Abstract
A high-yielding and practical synthesis of the bacterial cell division inhibitor PC190723 is described. The synthesis is completed in a longest linear sequence of five steps from commercially available starting materials and can be readily executed on a multi-gram scale.
The emergence of pathogenic bacteria that are resistant to current antibiotics requires the exploration of new methods for treating infections caused by these organisms. Recent efforts have included finding new ways to modify the structures of existing antibiotics as well as discovering and developing new leads and new molecular targets. One promising area of development involves inhibiting bacterial cell division.1 The central protein required for bacterial cytokinesis is FtsZ, a tubulin-like GTPase.2 Although FtsZ and tubulin share little sequence homology, X-ray crystallography revealed a high level of structural similarity.3 Tubulin’s function is effectively inhibited by a wide range of natural and synthetic inhibitors, some of which have become effective treatments for cancer (e.g. taxol). Binding sites for three potent tubulin inhibitors (colchicine, taxol, and Vinca alkaloids) have been elucidated by crystallography.4 In contrast, few selective inhibitors of FtsZ are known, and most exhibit modest potency as judged by inhibition of GTP hydrolysis.5 Moreover, there has been no direct elucidation (e.g. X-ray crystallography, NMR) of the binding sites for any FtsZ inhibitors.
We have initiated a program to synthesize and study naturally-occurring6 and synthetic inhibitors of FtsZ with a long term goal of elucidating their mechanism of inhibition. Researchers at Prolysis, Inc. recently described the activity of PC190723, a heterocyclic inhibitor of FtsZ that exhibits a high level of bacterial lethality against Gram-positive organisms and promising antimicrobial activity against Staphylococcus aureus infection in a mouse model.7 This compound represents an impressive optimization