Abstract
Long-QT syndrome (LQTS) is a congenital or drug-induced change in electrical activity of the heart that can lead to fatal arrhythmias. Mutations in 12 genes encoding ion channels and associated proteins are linked with congenital LQTS. With a computational systems biology approach, we found that gene products involved in LQTS formed a distinct functional neighborhood within the human interactome. Other diseases form similarly selective neighborhoods, and comparison of the LQTS neighborhood with other disease-centered neighborhoods suggested a molecular basis for associations between seemingly unrelated diseases that have increased risk of cardiac complications. By combining the LQTS neighborhood with published genome-wide association study data, we identified previously unknown single-nucleotide polymorphisms likely to affect the QT interval. We found that targets of U.S. Food and Drug Administration (FDA)–approved drugs that cause LQTS as an adverse event were enriched in the LQTS neighborhood. With the LQTS neighborhood as a classifier, we predicted drugs likely to have risks for QT effects and we validated these predictions with the FDA’s Adverse Events Reporting System, illustrating how network analysis can enhance the detection of adverse drug effects associated with drugs in clinical use. Thus, the identification of disease-selective neighborhoods within the human interactome can be useful for predicting new gene variants involved in disease, explaining the complexity underlying adverse drug side effects, and predicting adverse event susceptibility for new drugs.
INTRODUCTION
Phenotypic heterogeneity is the result of variations originating from genetic and environmental factors, as well as stochastic biomolecular events (1, 2). Before the sequencing of the human genome, it was evident that mutations in genes could be related to diseases (3, 4). Since the sequencing of the human genome and advances in high-throughput techniques, it is now clear that mutated gene products associated with disease phenotypes interact with other proteins to alter regulatory network behavior (5–7). Although compensatory mechanisms often allow these networks to remain robust to changes in a single component, mutations or gene variants, such as single-nucleotide polymorphisms (SNPs) or copy number variants that sufficiently alter the function of cellular components beyond a threshold, result in disease (8–10). Silent variations and mutations that do not lead to phenotypic changes can become unmasked through interactions of the organism with the environment (11). Networks that identify relationships among gene products that are responsible for phenotypic behavior can provide insight into the interaction between genes and the environment.
The construction of disease-centered networks of cellular interactions may also help explain the origins of variable responses to therapeutic or adverse effects of drugs. Drugs can be considered “environmental signals” because their targets often serve to link signaling networks to cellular machines and are responsible for the phenotypic changes (12, 13). If the adverse response to a drug produces a phenotype similar to that of an inherited disease, it is plausible that this drug acts on the same molecular pathways that are altered in the inherited disease. This line of reasoning leads to the hypothesis that identification of networks related to a clearly observable phenotype could be useful for understanding drug responses.
Long QT syndrome (LQTS) is a congenital or drug-induced change in electrical activity of the heart that can lead to fatal arrhythmias. LQTS is defined by a specific change (lengthening of the QT interval) in the electrocardiogram (ECG), and is thus a readily observable phenotype. Therefore, we analyzed the relationship between mutations in genes that lead to congenital LQTS and drugs that induce LQTS as an adverse event to test the hypothesis that network analysis could be useful for understanding drug responses.
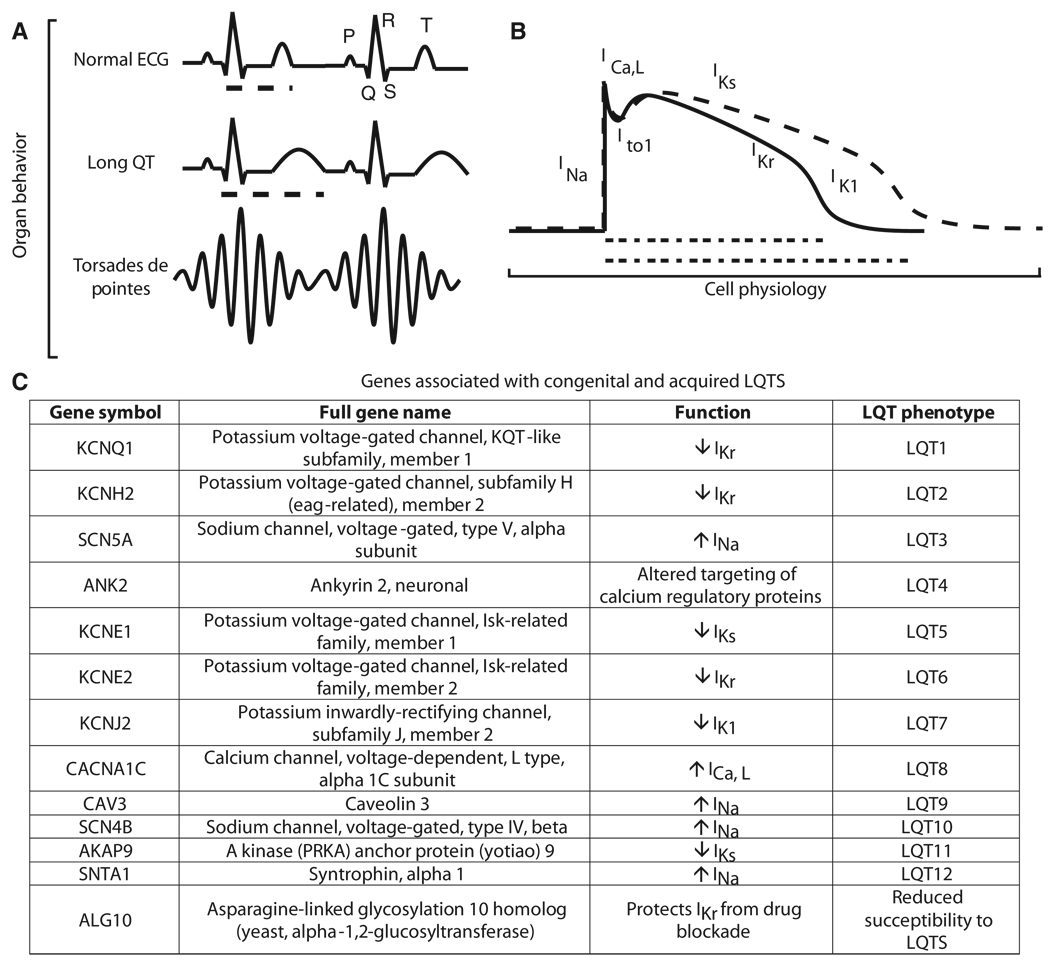
The ECG represents an integrated organismal measure of the electrical conduction system of the heart, and the different parts of an ECG pattern are labeled with individual letters (Fig. 1A). In a healthy heart, depolarization of cardiac atria generates the P wave of the ECG. This is followed by the Q, R, and S peaks representing the depolarization of the cardiac ventricles. The T wave represents the repolarization of the ventricles. The interval between the start of the Q peak and the end of the T wave is the QT interval. Changes in the QT interval are risk indicators for arrhythmias (Fig. 1A), which can be fatal. For example, torsades de pointes (TdP) is a potentially fatal arrhythmia associated with LQTS (14).
Fig. 1.
Multiscale relationships between mutated genes and QT interval. LQTS can be described across several organizational scales. (A) Organ level: LQTS symptoms result from disruptions of blood flow as a result of electrical disturbances of the heart that are observed clinically as a prolonged QT interval on an ECG. (B) Cell level: Changes in the QT interval arise from changes in the myocyte action potential leading to an increase in action potential duration. Normal and prolonged action potentials are shown as solid and dashed lines, respectively. The currents responsible for each phase are labeled. (C) Molecular level: The genes for ion channels that give rise to these currents and the proteins known to modulate these currents in LQTS are listed in the table. Known mutations in these genes allow clinical genetic testing to diagnose risk of LQTS. Dashed lines in (A) highlight the QT interval and in (B) highlight the action potential duration that gives rise to the QT interval.
Malfunction of specific ion channels in cardiac myocytes is often the cause of cardiac failure. At the cellular level, several major currents contribute to cardiac action potential (Fig. 1B) (14), and genetic mutations that alter the functional properties of the ion channels that produce these currents can alter the duration of the action potential and thus change the QT interval (Fig. 1B). Mutations in eight genes that encode ion channels and four genes encoding proteins known to interact with these channels have been associated with familial forms of LQTS (Fig. 1C). However, identical mutations can affect different members of a family to varying extents ranging from individuals with no observable disease to those that experience sudden cardiac death. This variability is indicative of complex interactions between mutated genes and other cellular components (15). This complex network could account for some of the variability in penetrance of LQTS mutations, as well as differences in susceptibility to acquired LQTS, which can be induced by certain drugs or metabolic disturbances (16).
Drugs used to treat noncardiovascular diseases can have cardiovascular risks as side effects (17). Many seemingly unrelated drugs, often used for noncardiac indications, cause QT interval prolongation and TdP as an adverse event (18). This rare, but dangerous, side effect occurs frequently enough that, in clinical practice, drugs with the risk of inducing acquired LQTS are avoided in patients with the congenital syndrome (18) and has also resulted in removal of drugs from the market. For example, the gastrointestinal drug cisapride was withdrawn from human usage because it could cause QT prolongation, leading to fatal arrhythmias (19), and the allergy medication terfenadine was withdrawn because of its propensity to cause LQTS (20). Although many drugs that cause LQTS interact with the product of the KCNH2 gene, the HERG potassium channel that regulates myocyte action potential, it would be useful to know whether other targets of drugs that cause QT prolongation are related to genes involved in LQTS. Because the degree of blockade of the HERG ion channel is not directly related to the risk of TdP and is not the only risk factor for TdP (21), it is likely that other genes associated with LQTS are targets of drugs that cause acquired LQTS.
