to the detoxification CYPs, they are clearly higher than their catalytic promiscuity. In effect, the location of Jinh values for the catalytically specific CYPs well above the diagonal in Figure 3 indicates that it is much ‘easier’ to inhibit a substrate selective enzyme, or one with intermediate promiscuity, than it is to provide an alternative substrate. Although it is intuitively expected that inhibition would be ‘easier’ than catalysis, the magnitudes of J inh values for the substrate specific enzymes are interestingly high. Because this ligand basis set spans a substantial fraction of chemical space, these results further suggest that this is likely to be a general behavior for CYPs.
Figure 3.
Plot of Jinh vs. Jcat for CYPs 3A4, 2D6, 2C8, 2C9, 2C19, 1A2, 26A1, 4F12. The gray diagonal line is for visualization only and represents a perfect correlation between Jcat and Jinh, The fitted line (r2 = 0.68, slope = 0.32) indicates that the negative trade-off of Jinh is nearly insignificant as the CYPs become more catalytically promiscuous. The shallow slope of the line is due to inherently high Jinh even for enzymes with low Jcat.
This, in turn, has very significant implications for the developing paradigm concerning the role of functional promiscuity in the evolution of new function from the existing pool of protein scaffolds. The line that is fit to the data, with r2 = 0.68 and slope 0.32 indicates that the negative trade-off of Jinh is much less than the line of equal negative trade-off (slope = 1). Each incremental increase in Jcat is accompanied by only a small increase in Jinh. However, more important than the shallow slope in this case, is the fact that the y-intercept is well above the origin. In fact, the Jinh is high even for catalytically specific CYPs and Jinh is relatively insensitive to Jcat as greater catalytic promiscuity is observed.
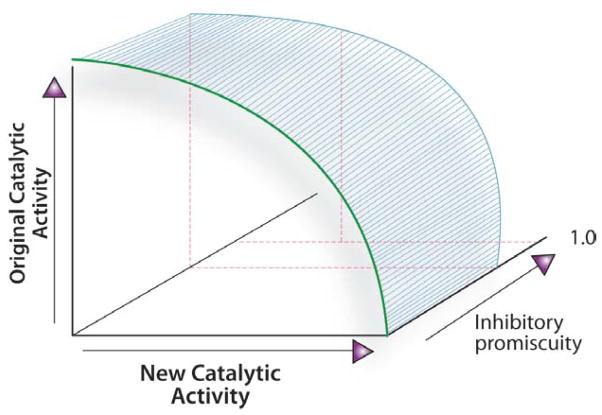
These results can be cast in the context of Figure 1. For the CYPs, the three dimensional plot of original activity vs. new activity vs. inhibitory promiscuity (Jinh) would include a curve that starts well-displaced along the Jinh axis. This curve only deviates slightly from a parallel trajectory as the Jcat increased and decreased again towards the new function. The initially high Jinh values changes only slightly as the Jcat values span the range from 0.16 to 0.89.
Discussion
Functional promiscuity of proteins has become increasingly recognized in the past few years, but the relationship between catalytic promiscuity and other physico-chemical traits is not yet understood. Here we utilize a quantitative index of the functional promiscuity of Cytochrome P450s, in order to advance our understanding the relationship of promiscuity ot other protein traits. We also consider the utility of CYPs as a model to examine the widespread suggestion that promiscuity is a useful property in the evolution of new protein function. For an extensive set of compounds with sufficient structural diversity, this index provides a useful tool for comparing catalytic promiscuity within a set of enzymes with common structural fold. Such an index facilitates testing of meaningful hypotheses to further advance our understanding of enzymatic promiscuity.
The results quantitatively support the well known, but ‘intuitive,’ notion that hepatic detoxification CYPs are more promiscuous than CYPs involved in specific biosynthetic pathways with defined substrates. The segregation of detoxification CYPs from biosynthetic CYPs along the diagonal of Figure 3 is striking. Equally apparent is the high degree of susceptibility toward inhibition, even for highly substrate specific isoforms. In fact, while the Jcat values span nearly the entire range of promiscuity, with CYP4F12 and CYP3A4 defining the lower and upper limits for this set of CYPs, the Jinh values vary only between 0.6 and 0.85. This supports the intuition that fewer chemical constraints must be satisfied to achieve inhibition than catalysis.