of the known cell-division inhibitor 3-methoxybenzamide (3MBA).8 3MBA shows activity in vivo at 10 mM (10,000 µM) and analysis of resistant mutants at the time of its discovery indicated that FtsZ was the target. Optimization of this modest lead revealed PC190723, which inhibits the GTPase activity of FtsZ with an IC50 of 0.154 µM. In the absence of direct evidence, analysis of the structure of FtsZ from resistant mutants suggests that this compound interacts with FtsZ at a site outside of the GTP binding pocket.7a In addition, a recent report documents stabilization of FtsZ polymers by 1.9 Herein we report a practical synthesis10 of this compound that will enable more detailed studies of this compound’s function and a broader understanding of the structural biology of FtsZ.
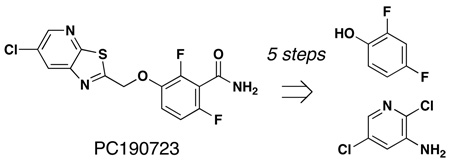
Retrosynthetic analysis reveals that PC190723 could be accessed by alkylation of phenol 3–5 with chloromethyl-substituted heterocycle 2 (Scheme 1). Phenols 3–5 are readily available from 2,4-difluorophenol by lithiation and acylation. The required chloroacetamide-substituted pyridine was prepared in two steps from commercially available 2,5-dichloro-3-aminopyridine 8.
Scheme 1.
Retrosynthetic analysis of PC190723 (1).
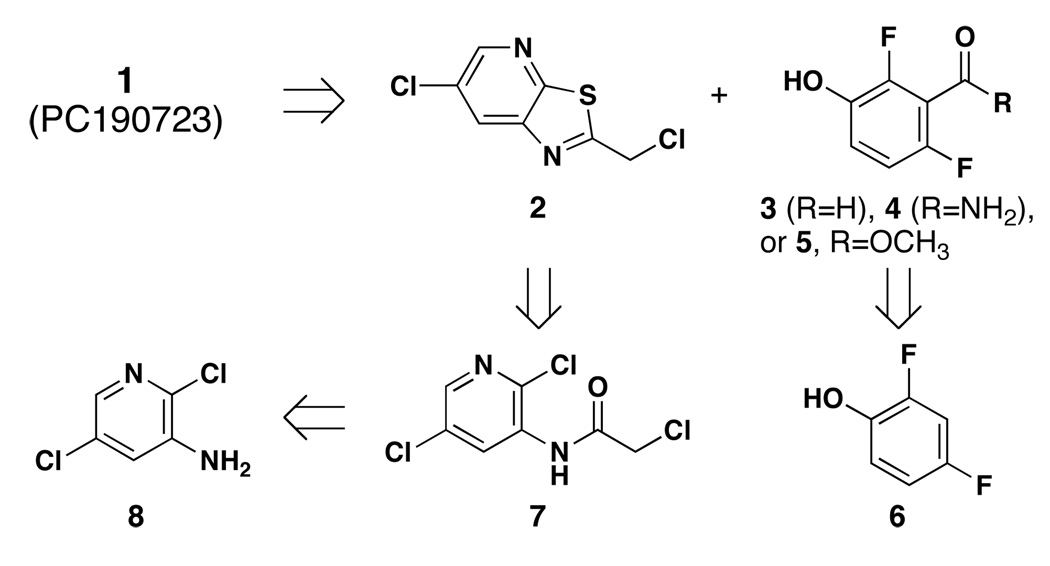
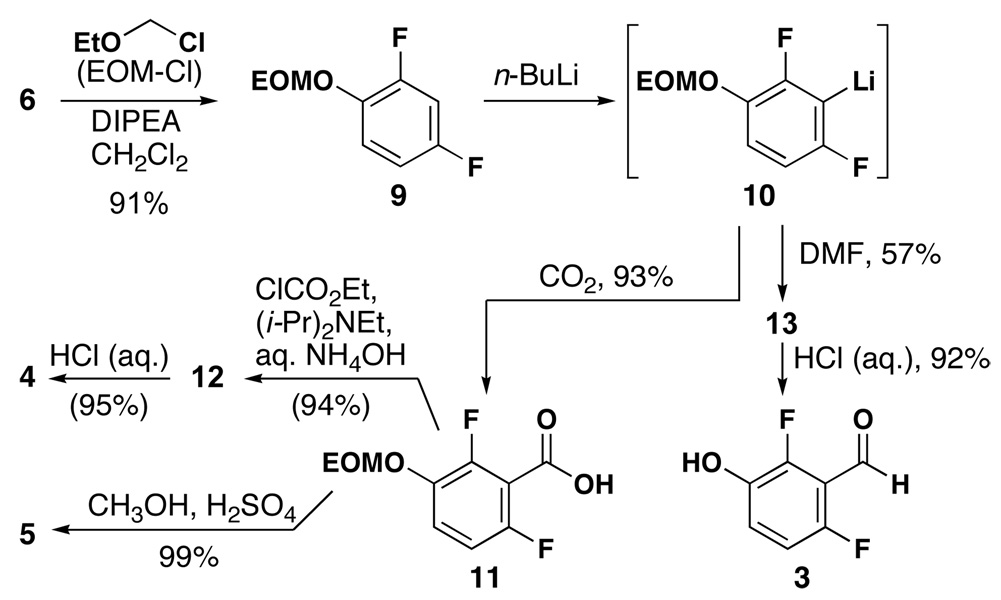
The phenol fragment of 1 could be introduced as either aldehyde 3, amide 4, or ester 5 (Scheme 2). Aldehyde 3 was prepared from difluorophenol in two steps.11 Protection with ethoxymethyl chloromethyl ether (EOM-Cl) followed by regioselective lithiation, acylation with DMF and EOM group removal provided aldehyde 3 in moderate yield. Quenching arylithium reagent12 10 with CO2 yielded acid 11, which was converted to the requisite amide 4 in two steps. Treatment of 11 with methanolic H2SO4 effected both esterification and deprotection to provide 5 in high yield.13
Scheme 2.
Synthesis of 2,6-difluoro-3-salicaldehyde 3, amide 4 and methyl ester 5.