LQTS can be considered a channelopathy related to altered cell signaling. An underlying defect in ion channel function decreases cardiac repolarization reserve, and then additional signals, such as QT-prolonging medications or adrenergic activation due to stress, can precipitate dangerous arrhythmias. The importance of cell signaling pathways is evident because the main treatment for patients with congenital LQTS is β-adrenergic blocking drugs (14).
We integrated protein-protein interactions with disease-associated genes to identify a signaling network that regulates the ion channels involved in LQTS. We show that such a network, based on known disease genes and protein-protein interactions, can be used in combination with clinical data sets, such as the U.S. Food and Drug Administration’s (FDA’s) Adverse Event Reporting System (AERS), to understand previously undetected relationships between drugs and adverse events.
RESULTS
Mean first-passage time scoring allows identification of LQTS neighborhood in protein-protein interaction space
We used network analysis (6, 22) to address two questions arising from the list of LQTS disease genes (Fig. 1C): What are the functional relationships among these gene products, and what is the relationship between the mutated gene products and other cellular components? Two approaches have been used to create protein-protein interaction networks that are associated with a disease. One approach requires manual identification of the components and interactions within the specific cell types that are responsible for the observed phenotypic behavior and then construction of a relatively small network of interactions among these components (23–25). Another approach starts with a large network of all known interactions in an entire organism, which is then limited by using the genes associated with the disease as “seed nodes” to identify regions within the global network that are preferentially situated in the interaction space near the seed nodes (22, 26, 27). We used the latter approach because it is not constrained by the existing limited knowledge of the disease or of the interactions within the specific cell type. Thus, we hypothesized that analysis of the entire human interactome would reveal not only the genomic underpinnings of LQTS, but also how drugs may cause this syndrome.
To identify a LQTS disease subnetwork, we started with the known LQTS gene products as seed nodes (Fig. 1C), which included the 12 genes with known mutations in congenital LQTS and one additional gene, ALG10, that reduces susceptibility to LQTS (14, 28). (Note that when referring to the genes as nodes in the network, the gene names are not italicized.) We constructed a global human protein-protein interaction network by integrating interactions from nine publicly available sources (fig. S1 and Materials and Methods). We then identified the LQTS gene product seed nodes in the global interaction network and used a random walk–based algorithm to identify other nodes proximal to these seed nodes. Random walk clustering algorithms identify functional subnetworks without overemphasizing connections through highly connected nodes and have been useful for predicting candidate disease genes (26, 29). For our random walk–based distance algorithm, we generated a module distance score calculated from “mean first-passage times” (MFPTs), which is the average number of steps a random walker takes to walk from a specified node to another specified node (30), to rank the nodes within the complete random walk network to identify those most likely to be relevant to LQTS. A score greater than 0 for a gene product implies a shorter MFPT from seed nodes than from nodes picked at random, indicating the gene falls within a network neighborhood of interest (see Materials and Methods for details of the calculations). From the MFPT for each node to each LQTS seed node, we calculated the module distance score for all 11,090 nodes in the global human protein-protein interaction network. The 1629 nodes that had a positive score, and all 9675 interactions between these nodes, were considered the “LQTS neighborhood” (Fig. 2 and table S1).
Fig. 2.
Identification of the long QT disease gene neighborhood. Gene products (11,090) in the global human interactome were ranked on the basis of their relative MFPT distance score with the 13 LQTS disease susceptibility genes as seed nodes. Selecting various score cutoffs allows neighborhoods of various sizes to be defined as shown in the nested circles. Leave-one-out (leave-seed-out) cross-validation results, shown in red on the left, demonstrates the rank of seed nodes if they were not included as seed nodes. Several other gene products associated with other arrhythmia syndromes or the pathogenesis of TdP, shown in blue on the right, fall within the LQTS neighborhood. The protein-protein interaction network formed with the interactions between the 13 seed nodes and top 42 ranked nodes is shown at the top, demonstrating the assignment of relative importance to nodes based on network topology.
To determine the validity of our MFPT ranking system in identifying the LQTS neighborhood, we performed “leave-one-out” cross-validation analysis (31). We left out one of the known LQTS genes from the seed list and identified a neighborhood on the basis of MFPT ranking. We then determined the rank of the excluded LQTS node within this neighborhood. The rank of the excluded seed nodes were high, always achieving a positive score (Fig. 2, left). In eight cases where the excluded seed node interacted directly with another seed node, the node that was left out was ranked within the top 1% of the complete integrated mammalian protein-protein interaction network. Of these, four ranked (KCNH2, KCNQ1, KCNE1, and KCNE2) in the top 10 nodes and another three (ALG10, AKAP9, and SNTA1) ranked in the top 50. This analysis demonstrates the utility of the MFPT-based neighborhood to accurately predict LQTS disease genes. SCN4B, which was three steps away (by shortest path) from any other seed node, had a positive score and would be included in the LQTS neighborhood when it is left out. In contrast, only 4% of the other nodes that were three steps away from the LQTS seed nodes achieved positive scores. Including only nodes with positive module distance scores, the LQTS neighborhood contained 1629 nodes, representing 14.7% of the entire protein-protein interaction network.
We compared our approach of constructing a LQTS neighborhood by MFPT ranking to networks constructed with the nearest-neighbor expansion method (figs. S2 and S3) (27).We also compared the network properties of networks created by MFPT ranking to the global interaction network. The 1629-node LQTS disease gene–centered neighborhood network was interconnected with an average of 11.9 neighbors per node, which was less than the average connectivity per node in the global network (14.3 neighbors per node). (In graph theory, the number of direct connections between a node and other nodes in the network is the node “degree,” so the average node degree in the LQTS subnetwork was 11.9, whereas in the global network it was 14.3.) In networks with different numbers of nodes (“cutoffs”), comparison of the average node degree of networks on the basis of MFPT ranking to the average node degree of networks formed by applying nearest-neighbor expansion showed that the networks formed by MFPT ranking had a lower average node degree than did those based on nearest-neighbor expansion (fig. S2). Additionally, shortest path length analysis (how many nodes it takes to walk from one node to another) showed that networks based on MFPT ranking included nodes that tended to be farther away from the seed list nodes compared to nodes in networks based on nearest-neighbor expansion (fig. S3). This demonstrates that the MFPT-based ranking was not biased toward highly connected nodes (hubs) and nodes indirectly connected to the seed nodes through these hubs. Thus, the LQTS neighborhood represented a local module within the global human interactome.
In addition to differences in network properties, the networks created by MFPT ranking compared to those created by the nearest-neighbor expansion method revealed differences in assigning disease relevance to individual genes between the two approaches. For instance, many second neighbors outranked some first neighbors, and many third neighbors outranked many second and first neighbors in the MFPT ranking network. For example, albumin, which is a hub with 173 direct neighbors in the integrated mammalian protein-protein interaction network, is a first neighbor of a seed node (SCN5A) but has a negative score and is excluded from the MFPT-based LQTS neighborhood. This is reasonable because, even though there is a direct interaction with a seed gene, it is likely not specifically related to LQTS or cardiac risk associated with LQTS. Inspection of the top 55 nodes from the LQTS neighborhood showed that there were several short paths between the seeds and that these contribute to prioritization of first neighbors. For example, nodes such as PRKACA that interacted directly with multiple seed nodes ranked higher than nodes that interact with fewer seed nodes (Fig. 2, top). Among the seed nodes themselves, KCNH2, encoding HERG, attains the highest score because it interacts directly with three other seed nodes. This observation is in agreement with the known importance of KCNH2 as a LQTS disease gene (14).
Since the start of the analysis in early 2007, two additional LQTS disease genes were identified, AKAP9 (32) and SNTA1 (33), both of which we ultimately incorporated into the seed list. Before we had incorporated AKAP9 into the seed list, an MFPT-based LQTS neighborhood independently predicted AKAP9 to be among the top 10 LQTS candidate genes. After incorporating AKAP9 into our seed list, SNTA1 was published as another gene that caused congenital LQTS when mutated. The leave-one-out cross validation showed that the MFPT-based LQTS neighborhood independently predicted SNTA1 to be among the top 50 LQTS disease gene candidates (Fig. 2). The LQTS neighborhood contained other examples of known modulators of QT interval and arrhythmogenesis (Fig. 2, right). The ranking of the genes encoding protein kinase A (PKA encoded by PRKCA) as first and protein kinase C (PKC encoded by PRKACA) as second are consistent with the importance of adrenergic signaling in LQTS (21). Several genes encoding adrenergic receptors ranked within the neighborhood: ADRA1D (rank 108), ADRA1A (267), ADRB1 (308), ADBR2 (311), ADRA1B (369), ADRA2A (588), ADRA2C (730), and ADRB3 (1139). The relative importance of β-adrenergic signaling and the major protein kinases downstream of these receptors is consistent with the use of drugs that block β-adrenergic receptors to reduce mortality in patients with congenital LQTS and with the observation that physical and emotional stresses are often triggers of TdP (14). NOS1AP (encoding nitric oxide synthase 1 adaptor protein), which modulates the QT interval (34), ranked 125. KCNA5, which encodes a potassium channel thought to be present only in the atria, is a risk modifier for cardiac arrest associated with KCNE1 (35), and we found KCNA5 within the LQTS neighborhood (Fig. 2, right). The results of the leave-one-out cross-validation and the presence of QT interval and arrhythmogenesis modulators in the LQTS MFPT neighborhood suggested that the MFPT ranking measure worked well for identifying additional LQTS-related genes.