Regardless of whether or not CYPs reflect general properties of other protein folds, one implication of the results is that inhibitory promiscuity is not an obligate negative trade-off in the progression of specific enzymes to promiscuous intermediates and further toward specific enzymes with new functions. This general conclusion is true despite the fact that more compounds in chemical space are likely to be inhibtors of an enzyme than a substate for that enzyme, as the data demonstrate. Inhibitory promiscuity may be a liability already for specific enzymes, prior to development of catalytic promiscuity, so relatively little additional cost is incurred, in terms of Jinh, when Jcat increases. If one considers catalytic promiscuity to be an evolutionary advantage for CYPs and inhibitory promiscuity to be the associated cost, then all mutations that confer catalytic promiscuity are potentially advantageous, owing to the inherently high starting point of CYP inhibitory promiscuity. In other words, any increase in P450 Jinh in a series of variants is nearly inconsequential when compared to the high initial value of Jinh. Although the positive correlation we observe in Figure 3 suggests a greater likelihood that catalytically promiscuous intermediates will exhibit inhibitory promiscuity, the results also suggest that inhibitory promiscuity does not necessarily represent a ‘new’ limit on the utility of such intermdiates, because its incremental increase among intermediates may be small compared to the beneficial catalytic promiscuity. Further studies of the type described here could reveal other parameters or properties that are negative trade-offs correlated with catalytic promiscuity, that do limit the evolutionary trajectory. Such properties could include thermal stability, susceptibility to proteases, aggregation, increased activity toward important metabolites, or others properties that may result from structural plasticity.
Other groups have examined the intriguing relationship between catalytic and inhibitory promiscuity, and also emphasize that evolution from promiscuous intermediates is dependent on the ‘ligand matrix’ (i.e., the collective interactions with potential inhibitors and alternative substrates) that such enzymes encounter (27). In that work, DNA shuffling was used to generate a library of variants from two parental enzymes. Clusters of variants with sufficiently similar catalytic properties were defined as a ‘quasi-species’. Individual proteins within a quasi-species may display preference towards one substrate over another or susceptibility towards a particular inhibitor, but have different amino acid sequences and hence can differ in other biophysical properties. Evolution from a quasi-species, rather than from a single protein sequence, ensures efficient sampling of sequence and functional space. Our results expand upon and complement the quasi-species concept in several ways: 1) just as with individual member proteins, we predict that the quasi-species that have the greatest catalytic promiscuity will also be those with the greatest inhibitory promiscuity; 2) nonetheless, all quasi-species will tend to inherit significant inhibitory promiscuity, so that a low Jinh is unlikely to strongly distinguish one quasi-species from another; 3) Jcat and Jinh provide global, model-free measures of evolutionarily relevant functional properties, which could, for example, be used to select first-generation variants for subsequent rounds of laboratory evolution. Clustering enzyme variants into quasi-species is analytically challenging in high-dimensional spaces, especially in the context of larger ligand matrices (e.g. the 65×65 matrix of the present work, compared to the 2×3 matrix of ref. 27).
The results summarized here provide potentially useful quantitative benchmarks for evolutionary models that are based on either quasi-species or individual proteins. We speculate that very low ratios of Jinh/Jcat may be a rare among members of any quasi-speices or individual intermediates if they derive from substrate specific enzymes; our results provide the first benchmarks for this ratio, and suggest that the upper limit is ~2 (CYP26A1), which differs only modestly from the lower limit of 1.2 (CYP2C8). The present results suggest the critical point that, in order to obtain a low ratio of Jinh/Jcat, very high values of Jcat are required (~0.8). Such high levels of Jcat, would require multiple mutations in a highly susbtrate specific enzyme, and could require a long evolutionary trajectory. Possibly, evolution would be more efficient by utilizing intermediates with lower J cat values, at the expense of higher Jinh/Jcat values.
Although these data refute the intuitive hypothesis that inhibitory promiscuity necessarily will be a negative trade-off for all inceases in catalytic promiscuity, it remains to be determined whether this is generally true of all enzyme familes. We chose CYPs as a model because, as a family, they are assumed to span a wide range of Jcat, which enhances our understanding of promiscuity at the extremes of this behavior. It is interesting to consider that the CYP scaffold is unique among protein folds, and is especially prone to high Jinh, thus making it an ideal scaffold for evolution of promiscuous detoxification enzymes. The CYP scaffold, in principle, could be uniquely biased toward promiscuity, with high Jinh/Jcat ratios even for isoforms with with low Jcat. This trait may have been selected evolutionarily to achieve the evolutionary end point for detoxification enzymes, with high Jcat. If this speculation is true, then our results can not be generalized to substrate specific enzymes that have lower starting Jinh values. Clearly, more studies of this type, with additional protein scaffolds, are required to understand the role of promiscuity in evolution.