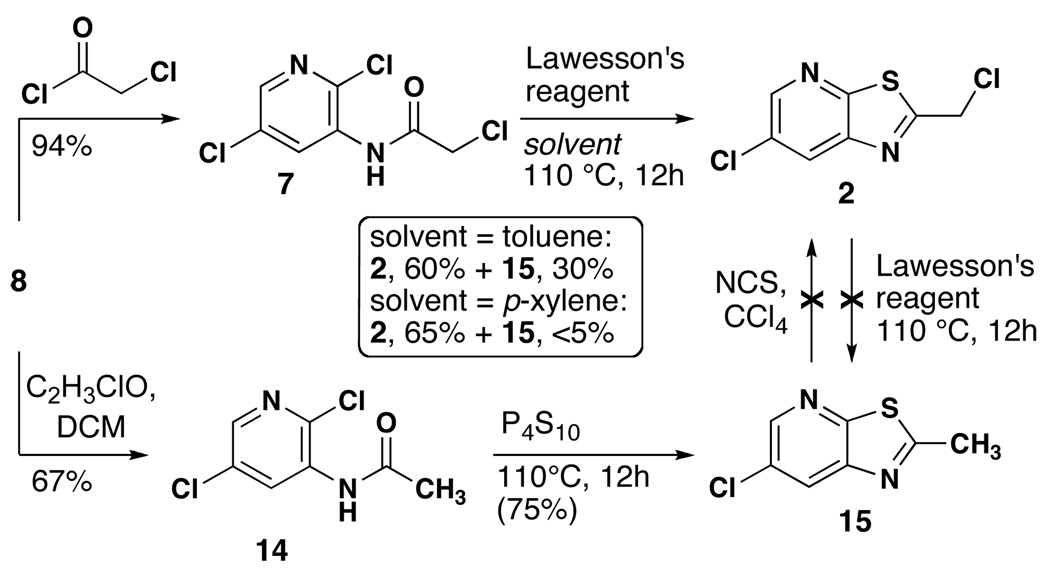
We initially explored several different methods for displacing the 2-chloro group of 8 with NaSH and a variety of nucleophilic sulfur sources. Ultimately this route was abandoned in favor of N-acylation followed by sulfurization of the amide with concomitant ring closure (Scheme 3). Acylation of 2,5-dichlo-3-aminopyridine proceeded smoothly under standard conditions. Sulfurization with Lawesson’s reagent14 in toluene provided the desired thiazole 2 as well as the dechlorinated product 15 as determined by NMR spectroscopy and mass spectrometry. Further proof of the identity of 15 was provided by synthesis of a reference sample. Reaction of 2 with Lawesson’s reagent did not produce 15 and the starting material was recovered unchanged, demonstrating that 2 is stable toward dechlorination. Attempts to convert 15 to 2 by radical chlorination were unsuccessful. Finally, switching from toluene to p-xylene for the reaction with Lawesson’s reagent increased the yield of 2 and only trace amounts of 15 were observed.
Scheme 3.
Synthesis of chloromethyl thiazoline 2.
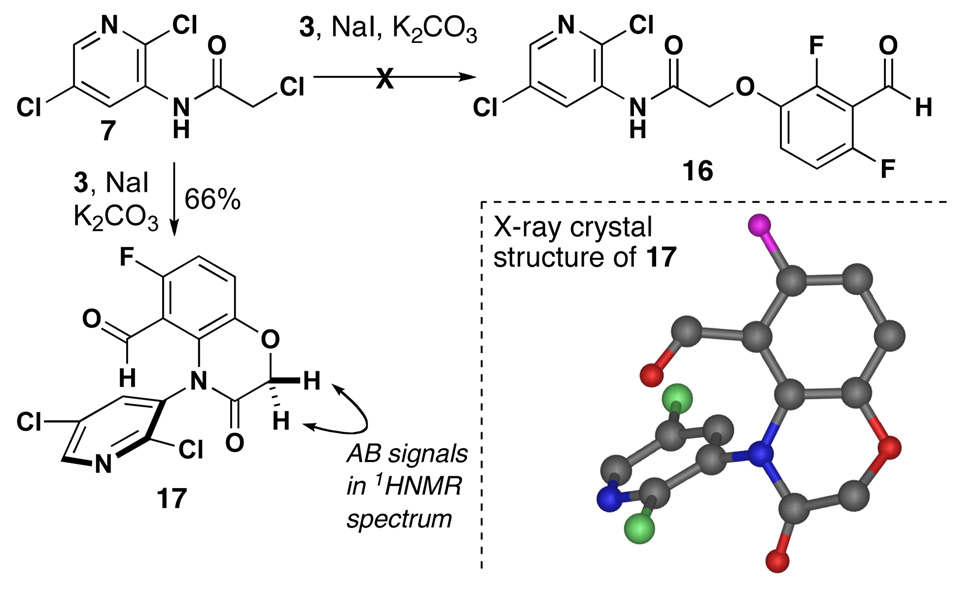
We attempted to solve the problem of dehalogenation during sulfurization by changing the order of events. In an attempt to alkylate 7 directly with aldehyde 3, an interesting cyclization occurred to produce lactam 17. Under the reaction conditions for alkylation, subsequent ring closure by SNAr displacement of the ortho fluorine could not be avoided.15 The structure of 17 was suspected based on the signals for diastereotopic methylene protons and confirmed by X-ray crystallographic analysis. Although alkylation of 7 with amide 4 did lead to the desired O-alkylation product (not shown), the subsequent sulfurization-cyclization resulted in a complex mixture of products.