The LQTS neighborhood is distinct from other disease networks and suggests relationships among diseases
To test whether the LQTS neighborhood was selective for LQTS, we generated three sets of random seed lists (fig. S4). The first set was made of 1000 random seed lists and each random seed list contained 13 nodes randomly picked from the global network. The second set contains 1000 seed lists of 13 nodes randomly chosen from the network where the connectivity degree of the nodes matched the connectivity degree of the LQTS seed nodes (four nodes with 2 to 3 neighbors, five nodes with 8 to 15 neighbors, and four nodes with 15 to 31 neighbors). This was done to reduce bias because the LQTS disease genes have more neighbors than average, most likely because they are extensively studied. To estimate the bias of starting with seed lists encoding proteins that are localized to the membrane and cytoplasm, the third set of random seed lists contained 2500 randomly generated seed lists of genes with the same distribution of Gene Ontology (GO) Cellular Component terms as those assigned to the LQTS seed nodes (36). The subnetworks created based on the GO-matched seed lists showed higher frequencies for including other cytoplasmic and membrane-associated proteins.
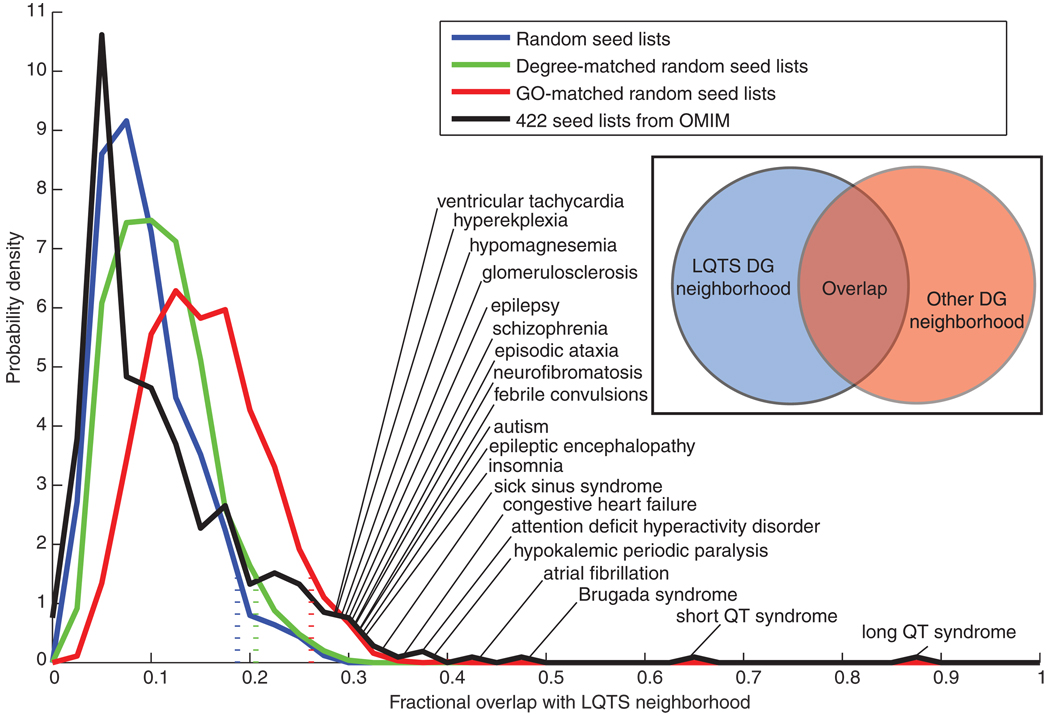
To compare the subnetworks created from the three categories of randomly created seed lists with the LQTS neighborhood, we calculated a fractional overlap between the networks using the Jaccard coefficient, which is the ratio of the intersection to the union of the genes in the subnetworks. For the subnetworks created from the completely random seed lists and the degree-matched seed lists, there was a 10% average overlap with the LQTS neighborhood with 95% of the subnetworks having less than 18.8% overlap. For the seed lists matched based on GO, there was on average a 15.6% overlap with 95% of the networks having less than 26.3% overlap with the LQTS neighborhood (Fig. 3). This analysis demonstrated that the LQTS neighborhood was a selective region within the human interactome.
Fig. 3.
Distribution of fractional overlap of other subnetworks with the LQTS neighborhood. The LQTS disease gene neighborhood was compared to disease gene neighborhoods generated from various control seed lists. These seed lists consisted of 13 randomly selected nodes, 13 randomly selected nodes for which their degree of connectivity matched the LQTS seed list, or 13 randomly selected nodes with GO annotation for the terms membrane and cytoplasm in a proportion matching those in the LQTS seed lists. Four hundred and twenty-two seed lists, with variable numbers of seeds, were created from the OMIM database. Each disease-related seed list was used to generate a neighborhood by selecting all nodes that had a positive score after MFPT ranking. The distribution of fractional overlap with the LQTS neighborhood, number of nodes shared by both neighborhoods divided by total number of unique nodes in either neighborhood, is shown for each set of random networks. Dashed lines indicate where the overlap threshold below which 95% of the random networks in each distribution falls. The 20 diseases showing the most overlap with LQTS are labeled. The “long QT syndrome” network with 87% overlap is based on the 11 seed genes that were in OMIM.
Because QT interval prolongation is a symptom of several seemingly unrelated diseases, we sought to rank other disease genes within the LQTS neighborhood. Using the Online Mendelian Inheritance in Man (OMIM) database, we identified 1398 nodes in the human interactome as encoded by disease-associated genes (37). Out of these disease-associated nodes, 290 were ranked within the LQTS neighborhood, demonstrating enrichment of known disease-associated genes within the LQTS neighborhood (the neighborhood that encompassed 14.7% of the entire human interactome contained 20.7% of the disease-associated genes). In some cases, this can be readily explained by known relationships among the diseases. For example, short-QT syndrome, a disorder characterized by an overly short QT interval, is caused by mutations in four of the same ion channel genes involved in causing LQTS (38, 39), which illustrates how gain-of-function and loss-of-function mutations in the same genes can have the opposite phenotypic effects.
To further explore the relationship between the LQTS neighborhood and other disease-centered neighborhoods, we used OMIM to generate seed lists for 422 diseases (table S2). To verify the accuracy of the MFPT ranking method, we performed leave-one-out analysis on all the 422 OMIM disease gene lists and also on an additional set of 19 disease gene lists used in a study by Chen et al. (26). We found that our MFPT ranking method classified candidate genes, as well as the method used by Chen et al. (26). We obtained a receiver operating characteristic (ROC) area under the curve (AUC) of 0.8, indicating that we matched their results and we did not require arbitrary parameter tuning (fig. S5). For the lists representing the full set of 422 OMIM diseases, which had on average fewer seed genes compared to the LQTS and the disease lists studied by Chen et al., the MFPT ranking classification performed well with an ROC AUC of 0.77 (fig. S5). For each of the 422 OMIM diseases, we calculated the fraction of the diseases genes that scored positively during the leave-one-out analysis (fig. S6A). We found that diseases can be grouped on the basis of how well their seed genes predict each other. For 160 diseases, more than 90% of the seed genes fall into the disease neighborhood when left off the seed list. However, for 117 diseases, less than 10% of the seed genes were recovered in this manner (table S2). In some cases, poorer ability to predict other members that had been left out of the seed list for specific diseases is probably due to paucity of known disease genes and interactions for these diseases (fig. S6B). An example of a disease with a large seed list (11 nodes) without interactions between them in the network is hemolytic anemia caused by physical damage to red blood cells. Our analysis suggests there may be a boundary of seed list size that allows recovery of all or none of the seed nodes during leave-one-out analysis. The fraction of seed node recovery from leave-one-out analysis for small starting seed lists tends to be all or none, because if one seed node is in proximity to the other seed node, the reciprocal relationship is often true. The overall average for recovery of seeds for diseases with only two seed nodes was 53% compared to the average recovery for the remaining diseases, which was 62%. Overall, seed lists of size greater than two outperform seed lists of size two. Beyond this cutoff, we did not find any correlation between seed list size and performance in leave-one-out analysis.
With the Jaccard coefficient, we compared the 422 disease-centered neighborhoods with the LQTS neighborhood (Fig. 3 and table S2). On average, there was only an 11.4% overlap between the LQTS neighborhood and all other disease neighborhoods, which was less overlap than when the LQTS neighborhood was compared to random networks, suggesting that many diseases arise from functionally distinct perturbations of the human interactome compared with the genes associated with LQTS. However, some diseases appeared to have a neighborhood much closer to the LQTS disease neighborhood when compared to the distribution of network overlap for randomly selected seed genes where only 5% of the GO-matched random networks had greater than 26.3% overlap with the LQTS neighborhood.