Finally, the hepatic CYPs studied here collectively account for the majority of human drug metabolism. Examples of CYP-dependent drug-drug interactions are numerous, wherein one drug inhibits the metabolism of a second. In many cases this severely restricts the utility of a drug, or at least limits its therapeutic application. Our results confirm that the susceptibility of CYPs to metabolic drug interactions is an inherent property of their structural scaffold, and is likely unavoidable.
Supplementary Material
Figure 4.
Schematized summary of the relationship between Jcat and Jinh for CYPs. The inhibitory promiscuity remains nearly constant, starting at high Jinh, with only marginal further increase as the enzymes progress toward higher Jcat values. As a result, promiscuity of inhibition is unlikely to be a significant negative trade-off as enzymes progress from specificity for the original activity toward promiscuity.
Acknowledgments
This work was supported by NIHGM 32165 (WMA) and an American Heart Association Postdoctoral Fellowship (AN).
Footnotes
Supporting Information Available. Table S1 includes the kcat, KM, kcat/KM, and k→0 values for the 65 substrates used for each CYP. This material is available free of charge via the internet at http://pubs.acs.org.
References
- 1.Wilhelm T, Nasheuer HP, Huang S. Physical and functional modularity of the protein network in yeast. Mol Cell Proteomics. 2003;2:292–298. doi: 10.1074/mcp.M300005-MCP200. [DOI] [PubMed] [Google Scholar]
- 2.Dunker AK, Cortese MS, Romero P, Iakoucheva LM, Uversky VN. Flexible nets. The roles of intrinsic disorder in protein interaction networks. FEBS J. 2005;272:5129–5148. doi: 10.1111/j.1742-4658.2005.04948.x. [DOI] [PubMed] [Google Scholar]
- 3.Sethi DK, Agarwal A, Manivel V, Rao KV, Salunke DM. Differential epitope positioning within the germline antibody paratope enhances promiscuity in the primary immune response. Immunity. 2006;24:429–438. doi: 10.1016/j.immuni.2006.02.010. [DOI] [PubMed] [Google Scholar]
- 4.Grubor NM, Hayes J, Small GJ, Jankowiak R. Cross-reactivity and conformational multiplicity of an anti-polycyclic aromatic hydrocarbon mAb. Proc Natl Acad Sci U S A. 2005;102:7453–7458. doi: 10.1073/pnas.0502540102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dimitrov JD, Lacroix-Desmazes S, Kaveri SV, Vassilev TL. Transition towards antigen-binding promiscuity of a monospecific antibody. Mol Immunol. 2007;44:1864–1873. doi: 10.1016/j.molimm.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 6.Jimenez R, Salazar G, Yin J, Joo T, Romesberg FE. Protein dynamics and the immunological evolution of molecular recognition. Proc Natl Acad Sci U S A. 2004;101:3803–3808. doi: 10.1073/pnas.0305745101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ekroos M, Sjogren T. Structural basis for ligand promiscuity in cytochrome P40 3A4. Proc Natl Acad Sci. 2006;103:13682–13687. doi: 10.1073/pnas.0603236103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Redinbo MR. Promiscuity: What protects us perplexes us. Drug Discovery Today. 2004;9:431–432. doi: 10.1016/S1359-6446(04)03087-9. [DOI] [PubMed] [Google Scholar]
- 9.Lewinson O, Adler J, Sigal N, Bibi E. Promiscuity in multidrug recognition and transport: the bacterial MFS Mdr transporters. Mol Microbiol. 2006;61:277–284. doi: 10.1111/j.1365-2958.2006.05254.x. [DOI] [PubMed] [Google Scholar]
- 10.O’Brien PJ, Herschlag D. Catalytic promiscuity and the evolution of new enzymatic activities. Chem Biol. 1999;6:R91–R105. doi: 10.1016/S1074-5521(99)80033-7. [DOI] [PubMed] [Google Scholar]