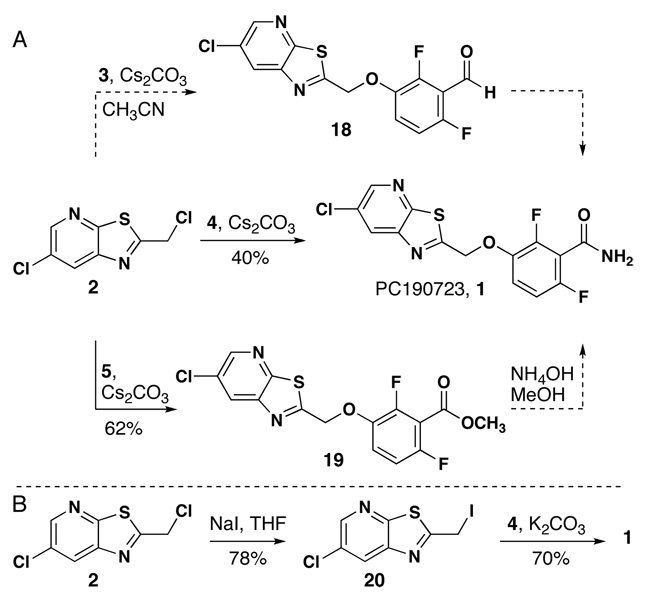
The synthesis of the PC190723 from chloromethyl thiazole 2 was completed in high yield after exploring three possible routes. When alkylation of aldehyde 3 with chloromethyl heterocycle 2 was attempted (Cs2CO3, THF or CH3CN), multiple products were observed. Reaction of amide 4 with 2 produced 1 in modest yield under mild conditions. Finally, we demonstrated that ester 5 underwent alkylation in higher yield (62%, unoptimized) relative to amide 4. Unfortunately, the resultant ester could not be converted to 1 by aminolysis under a variety of conditions.16 An alternative route is the conversion of chloride 2 to iodide 20 (78% yield),17 which alkylates 4 in good yield (70%). Although the reaction via 20 is one step longer, the overall yield is slightly higher (55%) when compared to the one-step process. Characterization data for 1/PC190723 (1HNMR, 13CNMR, 19FNMR, HRMS, IR) was consistent with published structure of PC190723.
In summary, we have demonstrated a practical synthesis of PC190723 in a longest linear sequence of five steps from 2,4-difluorophenol using a single protecting group. Although the synthesis requires metallation of an intermediate with n-BuLi, it is easily scaleable for the production of multi-gram quantities. A principle contrast between the previously reported route and ours is the use of low-cost starting materials, specifically avoiding 2,6-difluoro-3-methoxybenzamide and benzyloxyacetyl chloride. Access to this potent inhibitor of bacterial cell division will enable detailed studies of this essential process18 and pave the way for the development of new inhibitors that might serve as medicinal leads.
Experimental Section
1-Ethoxymethoxy-2,4-difluorobenzene (9)
2,4-difluorophenol (0.999 g, 7.68 mmol) was dissolved in 10 mL of dry CH2Cl2. (i-Pr)2NEt (1.58 mL, 9.22 mmol) was added and the mixture was cooled to 0 °C. This was followed by the addition of chloromethyl ethyl ether (9.86 mL, 9.26 mmol). After 2 h of stirring at room temperature, 3M NaOH (20 mL) was added and extracted 2x with CH2Cl2. The organic layers were combined and 1M HCl was added (20 mL) and extracted 2x with CH2Cl2. The organic layers were combined and washed with brine and dried over K2CO3. The solvent was evaporated in vacuo yielding a colorless oil (1.32 g, 91%): 1H NMR (300 MHz, CDCl3) δ 7.16 (td, J = 9.1, 5,4 Hz, 1H), 6.90-6.71 (m, 2H), 5.19 (s, 2H), 3.76 (q, J = 7.1 Hz, 2H), 1.22 (t, J = 7.1 Hz, 3H); 13C NMR (75 MHz, CDCl3) δ 157.4 (dd, J = 242.7, 10.5 Hz), 153.2 (dd, J = 248.8, 12.1 Hz), 141.7 (dd, J = 10.9, 3.6 Hz), 119.1 (dd, J = 9.4, 2.6 Hz), 110.6 (dd, J = 22.5, 3.9 Hz), 104.8 (dd, J = 26.8, 22.6 Hz), 95.0, 64.8, 14.9; Rf 0.851 in 30% EtOAc/hexanes; HRMS (ESI) m / z calcd for C9H10F2O2 (M - H)− 187.0574, found 187.0579.