We found that the OMIM-based disease neighborhoods with the most overlap with the LQTS neighborhood were cardiac disorders. The short QT syndrome neighborhood exhibited the highest overlap (64%). Brugada syndrome, which is another cardiac arrhythmia syndrome that causes ventricular tachycardias (40), was the neighborhood with the second highest overlap (48%). The atrial fibrillation (AF) neighborhood had a 43% overlap. This close relationship between AF and LQTS is consistent with the epidemiological observation that early-onset AF may be a long QT-related dysrhythmia (41). In total, 31 of the 422 diseases overlapped more than 25% with the LQTS neighborhood (Table 1). The top 20 diseases with overlapping gene sets are labeled in Fig. 3 and the complete list is in table S2. In addition to the three diseases mentioned, several other diseases related to cardiac arrhythmia, such as congestive heart failure, sick sinus syndrome, dilated cardiomyopathy, and ventricular tachycardia had greater than 25% overlap with the LQTS neighborhood. Ventricular tachycardias include catecholaminergic polymorphic ventricular tachycardia (CPVT), which predisposes patients to an arrhythmia similar to the LQTS-associated TdP. Hypomagnesemia, which can precipitate TdP, had an overlap of 29% with the LQTS neighborhood.
Table 1.
Overlap between disease neighborhoods. Diseases with gene neighborhoods that had a >25% overlap with the LQTS gene neighborhood are shown. Gray shading indicates cardiac diseases, yellow indicates neurological disorders. Bold indicates channelopathies.
| OMIM disease name | Neighborhood size |
Number seed |
Leave- one-out recovery |
Overlap with LQTS neighborhood (%) |
|---|---|---|---|---|
| LQTS | 1629 | 13 | 1 | 100 |
| Long QT* | 1561 | 11 | 1 | 87 |
| Short QT | 1409 | 3 | 1 | 64 |
| Brugada syndrome | 1680 | 3 | 1 | 48 |
| Atrial fibrillation | 1474 | 5 | 0.8 | 43 |
| Hypokalemic periodic paralysis | 1434 | 2 | 1 | 38 |
| Attention deficit hyperactivity disorder | 1713 | 3 | 1 | 37 |
| Congestive heart failure | 1609 | 2 | 1 | 35 |
| Sick sinus | 1452 | 2 | 0 | 33 |
| Insomnia | 1652 | 2 | 0 | 32 |
| Epileptic encephalopathy | 1703 | 3 | 0.67 | 31 |
| Autism | 1579 | 8 | 0.5 | 31 |
| Febrile convulsions | 1181 | 2 | 0 | 31 |
| Neurofibromatosis | 1666 | 3 | 0.33 | 31 |
| Episodic ataxia | 1249 | 4 | 0.5 | 31 |
| Schizophrenia | 1641 | 15 | 0.33 | 30 |
| Epilepsy | 1286 | 20 | 0.5 | 30 |
| Glomerulosclerosis | 1587 | 3 | 1 | 30 |
| Hypomagnesemia | 1522 | 5 | 0.4 | 29 |
| Hyperekplexia | 1205 | 3 | 0.67 | 29 |
| Ventricular tachycardia | 1432 | 3 | 0.67 | 29 |
| Anorexia nervosa | 1367 | 2 | 1 | 28 |
| Griscelli syndrome | 1432 | 3 | 1 | 28 |
| Nephrolithiasis | 1354 | 3 | 0 | 28 |
| Acromegaly | 1191 | 2 | 1 | 28 |
| Obsessive compulsive disorder | 1404 | 3 | 0.67 | 27 |
| Fraser syndrome | 1130 | 2 | 1 | 27 |
| Becker muscular dystrophy | 1281 | 2 | 0 | 27 |
| Myotonia congenita | 1378 | 2 | 0 | 26 |
| Adenocarcinoma of lung | 1714 | 3 | 1 | 26 |
| Dilated cardiomyopathy | 1411 | 26 | 0.88 | 25 |
| Malignant hyperthermia susceptibility | 1242 | 2 | 1 | 25 |
Based on OMIM disease-associated genes for the seed list.
Several of the 31 diseases with neighborhoods with greater than 25% overlap with the LQTS neighborhood do not have obvious cardiac implications. For example, hypokalemic periodic paralysis primarily affects skeletal muscle, and three examples of diseases that are primarily neurological are autism, schizophrenia, and attention deficit hyperactivity disorder (ADHD) (Table 1). However, autism is a symptom associated with a particular LQTS, Timothy syndrome, and periodic paralysis is associated with another LQTS, Andersen-Tawil syndrome (42, 43). The overlap of these syndromes’ neighborhoods with the LQTS neighborhood may relate to the shared symptoms of specific forms of LQTS. The schizophrenia neighborhood also had a high degree of overlap with the LQTS neighborhood (30%). Acute psychosis can lead to increased QT variability in these patients (44), which has been attributed to high sympathetic activity affecting the heart. This observation can potentially be explained by the importance of adrenergic signaling in determining the QT interval and regulating neuronal activity. It would be useful to understand if some of the same factors that predispose patients to schizophrenia also predispose patients to LQTS. The overlap between the LQTS and schizophrenia neighborhoods suggests that there could be a strong relationship between these diseases. This is especially important because many of the pharmaceutical treatments for schizophrenia can also affect the QT interval and increase cardiovascular risk (45). This issue is also important for the treatment of ADHD, which also has a high network overlap with the LQTS neighborhood (37%). Recent debate has questioned whether cardiac screening should be required before ADHD treatment (46). The overlap between the LQTS and ADHD neighborhood suggest that cardiac risk in ADHD patients might have a genetic underpinning, and understanding which patients are at risk could potentially allow for safer prescribing practices in treating children with ADHD.
Several of the diseases with high degree of network overlap with LQTS can be classified as channelopathies and, like LQTS, these diseases are frequently caused by dysfunctional ion channels or altered regulation of ion channels (Table 1). This trend toward network overlap with diseases related to other ion channels suggests that the human interactome contains a core ion channel regulatory subnetwork responsible for global cellular ion homeostasis. The idea that alterations in the ion channel regulatory subnetwork that may alter susceptibility to both neuronal and cardiac disease is consistent with work that has linked certain variants of LQTS and risk of certain types of epilepsy (47) and work that has suggested a connection between the QT interval and depression (48). It is also possible that LQTS phenotypes and susceptibility to TdP may be affected by variations in the behavior of neuronal tissues. Cardiac innervation, especially by sympathetic neurons, plays an important role in arrhythmogenesis, and several of the LQTS disease genes are expressed in both heart and neuronal tissues (49–51). Our network-based approach to understanding disease suggests how various pathophysiologies are interrelated. Such analysis may provide explanations for the degree and variability of diseases occurring together in the population. We hypothesize that genes associated with a disease (disease A) that fall within proximity to another disease neighborhood (disease B) would affect the risk and susceptibility to symptoms related to disease B.
LQTS neighborhood disease gene ranking allows identification of QT-related polymorphisms
Genome-wide association studies (GWAS) have looked for SNPs that can affect the QT interval. Whereas these studies mostly recovered variations in genes previously known to be involved in LQTS, several previously unassociated loci were suggested in two studies (52, 53) (Table 2). Between these two GWAS, 11 distinct loci were reported as significantly associated with the duration of the QT interval. Four of these loci are located on chromosomes near five of the known LQTS-associated genes. Five of the remaining loci appeared in both GWAS studies, of which four are located within 150 kilobases (kb) of genes that were highly ranked in the LQTS neighborhood (Table 2). The predictive capability of our LQTS neighborhood is limited by existing knowledge about a gene product’s protein interactions. For example, GINS3, which was identified in both GWAS as associated with the duration of the QT interval, was identified as a regulator of repolarization reserve in a drug-sensitized zebrafish screen (54). GINS3 could not be predicted by our network, because there were no known protein-protein interactions involving GINS3 in any of the protein interaction databases used to construct our human interactome.
Table 2.
Loci identified in QT-interval GWAS or by LQTS gene neighborhood ranking. Genes within the LQTS gene neighborhood are marked in bold. Other genes that are within 150 kb, but were not in the LQTS gene neighborhood, are not bold. A negative value in the LQTS MFPT score means the gene fell outside the LQTS neighborhood. NA means the gene did not have any protein interactions and thus could not be scored. Gray rows were not found to be significant in either of the two GWAS (52, 53), but fell in intronic or coding regions of one of the 150 highest-ranked LQTS neighborhood genes and had an association strength of P < 5 × 10−6 in the QTSCD GWAS.