- 11.Khersonsky O, Roodveldt C, Tawfik DS. Enzyme promiscuity: evolutionary and mechanistic aspects. Curr Opin Chem Bio. 2006;l10:498–508. doi: 10.1016/j.cbpa.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 12.James LC, Tawfik DS. Conformational diversity and protein evolution--a 60-year-old hypothesis revisited. Trends Biochem Sci. 2003;28:361–368. doi: 10.1016/S0968-0004(03)00135-X. [DOI] [PubMed] [Google Scholar]
- 13.Poelarends GJ, Veetil VP, Whitman CP. The chemical versatility of the beta-alpha-beta fold: catalytic promiscuity and divergent evolution in the tautomerase superfamily. Cell Mol Life Sci. 2008;65:3606–3618. doi: 10.1007/s00018-008-8285-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yew WS, Fedorov AA, Fedorov EV, Rakus JF, Pierce RW, Almo SC, Gerlt JA. Evolution of enzymatic activities in the enolase superfamily: L-fuconate dehydratase from Xanthomonas campestris. Biochemistry. 2006;45:14598–14608. doi: 10.1021/bi061687o. [DOI] [PubMed] [Google Scholar]
- 15.Glasner ME, Gerlt JA, Babbitt PC. Mechanisms of protein evolution and their application to protein engineering. Adv Enzymol Relat Areas Mol Biol. 2007;75:193–239. doi: 10.1002/9780471224464.ch3. [DOI] [PubMed] [Google Scholar]
- 16.Lairson LL, Watts AG, Wakarchuk WW, Withers SG. Using substrate engineering to harness enzymatic promiscuity and expand biological catalysis. Nat Chem Biol. 2006;2:724–728. doi: 10.1038/nchembio828. [DOI] [PubMed] [Google Scholar]
- 17.Mannervik B, Runarsdottir A, Kurtovic S. Multi-substrate-activity space and quasi-species in enzyme evolution: Ohno’s dilemma, promiscuity and functional orthogonality. Biochem Soc Trans. 2009;37(Pt 4):740–744. doi: 10.1042/BST0370740. [DOI] [PubMed] [Google Scholar]
- 18.Glasner ME, Gerlt JA, Babbitt PC. Evolution of enzyme superfamilies. Curr Opin Chem Biol. 2006;10:492–497. doi: 10.1016/j.cbpa.2006.08.012. [DOI] [PubMed] [Google Scholar]
- 19.Nath A, Atkins WM. A quantitative index of substrate promiscuity. Biochemistry. 2008;47:157–166. doi: 10.1021/bi701448p. [DOI] [PubMed] [Google Scholar]
- 20.Khersonsky O, Tawfik DS. Enzyme Promiscuity: A mechanistic and evolutionary perspective. Annu Rev Biochem. 2010;79:471–505. doi: 10.1146/annurev-biochem-030409-143718. [DOI] [PubMed] [Google Scholar]
- 21.Guengerich FP. Cytochrome p450 and chemical toxicology. Chem Res Toxicol. 2008;21:70–83. doi: 10.1021/tx700079z. [DOI] [PubMed] [Google Scholar]
- 22.Pikuleva IA. Cholesterol-metabolizing cytochromes P450: implications for cholesterol lowering. Expert Opin Drug Metab Toxicol. 2008;4:1403–1414. doi: 10.1517/17425255.4.11.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karlgren M, Miura S, Ingelman-Sundberg M. Novel extrahepatic cytochrome P450s. Toxicol Appl Pharmacol. 2005;207(2 Suppl):57–61. doi: 10.1016/j.taap.2004.12.022. [DOI] [PubMed] [Google Scholar]
- 24.Nath A, Zientek MA, Burke BJ, Jiang Y, Atkins WM. Specificity and Promiscuity in Small-Molecule Inhibition of Cytochrome P450 Isoforms. Drug Metab Dispos. 2010;38:2195–203. doi: 10.1124/dmd.110.034645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Obach RS, Reed-Hagen AE. Measurements of Michaelis constants for cytochrome P450-mediated biotransformation reactions using a substrate depletion approach. Drug Metab Dispos. 2002;30:831–837. doi: 10.1124/dmd.30.7.831. [DOI] [PubMed] [Google Scholar]
- 26.Nath A, Atkins WM. A theoretical validation of the substrate depletion approach to determining kinetic parameters. Drug Metab Dispos. 2006;34:1433–1435. doi: 10.1124/dmd.106.010777. [DOI] [PubMed] [Google Scholar]
- 27.Mannervik B, Runarsdottir A. The quest for molecular quasi-species in ligand-activity space and its application to directed enzyme evolution. FEBS Lett. 2010;584(12):2565–71. doi: 10.1016/j.febslet.2010.04.024. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.