3-Ethoxymethoxy-2,6-difluorobenzoic acid (11)
Compound 9 (3.93 g, 20.9 mmol) was dissolved in dry THF (80 mL) and n-BuLi (10 mL, 25.0 mmol, 2.5M in hexanes) was added dropwise at −78 °C. After stirring at −78 °C for 2 h, the mixture was poured onto freshly crushed CO2 followed by the addition of another layer of freshly crushed CO2. The mixture was allowed to warm to room temperature overnight. Water was added to the mixture (50 mL) and extracted with EtOAc. The aqueous layer was acidified to pH 1 and extracted immediately with CH2Cl2. The organic layers were combined and dried over Na2SO4. The solvent was evaporated in vacuo, yielding a white solid (4.51 g, 93%): mp 154-154 °C; 1H NMR (400 MHz, CDCl3) δ 7.37 (m, 1H), 6.91 (dd, J = 10.1, 8.3 Hz, 1H), 5.25 (s, 2H), 3.80 (q, J = 7.1 Hz, 2H), 1.25 (t, J = 7.1 Hz, 3H); 13C NMR (100 MHz, CDCl3) δ 166.4, 155.5 (dd, J = 253.5, 4.4 Hz), 151.9 (dd, J = 260.0, 5.9 Hz), 142.2 (dd, J = 11.0, 3.7 Hz), 122.2 (dd, J = 9.9, 3.2 Hz), 111.6 (dd, J = 23.1, 4.4 Hz), 110.5 (dd, J = 17.7, 14.5 Hz), 95.1, 65.0, 15.1; IR (neat) 3536, 3476, 1671 cm−1; Rf 0.340 in 50% EtOAc/hexanes ; HRMS (ESI) m / z calcd for C10H10F2O4 (M - H)− 231.0469, found 231.0479.
3-Ethoxymethoxy-2,6-Difluoro-benzamide (12)
Compound 11 (1.20 g, 5.18 mmol) was dissolved in 50 mL of dry CH2Cl2 followed by the addition of (i-Pr)2NEt (0.976 mL, 5.70 mmol). Ethyl chloroformate (0.600 mL, 6.30 mmol) was added at 0 °C. After 1.5 h at 0 °C, 6 mL of 12M NH4OH was added and stirred for an additional 30 min. The reaction was monitored by TLC. After 3 h, the solvent was evaporated in vacuo and then 1M HCl (30 mL) was added to the mixture and extracted 2x with ethyl acetate. The organic layers were combined, washed with brine and concentrated. The crude mixture was purified by flash chromatography (40 to 70% EtOAc/hexanes). The solvent was evaporated in vacuo yielding a white solid (1.13 g, 94%): mp 87.4–87.5 °C; 1H NMR (400 MHz, CDCl3) δ 7.27 (td, J = 9.4, 5.3, 1H), 6.98 – 6.80 (m, 1H), 6.33 (brs, 1H), 6.06 (brs, 1H), 5.23 (s, 2H), 3.78 (q, J = 7.1, 2H), 1.23 (t, J = 7.1, 3H); 13C NMR (75 MHz, CDCl3) δ 162.7 , 154.2 (dd, J = 247.3, 5.2), 150.5 (dd, J = 253.9, 7.2), 142.0 (dd, J = 11.1, 3.4), 120.3 (d, J = 9.3), 114.4-113.8 (m), 111.3 (dd, J = 23.5, 3.6), 94.9 , 64.9, 15.1; IR (neat) 3381, 3198, 1656 cm−1; Rf 0.409 in 60% EtOAC/hexanes; HRMS (ESI) m / z calcd for C10H11F2NO3 (M + H) + 232.0785, found 232.0765.
2,6-Difluoro-3-hydroxy-benzamide (4)
The EOM-protected amide 12 (0.875 g, 3.78 mmol) was dissolved in a 50:50 mixture of 6M HCl and MeOH (30 mL total volume). The reaction was monitored by TLC. After ~2 h, the solvent was evaporated in vacuo yielding an amorphous white solid (0.622 g, 95%): 1H NMR (400 MHz, DMSO-d) δ 9.96 (brs, 1H), 8.09 (brs, 1H), 7.79 (brs, 1H), 6.93 (m, 2H); 13C NMR (75 MHz, DMSO-d) δ 162.5, 151.3 (dd, J = 238.6, 6.3), 147.6 (dd, J = 245.2, 8.6), 142.1 (d, J = 14.9), 118.1 (dd, J = 9.0, 3.6), 117.1 (dd, J = 24.6, 20.7), 111.6 (dd, J = 22.8, 3.4); IR (neat) 3433, 3371, 1687 cm−1; Rf 0.185 in 50% EtOAc/hexanes; HRMS (ESI) m / z calcd for C7H5F2NO2 (M + H)+ 174.0366, found 174.0354. 1H NMR spectrum was in accordance with the literature10.