| SNPs (GWAS study or LQTS neighborhood ranking)* |
Genes within 150 kb of SNP (LQTS MFPT score, distance to SNP in base pairs) |
|
|---|---|---|
| Chromosome 1 | ||
| RNF207(NA, 0) | ||
| ICMT(−5.9 × 10−6, 1883) | ||
| C1orf211(NA, 18,501) | ||
| RPL22(−0.0026, 19,690) | ||
| rs846111(Both) | HES3(NA, 24,892) | |
| GPR153(NA, 28,044) | ||
| CHD5(NA, 39,186) | ||
| ACOT7(−0.00015, 44,962) | ||
| KCNAB2(0.0014, 118,846) | ||
| rs16857031(QTGEN) | NOS1AP(0.0043, 0) | |
| rs12143842(Both) | OLFML2B(NA, 40,245) | |
| rs12029454(QTGEN) | ATF6(−0.00039, 105,037) | |
| ATP1B1(−0.00020, 0) | ||
| rs10919071(QTSCD) | NME7(−0.00024, 2286) | |
| Chromosome 2 | ||
| rs13394655 (LQTS) | SLC8A1 (0.0059, 0) | |
| Chromosome 3 | ||
| SCN5A(0.082, 0) | ||
| rs11129795(QTSCD) | ENDOGL1(NA, 22,721) | |
| rs12053903(QTGEN) | ACVR2B(−0.0012, 54,529) | |
| XYLB(NA, 132,695) | ||
| SCN10A(0.00051, 145,444) | ||
| Chromosome 6 | ||
| rs11970286(QTSCD) | C6orf204(NA, 0) | |
| rs11756438(QTGEN) | SLC35F1(NA, 41,534) | |
| PLN(0.00034, 112,044) | ||
| Chromosome 7 | ||
| KCNH2(0.20, 4186) | ||
| NOS3(0.0061, 50,284) | ||
| ABP1(−0.00023, 63,782) | ||
| ATG9B(NA, 71,441) | ||
| ABCB8(−7.6 × 10−5, 87,647) | ||
| rs4725982(QTGEN) | ACCN3(0.0012, 107,742) | |
| rs2968864(QTGEN) | CDK5(0.0047, 113,036) | |
| rs2968863(QTSCD) | SLC4A2(0.00052, 118,794) | |
| TMEM176A(NA, 119,953) | ||
| TMEM176B(NA, 123,713) | ||
| FASTK(−0.00027, 135,845) | ||
| TMUB1(NA, 140,309) | ||
| AGAP3(NA, 145,091) | ||
| Chromosome 11 | ||
| KCNQ1(0.17, 0) | ||
| rs12576239(QTGEN) | TRPM5(NA, 40,527) | |
| rs2074238(QTGEN) | TSSC4(0.00027, 59,696) | |
| rs12296050(QTSCD) | CD81(−0.00024, 66,153) | |
| TSPAN32(0.00029, 145,372) | ||
| Chromosome 16 | ||
| LITAF(−0.00022, 10,430) | ||
| rs8049607(Both) | SNN(NA, 70,548) | |
| TXNDC11(−0.00043, 81,190) | ||
| LOC400499(NA, 106,141) | ||
| CNOT1(−0.00049, 0) | ||
| SETD6(NA, 13,685) | ||
| SNORA46(NA, 15,165) | ||
| rs37062(QTGEN) | NDRG4(NA, 19,714) | |
| rs7188697(QTSCD) | SNORA50(NA, 26,462) | |
| SLC38A7(NA, 78,120) | ||
| GOT2(0.00048, 118,857) | ||
| GINS3(NA, 127,189) | ||
| Chromosome 17 | ||
| LIG3(−0.00030, 0) | ||
| RAD51L3(−0.00062, 11,749) | ||
| RFFL(NA, 11,749) | ||
| rs2074518(QTGEN) | ZNF830(NA, 34,176) | |
| CCT6B(−0.00011, 35,875) | ||
| FNDC8(NA, 124,249) | ||
| NLE1(−0.00013, 133,986) | ||
| rs9910577 (LQTS) | PRKCA (0.053, 0) | |
| Chromosome 21 | ||
| KCNE1(0.11, 0) | ||
| LOC388820(NA, 25,415) | ||
| rs1805128(QTGEN) | FAM165B(−0.00014, 46,605) | |
| RCAN1(−9.3 × 10−5, 67,104) | ||
| KCNE2(0.11, 78,239) |
“Both” refers to the identification of the SNP in each GWAS.
We asked if the LQTS neighborhood ranking method could identify additional SNPs from the QT sudden cardiac death (QTSCD) GWAS data that may be relevant to LQTS. We found that 21,651 of the 2,458,933 SNPs scored in the QTSCD GWAS were located in coding regions or introns of either the 13 LQTS seed genes or the top 150 ranked LQTS neighborhood genes. This subset represents less than 1% of all the SNPs in the study. We looked for SNPs with association P values less than 5 × 10−6, which is 100 times greater than the cutoff for genome-wide significance. This, of course, identified the same SNPs that were identified as having genome-wide significance and identified two additional signals, rs9910577 and rs13394655 (Table 2). The first, rs9910577, is located in introns of PRKCA, the gene encoding PKCα, and the second, rs13394655, is located in the introns of SLC8A1, the gene encoding the cardiac sodium-calcium exchanger, which are respectively the 1st and 94th ranked genes in the LQTS neighborhood (fig. S7). Thus, polymorphisms in these genes may affect QT interval duration in humans.
LQTS neighborhood genes are selectively enriched for targets of drugs associated with acquired long-QT syndrome
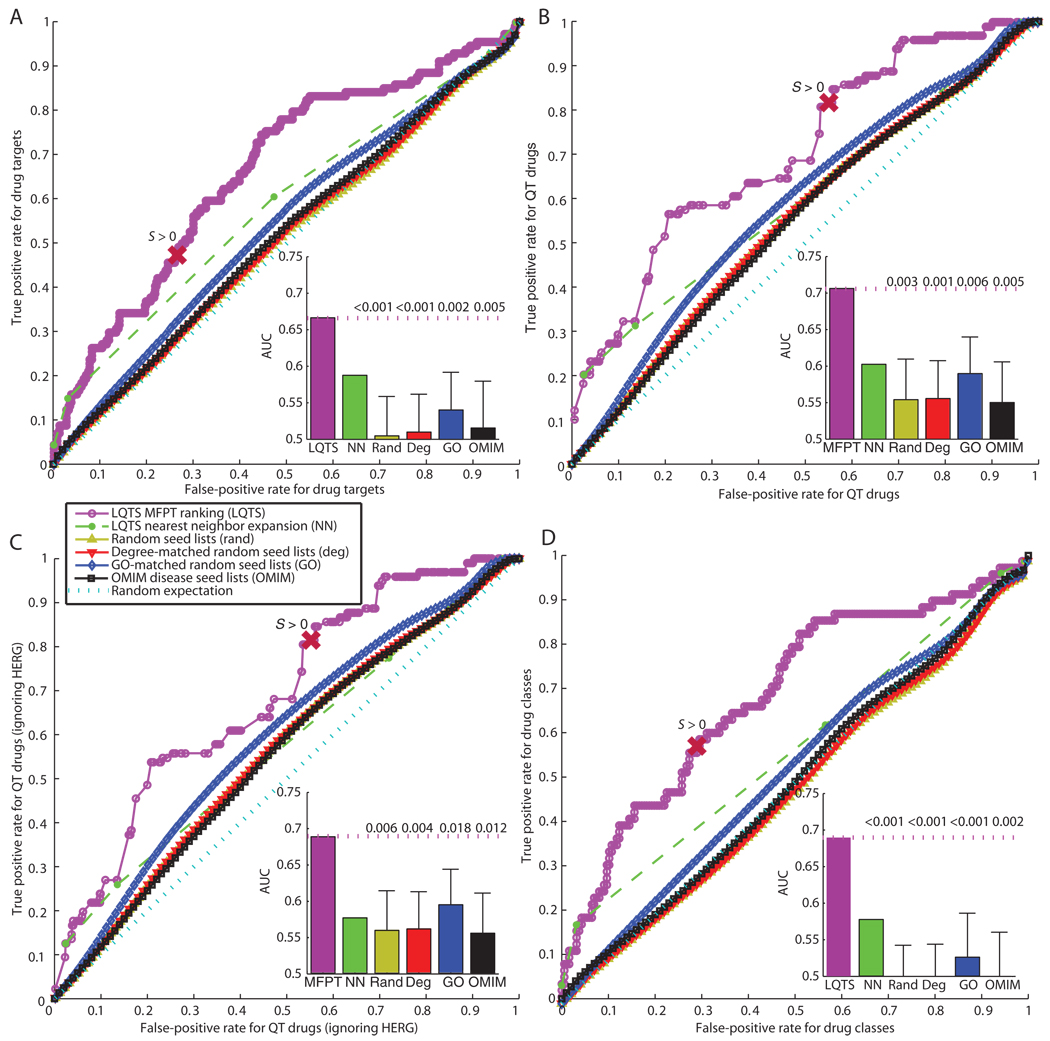
We wanted to determine if the LQTS neighborhood could be used to classify drugs associated with acquired LQTS or TdP. We identified the proteins (gene products) that were both nodes in the human interactome and drug targets in DrugBank (fig. S8 and table S3). Most drugs were reported to have only one or two targets in DrugBank and most targets were only targeted by one or two drugs (fig. S8). Then, we used ROC curve analysis to investigate if genes encoding the targets of drugs that were associated with the side effects of acquired LQTS and TdP ranked higher within the LQTS neighborhood than did genes encoding proteins that were targeted by other drugs. We examined drug targets, individual drugs, and classes of drugs. In this ROC analysis, the AUC provides a single value that assesses the quality of a binary classifier by representing the probability that a given QT-prolonging drug target, QT-prolonging drug, or QT-prolonging drug class will rank higher than drug targets, drugs, or drug classes not associated with the QT prolongation, respectively. A random classifier would have an AUC of 0.5 and a perfect classifier would have an AUC of 1. The LQTS neighborhood used as a classifier gave an AUC of 0.67 for classification of QT-prolonging drug targets (Fig. 4A). The statistical significance of this 0.67 AUC was determined by counting the fraction of control neighborhoods, generated with the previously mentioned random seed lists (P = 0.002 based on the GO-matched networks, which gave the next highest AUC values).
Fig. 4.
LQTS neighborhood rankings classify QT-prolonging drug targets and drugs. ROC curves for MFPT-ranked nodes using the LQTS seed list, nearest-neighbor expansion from the LQTS seed list (NN), and the mean ROC curves for the various random seed list distributions are shown. The point on the ROC curves that represents the cutoff for the LQTS neighborhood is labeled with S > 0. Insets represent AUCs with mean and SD shown for the random network distributions. The numbers above the dotted line in the insets is the fraction of each distribution that had an AUC greater than or equal to the AUC of the LQTS neighborhood. In all analyses, the LQTS MFPT ranking significantly outperformed all other distributions (P values are indicated in insets). (A) Classification of drug targets. For every possible cutoff, a true positive rate (the fraction of targets of QT-prolonging drugs that are in the indicated neighborhood) is plotted against the false-positive rate (the fraction of all other drug targets that are in the indicated neighborhood). The AUC represents the probability that a given target of a QT-prolonging drug will rank higher than a given drug target that is not targeted by a QT-prolonging drug. (B) Classification of drugs. For every possible cutoff, a true positive rate (the fraction of QT-prolonging drugs that have a target in the neighborhood) is plotted against the false-positive rate (the fraction of the remaining drugs that have a target in the neighborhood). The AUC represents the probability that QT-prolonging drugs target higher-ranked targets than other drugs not associated with QT events. (C) Classification of drugs excluding HERG, the target encoded by KCNH2. The AUC represents the probability that QT-prolonging drugs target higher-ranked targets (other than HERG) than other drugs not associated with QT events. (D) Classification of drug classes. ROC analysis was performed with drug classes defined by grouping all drugs that share a common highest-ranked target, on the basis of the targets’ module distance scores, and including only these targets in the rankings. The AUC represents the probability that a given drug class containing QT-prolonging drugs target higher-ranked targets than drug classes not associated with QT-prolonging drugs.
Because drugs can have multiple targets, only some of the targets of the QT-prolonging drugs are likely to be related to the pathogenesis of arrhythmias; other targets could relate to the other effects of the drug. Therefore, enrichment of drug targets in the LQTS neighborhood could underestimate the actual targets of QT-prolonging drugs that mediate the QT-prolonging effects. Therefore, we ranked drugs on the basis of their highest-scoring targets in the LQTS neighborhood and assessed if the QT-prolonging drugs ranked higher than other drugs. We found that the AUC of this ROC curve was 0.71 (P = 0.006 compared to the GO-matched networks) (Fig. 4B), suggesting that QT-prolonging drugs generally target higher-ranked nodes in the LQTS neighborhood compared to drugs that are not known to prolong the QT interval. Because most LQTS-associated drugs interact with the HERG potassium channel (the product of KCNH2), we asked if this was the only target contributing to the observed enrichment. Only 2 of the 81 QT-prolonging drugs with targets in the neighborhood had KCNH2 as their only target in the neighborhood. When we removed the drug-KCNH2 interactions from the analysis, the enrichment results were not significantly affected (Fig. 4C).
To determine if the enrichment for targets of QT-prolonging drugs in the LQTS neighborhood was due to multiple drugs targeting a single protein, we assigned each drug to a drug class by grouping drugs together if they share the same highest-scoring target in the neighborhood. We made the assumption that this target represented the most likely mechanism for the drug to modify the properties of the QT interval. Because most QT-prolonging drugs target the product of KCNH2, KCNH2 was ignored as a target while drugs were being grouped into 253 classes of which 67 contained QT-prolonging drugs. Even when excluding the HERG channel as a drug target, the LQTS neighborhood performed significantly better than chance, achieving an ROC AUC of 0.70 for classification of drug classes (Fig. 4D) (P = 0.001 compared to the GO-matched networks).
LQTS neighborhood ranking allows prediction and explanation of unexplained adverse drug events
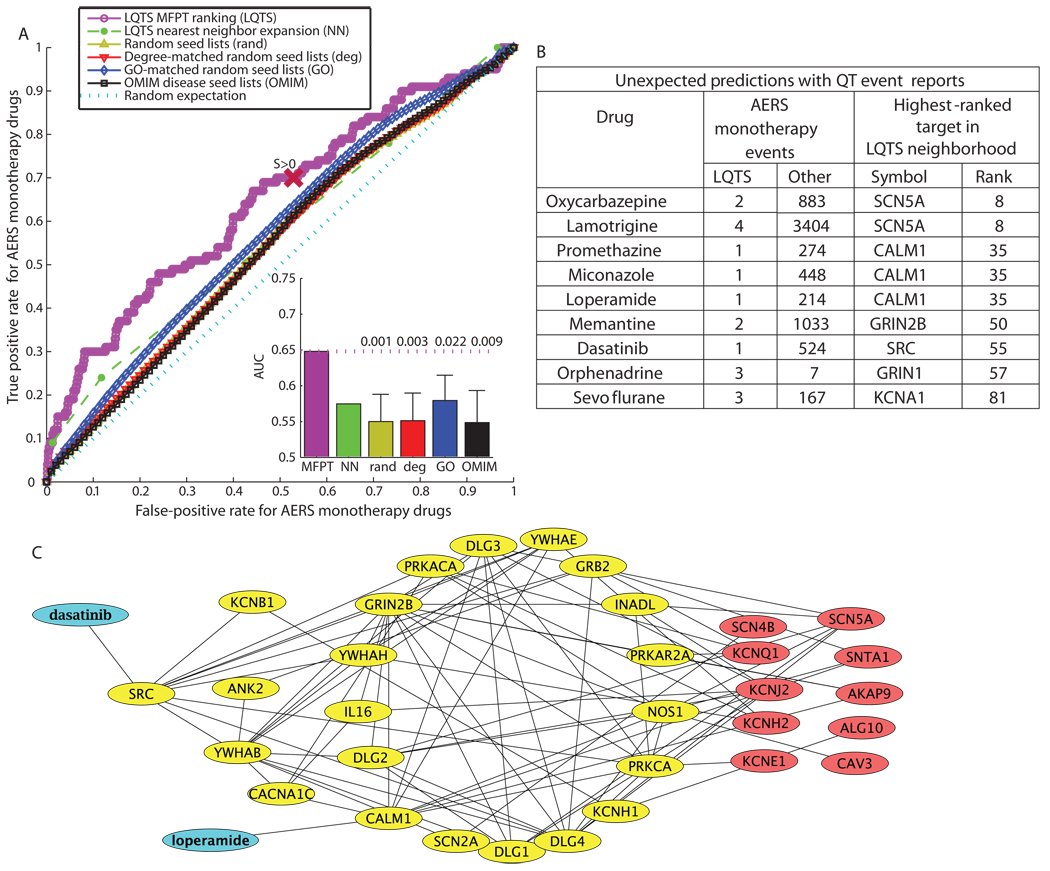
We used the FDA AERS database as a test set to predict if the LQTS neighborhood could predict whether drugs have potential for arrhythmia-related risks. The AERS database contains reported adverse events for patients and all of their medications at the time of the adverse event. Previous studies have calculated the relative risk for drugs to cause severe adverse events (55). However, these studies did not combine protein interaction data with the clinical data set to improve predictions and generate hypotheses about the mechanisms of the side effects.
To simplify the identification of specific drugs and targets that ranked highly in the LQTS neighborhood and are associated with QT event reports in AERS, we focused on adverse event reports associated with monotherapies, which are adverse events associated with only a single drug. Excluding the known QT-prolonging drugs, there were 654 drugs associated with monotherapy case reports of which 363 had targets in the LQTS neighborhood. Out of the 654 monotherapy drugs, 100 were associated with QT events. Seventy of these QT-associated drugs had targets in the LQTS neighborhood. ROC analysis illustrates this enrichment, demonstrating the utility of a classifier for drugs that may cause LQTS with an AUC of 0.65 (P = 0.02 compared to GO-matched networks) (Fig. 5A). From this analysis, we predict that several of the drugs that have targets in the LQTS neighborhood and also have event reports of QT-related symptoms in AERS might warrant closer clinical investigation for a potential risk of causing LQTS or TdP adverse events (Fig. 5B). Several of the high-ranking drugs (ranked within the top 60 on the basis of the rank within the LQTS neighborhood of their targets) did not have previously annotated toxicity of arrhythmias in DrugBank (56, 57) or Arizona Center for Education and Research on Therapeutics (CERT) QT drugs lists (18, 58) (Fig. 5B and table S3), although the FDA AERS reports indicated that they are associated with LQTS events (Fig. 5B). We further explored why these drugs ranked highly by tracing the connections from these drugs through their targets in the LQTS neighborhood to the LQTS disease seed genes. The identification of such potential signaling pathways provides initial insights. For example, two drugs, dasatinib and loperamide, used to treat different pathophysiologies, cancer and severe diarrhea can have QT prolongation as an adverse event and can be connected to the LQTS disease genes through the LQTS neighborhood (Fig. 5C). The paths can be short; for example, loperamide can be connected to KCNH2 through CALM1 (one step), and dasatinib can be connected to KCNQ1 through PRKACA and SRC (two steps), or more convoluted. Such a tracking exercise provides hypotheses about how these drugs might affect the QT interval and increase TdP risk, which can be used to design experiments in animal models or combined with whole genome information to identify “at-risk” patients.
Fig. 5.