2-Chloro-N-(2,5-dichloro-pyridin-3-yl)-acetamide (7)
3-amino-2,5-dichloropyridine 8 (1.02 g, 6.25 mmol) was dissolved in dry CH2Cl2 (50 mL). The mixture was cooled to 0 °C and chloroacetyl chloride (0.75 mL, 9.43 mmol) was added dropwise. This was followed by the addition of (i-Pr)2NEt (1.6 mL, 9.35 mmol). The reaction was monitored by TLC. After 3 h, the solvent was evaporated in vacuo and the crude mixture was purified by flash chromatography (30 to 50% EtOAc/hexanes) yielding an off-white solid (1.44 g, 96%): mp 121–122 °C; 1H NMR (300 MHz, CDCl3) δ 8.94 (s, 1H), 8.82 (d, J = 2.4, 1H), 8.14 (d, J = 2.4, 1H), 4.26 (s, 2H); 13C NMR (75 MHz, CDCl3) δ 164.7, 143.3, 137.9, 131.6, 131.4, 128.1, 42.7; IR (neat) 3366, 2946, 1682 cm−1; Rf 0.833 in 50% EtOAc/hexanes; HRMS (ESI) m / z calcd for C7H5Cl3N2O (M + H)+ 238.9545, found 238.9512.
6-Chloro-2-chloromethyl-thiazolo[5,4-b]pyridine (2)
Compound 7 (0.561 g, 2.34 mmol) was dissolved in dry toluene (25 mL) and Lawesson’s reagent (0.946 g, 4.68 mmol) was added. The reaction mixture was refluxed for 12 h at 110 °C. The reaction was monitored by mass spectroscopy for disappearance of starting material. After ~12 h, the color of the reaction mixture turned slightly red. At this point, the reaction was stopped and cooled to room temperature. The crude mixture was passed through a plug of silica gel and washed with CH2Cl2. The solvent was evaporated in vacuo and the crude mixture purified by flash chromatography (30 to 50% EtOAc/hexanes) yielding a solid (0.307 g, 60%): mp 89–90 °C; 1H NMR (600 MHz, CDCl3) δ 8.58 (d, J = 2.1, 1H), 8.23 (d, J = 2.1, 1H), 4.92 (s, 2H); 13C NMR (150 MHz, CDCl3) δ 170.4, 156.6, 146.8, 146.5, 130.3, 130.1, 42.4; IR (neat) 3038, 1736, 1682 cm−1; Rf 0.833 in 50% EtOAc/hexanes; HRMS (ESI) m / z calcd for C7H4Cl2N2S (M + H)+ 218.9850, found 218.9839.
3-({6-Chloropyrido[3,2-d][1,3]thiazol-2-yl}methoxy)-2,6-difluoro- benzamide (1)
Compound 4 (0.250 g, 1.44 mmol) was dissolved in 20 mL of dry CH3CN and Cs2CO3 (0.94 g, 2.88 mmol) was added. Compound 2 (0.211 g, 0.963 mmol) was added to the mixture and stirred at 65 °C for ~3 h. The solution was filtered, concentrated in vacuo and purified by flash chromatography (30 to 70% EtOAc/hexanes) yielding an off-white solid (0.135 g, 40%): m.p. 212.5–214.5 (lit10. 218°C); Rf 0.333 in 50% EtOAc/hexanes; 1H NMR (600 MHz, DMSO-d) δ 8.73 (d, J = 2.2, 1H), 8.68 (d, J = 2.2, 1H), 8.18 (brs, 1H), 7.90 (brs, 2H), 7.41 (m, 2H), 7.12 (m, 2H), 5.73 (s, 2H); 13C NMR (100 MHz, DMSO-d) δ 171.5, 161.8, 156.2, 154.3–154.4 (dd, J = 235.49, 7.0), 152.1-152.0 (dd, J = 239.7, 9.0), 149.9 (d, J = 8.3), 147.4 (d, J = 8.3Hz), 142.5 (d, J = 10.9), 130.6, 129.9, 117.4 (dd, J = 41.7, 17.0), 117.0 (d, J = 8.8), 111.9 (d, J = 23.1), 69.6; 19F NMR (282 MHz, DMSO-d) δ -123.4, -134.1; IR 3393, 3198, 1671 cm−1; HRMS (ESI) m / z calcd for C14H8ClF2N3O2S (M + H)+ 356.0072, found 356.0051. 1H NMR spectrum was in accordance with the literature10.
3-({6-Chloropyrido[3,2-d][1,3]thiazol-2-yl}methoxy)-2,6-difluoro- benzamide (1)
Compound 4 (0.100 g, 0.579 mmol) was dissolved in 20 mL of dry CH3CN and K2CO3 (0.241 g, 1.73 mmol) was added. Iodide 20 (0.120 g, 0.386 mmol) was added and stirred at 65 °C for ~3 h. The solution was filtered, concentrated in vacuo and purified by flash chromatography (30 to 70% EtOAc/hexanes) yielding an off-white solid (0.097 g, 70%). 1H NMR spectrum was in accordance with the literature10.
4-(2,5-Dichloro-pyridin-3-yl)-6-fluoro-3-oxo-3,4-dihydro-2H-benzo[1,4]oxazine-5-carbaldehyde (17)
Compound 7 (0.314 g, 1.31 mmol) was dissolved in dry DMF (15 mL) and NaI (0.197 g, 1.31 mmol) was added and stirred at 45 °C for 40 min. Then compound 3 (0.249 g, 1.57 mmol) and K2CO3 (1.0 g, 7.23 mmol) were added and the mixture was stirred for 2–3 h. The solution was filtered, concentrated in vacuo and purified by flash chromatography (30 to 50% EtOAc/hexanes) yielding a white solid (0.295 g, 66%); mp 190–191 °C; Rf 0.833 in 50% EtOAc/hexanes; 1H NMR (300 MHz, CDCl3) δ 9.90 (s, 1H), 8.34 (d, J = 2.4, 1H), 7.90 (d, J = 2.4, 1H), 7.37 (dd, J = 9.0, 5.1, 1H), 7.00 (m, 1H), 4.82 (d, J = 14.3, 1H), 4.63 (d, J = 14.3, 1H); 13C NMR (75 MHz, CDCl3) δ 185.2 (d, J = 10.6), 164.3, 162.7, 159.3, 148.0, 145.5 (d, J = 14.7), 141.3, 132.9, 130.5, 129.2, 123.4 (d, J = 10.5), 115.9 (d, J = 11.3), 113.2 (d, J = 23.8), 68.8; 19F NMR (282 MHz, CDCl3) δ -125.0; IR (neat) 1724, 1694 cm−1; HRMS (ESI) m / z calcd for C14H7Cl2FN2O3 (M + H)+ 340.9896, found 340.9897.
Supplementary Material
Scheme 4.
Attempted alkylation of 7 with phenol 3.
Scheme 5.
A) Final alkylation to produce 1. B) Improved alkylation yield from conversion of chloride 2 to iodide 20.
ACKNOWLEDGMENT
This research was supported by startup funds from UC Davis, the UC Davis Committee on Research and the National Institute of Allergy and Infectious Disease (R56AI80931-01 and R01AI080931-01). Nohemy A. Sorto thanks Proctor & Gamble and Bristol-Myers Squibb for graduate research fellowships.
Footnotes
Supporting Information Available: Experimental procedures for 3, 5, 13–15, and 19. Spectra (1HNMR, 13CNMR, and 19F) for all new compounds spectra and the crystallographic information file (.cif) for 17. In addition, complete references 7a, 7b and 10 are provided. (This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.a) Kapoor S, Panda D. Expert Opin. Ther. Targets. 2009;13:1037–1051. doi: 10.1517/14728220903173257. [DOI] [PubMed] [Google Scholar]; b) Vollmer W. Chem. Biol. 2008;15:93–94. doi: 10.1016/j.chembiol.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 2.a) Romberg L, Levin PA. Ann. Rev. Microbiol. 2003;57:125–154. doi: 10.1146/annurev.micro.57.012903.074300. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Margolin W. Nat. Rev. Mol. Cell Biol. 2005;6:862–871. doi: 10.1038/nrm1745. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Dajkovic A, Lutkenhaus J. J. Mol. Microbiol. Biotechnol. 2006;11:140–151. doi: 10.1159/000094050. [DOI] [PubMed] [Google Scholar]; d) Callaway E. Nature. 2008;451:124–126. doi: 10.1038/451124a. [DOI] [PubMed] [Google Scholar]
- 3.a) Lowe J. J. Struct. Biol. 1998;124:235–243. doi: 10.1006/jsbi.1998.4041. [DOI] [PubMed] [Google Scholar]; b) Leung AK, Lucile White E, Ross LJ, Reynolds RC, DeVito JA, Borhani DW. J. Mol. Biol. 2004;342:953–970. doi: 10.1016/j.jmb.2004.07.061. [DOI] [PubMed] [Google Scholar]; c) Oliva MA, Trambaiolo D, Lowe J. J. Mol. Biol. 2007;373:1229–1242. doi: 10.1016/j.jmb.2007.08.056. [DOI] [PubMed] [Google Scholar]