The use of the LQTS neighborhood to classify and explain FDA adverse event reports. (A) ROC analysis tests whether the LQTS neighborhood is more effective than the random neighborhoods for predicting if a drug, not previously reported as causing LQTS, has the risk of the side effect. Insets represent AUCs with mean and SD shown for the random network distributions. The numbers above the dotted line in the insets is the fraction of each distribution that had an AUC greater than or equal to the AUC found using the LQTS neighborhood. The point on the ROC curves that represent the cutoff for the LQTS neighborhood is labeled with S > 0. (B) The table lists selected drugs that were not previously annotated as prolonging the QT interval or causing TdP, but are associated with FDA AERS event reports of QT prolongation or TdP. (C) Tracing back to the LQTS seed nodes (red) from two drugs (blue), the Src inhibitor dasatinib (a cancer drug) and loperamide (an antidiarrheal) that interacts with calmodulin (CALM1), generates a subnetwork that suggests different ways in which these drugs used to treat different pathophysiologies could affect QT intervals.
Independent predictions from network analysis match experimental and clinical observations
The list of 13 LQTS-associated disease genes and the human interactome were the only two data sets used to generate the LQTS disease gene neighborhood. Despite the minimal disease-specific information, the resulting LQTS neighborhood contained information about other LQTS genes and drug targets for drugs that cause acquired LQTS. The top two predicted genes in the LQTS disease-gene neighborhood were those encoding PKA and PKC. Matavel et al. demonstrated that PKA and PKC can partially rescue certain long QT phenotypes (59). Additionally, we identified SNPs in PKCα using the QTSCD genome-wide association study (fig. S7 and Table 2).
We evaluated the relative rankings of kinases targeted by 15 inhibitors for the treatment of various cancers (table S4), because clinical information was readily available for these drugs. We included 9 inhibitors listed in DrugBank and 6 other kinase inhibitors (60). The results suggested that drugs targeting phosphatidylinositol 3-kinases generally ranked lower and were not expected to have QT-related side effects (table S4). With the FDA AERS database, we identified drugs (dasatinib, sorafenib, imatinib, and gefitinib) that were not known to cause QT prolongation, yet were associated with reports of QT prolongation. Analysis of the ranking of their targets in the LQTS neighborhood suggested that Src, PDGFR (platelet-derived growth factor receptor), or EGFR (epidermal growth factor receptor) inhibition may affect risk of LQTS (Table 3). This prediction is supported by the work of Zhang et al., who reported that both EGFR and Src inhibition decrease HERG function (61).
Table 3.
Identification of putative QT-relevant targets of kinases with AERS QT events, but not classified as QT drugs. All targets, including those ranked above 1629 and thus outside the LQTS neighborhood, are shown. Bold indicates the highest-ranking LQTS targets for each drug (the target of each drug with the largest module distance score compared to its other targets).
| Drug | AERS QT events |
Total AERS events |
Targeted kinases |
Target rank |
|---|---|---|---|---|
| Dasatinib | 1 | 524 | SRC | 42 |
| PDGFRB | 96 | |||
| FYN | 226 | |||
| YES1 | 478 | |||
| KIT | 1182 | |||
| ABL2 | 1550 | |||
| ABL1 | 3093 | |||
| EPHA1 | 5197 | |||
| EPHA2 | 6570 | |||
| LCK | 7562 | |||
| Sorafenib | 4 | 1620 | PDGFRB | 96 |
| BRAF | 333 | |||
| RAF1 | 491 | |||
| KIT | 1182 | |||
| KDR | 2392 | |||
| FLT3 | 2664 | |||
| FLT4 | 6442 | |||
| Imatinib | 4 | 2691 | PDGFRB | 96 |
| PDGFRA | 107 | |||
| RET | 319 | |||
| NTRK1 | 573 | |||
| KIT | 1182 | |||
| CSF1R | 1553 | |||
| ABL1 | 3093 | |||
| DDR1 | 6743 | |||
| Sutent | 0 | 0 | PDGFRB | 96 |
| KIT | 1182 | |||
| KDR | 2392 | |||
| Gefitinib | 1 | 1372 | EGFR | 105 |
We also noted that several heterotrimeric guanosine triphosphate–binding protein (G protein)–coupled receptors ranked highly in the LQTS neighborhood and suggested that the activity of these receptors, in addition to HERG blockade, may account for the severity of QT side effects for various drugs. Keiser et al. (62) reported that motilium, an antiemetic dopamine receptor antagonist and known HERG blocker, also interacts with adrenergic receptors α-1D, α-1A, and α-1B, which may contribute to the cardiovascular side effects of this drug. Out of all 11,090 nodes ranked in the integrated protein-protein interaction network, these receptors were ranked 108, 267, and 369, respectively, in the LQTS neighborhood. Therefore, our LQTS neighborhood independently predicted several experimentally reported and clinically relevant observations.
DISCUSSION
The analysis of the LQTS neighborhood provides a new perspective of the pathogenesis and susceptibly of LQTS, as well as broader insights into drug-induced pathophysiologies. This analysis provides clues to why LQTS can be a side effect of so many different classes of drugs. Furthermore, examination of the LQTS neighborhood may explain why the degree of HERG blockade is not directly related to the severity of the syndrome, while confirming the importance of the HERG ion channel in acquired LQTS. Most gene products involved in congenital LQTS are not direct drug targets. However, these proteins are located within a region of the human interactome that is targeted by many drugs, which implies that many drugs have targets that interact directly or indirectly with the LQTS-associated proteins. Therefore, many of these drugs could potentially affect the function of these gene products indirectly, leading to acquired LQTS. The function of the HERG ion channel is inhibited by many QT-prolonging drugs and the importance of HERG was confirmed because it was the highest-ranked “seed node.” However, drugs related to LQTS have multiple targets within the LQTS neighborhood, and the modulation of multiple QT-interval regulatory proteins may explain the variable degree of risk associated with different LQTS-associated medications and the independence of this risk on the degree of HERG blockade.
Integrating disease genes and drug actions through protein-protein interactions can serve as the basis for multiscale mechanistic understanding of pathophysiology and drug action by bridging the gap between the molecular understanding of genes involved in disease and the cellular behavior elicited by changes in the activities of these genes or their encoded products. This type of analysis can reveal potential compensatory mechanisms that may explain why genotypic variation does not always manifest at the phenotypic level. There is an underlying interaction network that connects the genes and their products that are responsible for the phenotype. When the phenotype is related to a disease or to an adverse event, the network can reveal new targets for treatment of the disease or explanations for pathogenesis of the adverse event (fig. S9).
Systems biology approaches enable the integration of large data sets to identify emergent properties and behaviors. We used an integrated protein-protein interaction network to identify genes encoding products that can potentially regulate cardiac myocyte action potentials. We chose a global network, representing a composite of multiple cell types. A study focused solely on genes known to be expressed in myocytes would have missed important relationships that we identified, such as the overlap between neuronal disease networks with the LQTS neighborhood and how this can explain patients’ symptoms that are not directly related to cardiac defects, as well as some of the cardiac risks associated with noncardiac diseases. Cell type–specific networks will also be useful, especially in the design of new drugs or drug variants intended to evoke more selective effects. Integration of biochemical network information with clinical data sets enhanced signal detection from drug adverse event surveillance data sets such as AERS. We anticipate that approaches such as the one we demonstrated here will be useful in the analysis of electronic medical records and the next generation of prospective monitoring tools, such as the FDA’s Sentinel Network (63).
Because the links in the human interactome used in our study lack directionality (source or target) and sign (activation or inhibition), this analysis can suggest proximity to the LQTS seed genes but cannot predict if a gene or a drug would have a pro-arrythmic or anti-arrhythmic effect. Such information is likely to be useful for improving the predictions. Despite this limitation, we predicted that polymorphisms and mutations in highly ranked genes play a role in regulation of repolarization reserve and modulate drug-induced TdP and LQTS risk.
We found that QT-prolonging drugs were enriched for targets in the LQTS neighborhood. Other drugs, which are not labeled as QT-prolonging drugs but are known to cause QT-related side effects, also target high-ranked proteins in the LQTS neighborhood. For example, inhalation anesthetics are not on the Arizona CERT list because they are not used in an outpatient setting. However, they targeted various highly ranked nodes in the neighborhood and are well known to cause TdP in some patients (64).
The development of directed, sign-specified disease gene–drug target networks should improve disease gene and adverse event prediction. A drug may target multiple proteins within the neighborhood where such targets have opposing effects. For example, verapamil, which blocks both the HERG channel and a calcium channel, has a lower risk than expected from targeting the HERG channel alone (65). It is clear that these other targets can affect the arrhythmogenic risks of drugs and that HERG blockade alone, although involved in most cases of drug-induced LQTS, does not account for all the risks of LQTS. Therefore, screening drug for an interaction with just HERG can either overrepresent or underrepresent the true risk of drug-induced LQTS.
As the cost of genomic sequencing drops and genome characterization becomes a common diagnostic tool, physicians will have more information for making better diagnoses. However, we currently lack the interpretive framework to use such information to its full potential. Systems pharmacology approaches, such as the one presented here, will enable interpretation of large genomic data sets to generate hypotheses that could guide clinical studies in the future to determine risk of adverse drug effects in individual patients for clinical management. Candidate genes suggested in this analysis can be screened for mutations in patients with no known cause of LQTS to more rapidly identify causative mutations. Further work can delineate targets in the LQTS neighborhood into positive and negative regulators of the QT-interval and TdP susceptibility, as well as categorize SNPs and other mutations that influence patient risk. Similar approaches can be applied to other drug-induced side effects related to diseases that result from the interactions between pharmaceutical and a complex biological network. There are often several pharmaceutical options available for a particular pathophysiology and it will become increasingly possible to select the most effective treatment with the lowest adverse event risk based on the patient’s personal genetic background. Such application of personalized medicine will allow greater safety and efficacy of both existing and new pharmaceuticals.