- 4.a) Lowe J, Li H, Downing KH, Nogales E. J. Mol. Biol. 2001;313:1045–1057. doi: 10.1006/jmbi.2001.5077. [DOI] [PubMed] [Google Scholar]; b) Gigant B, Wang C, Ravelli RB, Roussi F, Steinmetz MO, Curmi PA, Sobel A, Knossow M. Nature. 2005;435:519–522. doi: 10.1038/nature03566. [DOI] [PubMed] [Google Scholar]; c) Gigant B, Wang C, Ravelli RB, Roussi F, Steinmetz MO, Curmi PA, Sobel A, Knossow M. Nature. 2005;435:519–522. doi: 10.1038/nature03566. [DOI] [PubMed] [Google Scholar]
- 5.a) Vollmer W. Appl. Microbiol. Biotechnol. 2006;73:37–47. doi: 10.1007/s00253-006-0586-0. [DOI] [PubMed] [Google Scholar]; b) Jaiswal R, Beuria TK, Mohan R, Mahajan SK, Panda D. Biochemistry. 2007;46:4211–4220. doi: 10.1021/bi602573e. [DOI] [PubMed] [Google Scholar]; c) Domadia PN, Bhunia A, Sivaraman J, Swarup S, Dasgupta D. Biochemistry. 2008;47:3225–3234. doi: 10.1021/bi7018546. [DOI] [PubMed] [Google Scholar]; d) Beuria TK, Singh P, Surolia A, Panda D. Biochem. J. 2009;423:61–69. doi: 10.1042/BJ20090817. [DOI] [PubMed] [Google Scholar]; e) Plaza A, Keffer JL, Bifulco G, Lloyd JR, Bewley CA. J. Am. Chem. Soc. 132:9069–9077. doi: 10.1021/ja102100h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.a) Urgaonkar S, La Pierre HS, Meir I, Lund H, RayChaudhuri D, Shaw JT. Org. Lett. 2005;7:5609–5612. doi: 10.1021/ol052269z. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Kim MB, Shaw JT. Org. Lett. 2010:3324–3327. doi: 10.1021/ol100929z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.a) Haydon DJ, et al. Science (Washington, DC, U. S.) 2008;321:1673–1675. [Google Scholar]; b) Czaplewski LG, et al. Bioorg. Med. Chem. Lett. 2009;19:524–527. doi: 10.1016/j.bmcl.2008.11.021. [DOI] [PubMed] [Google Scholar]
- 8.Ohashi Y, Chijiiwa Y, Suzuki K, Takahashi K, Nanamiya H, Sato T, Hosoya Y, Ochi K, Kawamura F. J. Bacteriol. 1999;181:1348–1351. doi: 10.1128/jb.181.4.1348-1351.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Andreu JM, Schaffner-Barbero C, Huecas S, Alonso D, Lopez-Rodriguez ML, Ruiz-Avila LB, Nunez-Ramirez R, Llorca O, Martin-Galiano AJ. J. Biol. Chem. 2010;285:14239–14246. doi: 10.1074/jbc.M109.094722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.During the preparation of this manuscript, a report from Prolysis included a synthesis of 1 and analogs prepared by a closely related route: Haydon DJ, et al. J. Med. Chem. 2010;53:3927–3936. doi: 10.1021/jm9016366.
- 11.Chen GT, King M, Gusovsky F, Creveling CR, Daly JW, Chen BH, Nie JY, Kirk KL. J. Med. Chem. 1993:3947–3955. doi: 10.1021/jm00076a024. [DOI] [PubMed] [Google Scholar]
- 12.Marzi E, Gorecka J, Schlosser M. Synthesis. 2004:1609–1618. [Google Scholar]
- 13.Yamaguchi Y, Menear K, Cohen N-C, Mah R, Cumin F, Schnell C, Wood JM, Maibaum J. Bioorg. Med. Chem. Lett. 2009:4863–4867. doi: 10.1016/j.bmcl.2009.05.128. [DOI] [PubMed] [Google Scholar]
- 14.2,4-Bis(4-methoxyphenyl)-1,3,2,4-dithiadiphosphetane 2,4-disulfide.
- 15.For a related two-step transformation of an α-bromoamide, see: Coutts IGC, Southcott MR. J. Chem. Soc. Perkin Trans. 1990;1:767–771.
- 16.Attempted aminolysis reagents included NH4OH with various co-solvents, NH3/methanol, and LiNH2 in THF generated from NH4Cl and n-BuLi.
- 17.Williams DR, Berliner MA, Stroup BW, Nag PP, Clark MP. Org. Lett. 2005;7:4099–4102. doi: 10.1021/ol051345v. [DOI] [PubMed] [Google Scholar]
- 18.Research quantities of this compound are available on request. Please contact JTS by (shaw@chem.ucdavis.edu).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.