MATERIALS AND METHODS
Background network construction
We constructed a large-scale human protein-protein interaction network by integrating publicly available interactions from nine sources (66–75) (fig. S1A). All data sets were downloaded on 8 December 2008 unless otherwise noted. Information from the National Center for Biotechnology Information (NCBI) and Uniprot was used to construct a lookup table from various accession numbers to Entrez Gene Symbols for all mammalian species. Data from NCBI homologene, Jackson Labs Mouse informatics, and NCBI gene history were used to construct a lookup table that matched each mammalian gene to its human ortholog. Curated data from Jackson labs took precedence over predicted orthology from homologene. These lookup tables were used to process the protein interaction data from BioGRID (67), DIP (74), HPRD (73), Intact (70), MINT (68), MIPS (72), PDZBase (66), Phospho.Elm (69), and Reactome (75). Each protein from a mammalian species was matched to its gene and subsequently to its human ortholog (71, 76). With a breadth-first search, all interactions that were not reachable from KCNH2 were discarded from the network to leave a large fully connected cluster (one island). This global human protein-protein interaction network represented a composite of all cell types and tissues, which as of 8 December 2008 was a large fully connected island of 11,090 nodes (gene products with human gene ID) and 79,530 edges (interactions) based on 23,423 distinct literature references (PubMed IDs).
Neighborhood ranking
Using custom c code, we calculated a matrix of MFPT between all pairs of nodes in a network with the algorithm of Kemeny and Snell (30). We defined a module distance score, Sj, for each node in the protein-protein interaction (PPI) network as the difference in average of the MFPTs to the node when starting on a non–seed node compared to starting on a seed node, normalized to the average MFPT to reach the node from a random start.
| (1) |
Here C is the set of nodes that fall in the seed lists and C′ is the set of nodes reachable from random walks from the seed list. Therefore, a score greater than zero implies that node j falls closer on average to the seed nodes than it does on average to the rest of the network.
Network generation was performed on the LQTS seed list and each of the control seed lists by scoring all nodes in the network and using a cutoff score of zero to define the neighborhood. Drugs were ranked by assigning each drug the score of its highest-scoring target. Classes were then constructed by combining all drugs that shared the same highest-scoring target into a single class, excluding the HERG ion channel. If a class contained at least one QT drug, this class was considered a QT drug class.
Control seed lists
Distributions of control networks were generated from random seed lists. Each set of random networks was based on one of three null hypotheses. All random seed lists had 13 seed nodes (like the LQTS seed list). The first set of 1000 random seed lists was generated by randomly selecting seed nodes uniformly out of the background network. The second set of 1000 random seed lists was generated by selecting a random seed node matched by degree to each of the LQTS seed nodes. The degree, number of first neighbors, for each node in the network was log binned by taking the floor integer of the base 2 logarithm of the degree of each node. For each LQTS seed node, a random node was selected from the same degree bin to match it in the randomized seed list. The third set of 2500 GO-matched seed lists were created on the basis of the GO terms for membrane (GO: 0016020) and cytoplasm (GO: 0005737). This provided a seed list with the same localization characteristics of the LQTS seed list without overconstraining the random seeds to the original LQTS seed nodes. Each node in the network was grouped as having neither term, both terms, just cytoplasmic, or just membrane. For each LQTS seed node, a random node of the same type was chosen to match it in the randomized seed list.
Network overlap
For all the control neighborhoods and disease neighborhoods, the number of genes in common with the LQTS neighborhood was counted. This was divided by the number of unique genes that occurred in either neighborhood to calculate the Jaccard coefficient. A histogram was generated by counting the number of networks that had overlap with the LQTS neighborhood by binning the data into 40 equally spaced bins. The counts of networks in each bin were divided by the total number of networks in the distribution and the bin width (0.025) to generate a probability density. The area under segments of this curve represents the probability that a randomly chosen network overlaps with the LQTS neighborhood within the magnitude spanned by the segment.
QTSCD GWAS analysis
The SNPs and association strengths from the QTSCD GWAS were obtained from A. Chakravarti (Johns Hopkins University School of Medicine, Baltimore, MD). The genomic locations of all SNPs and RefSeq genes were taken from the February 2009 release of the human genome, downloaded from the UCSC genome browser. The SNPs that fell between annotated transcriptional start and end points of genes that ranked among the top 150 nodes in the LQTS neighborhood were identified.
Drug target analysis
We labeled the proteins (gene products) that were nodes in the human interactome and were drug targets in DrugBank (fig. S8 and table S3) (56, 57). Nodes targeted by drugs listed on the Arizona CERT LQTS Drug List were also labeled as having a known risk of QT-related side effects (77). There are 951 FDA-approved small-molecule drugs in DrugBank (not classified as nutraceuticals) that have 518 different protein targets in the human interactome. Ninety-nine of these drugs, found on the Arizona CERT QT drug list, target 114 proteins (58, 78).
AERS database
The AERS database was processed with Perl code to match the event reports to drugs found in DrugBank. Out of the 1.5 million adverse event reports in the database, we focused on the 5168 reports of prolonged electrocardiographic QT intervals or TdP. The distribution of drugs associated with adverse events follows an exponential distribution (fig. S10). Drugs that had at least one target in the LQTS neighborhood, and were thus predicted to have an effect on QT interval, were present in 61.7% of all the reports in the database where these reports include 92% of the QT event reports. Many of the predictions are drugs previously associated with acquired LQTS. Of the 5091 QT event reports, 78% involved at least one previously known QT-prolonging drug. Out of the 1123 remaining QT event reports, 78% had targets in the LQTS neighborhood (fig. S11). As such, the predictions of drugs not previously known to be involved in the regulation of the QT interval can account for a large number of these QT event reports. There are 1221 drugs that are associated with event reports where the drug was used alone. These were labeled as monotherapy drugs; 185 monotherapy drugs were associated with a monotherapy event report of QT prolongation or TdP. Excluding the known QT-prolonging drugs from these lists produced a total of 1113 monotherapy drugs of which 129 were associated with reports of QT prolongation. Excluding all drugs in DrugBank annotated as nutraceuticals or not annotated as FDA approved or small molecules left 654 monotherapy drugs of which 100 were associated with QT events and were used for further analysis.
ROC analysis
For every possible rank threshold cutoff (starting from the top node through all 11,090 nodes) in the ranked list of nodes, the true positive rate was plotted against the false-positive rate with custom Perl code. A true positive rate could be the fraction of targets of QT-prolonging drugs within the LQTS disease gene neighborhood, the fraction of QT-prolonging drugs with at least one target in the neighborhood, or the fraction of QT-prolonging drug classes within the neighborhood. We defined drug classes by grouping all drugs that shared a common highest-ranked target other than the HERG ion channel. The corresponding false-positive rate is the fraction of the remaining drug targets, remaining drugs, or remaining drug classes within the neighborhood. The AUC was calculated by summing the area of rectangles between each point in the ROC curve. This process was repeated for the LQTS neighborhood ranking, as well as the 4500 random networks and all OMIM disease networks. Statistical significance of the AUC was calculated by counting the fraction of each control network set that had an AUC greater than or equal to the LQTS neighborhood’s AUC. In most cases, the largest P values were associated with the GO-matched random networks.
Supplementary Material
Footnotes
SUPPLEMENTARY MATERIALS
www.sciencesignaling.org/cgi/content/full/3/118/ra30/DC1
Fig. S1. Overview of data integration and analysis.
Fig. S2. Comparison of node connectivity in networks created by nearest-neighbor expansion or through module distance score based on MFPT.
Fig. S3. Average shortest path to seed node as a function of network size.
Fig. S4. Network size distribution.
Fig. S5. ROC curve for classification of disease genes.
Fig. S6. Network modular versus nonmodular diseases: Leave-one-out recovery rate distribution.
Fig. S7. The use of the LQTS neighborhood rankings to identify SNPs in the QTSCD GWAS.
Fig. S8. The distribution of drug targets.
Fig. S9. Methodology for integrating systems pharmacology and personalized medicine.
Fig. S10. The distribution of number of drugs associated with each adverse event report in AERS.
Fig. S11. Fraction of AERS events involving neighborhood drugs.
Table S1. Ranked nodes in the LQTS neighborhood.
Table S2. Table of diseases and network overlap and recovery rate.
Table S3. Drugs used to evaluate the LQTS neighborhood.
Table S4. Ranking of kinase inhibitors based on their targets in LQTS neighborhood.
REFERENCES AND NOTES
- 1.Blake WJ, Balázsi G, Kohanski MA, Isaacs FJ, Murphy KF, Kuang Y, Cantor CR, Walt DR, Collins JJ. Phenotypic consequences of promoter-mediated transcriptional noise. Mol. Cell. 2006;24:853–865. doi: 10.1016/j.molcel.2006.11.003. [DOI] [PubMed] [Google Scholar]
- 2.Raj A, van Oudenaarden A. Nature, nurture, or chance: Stochastic gene expression and its consequences. Cell. 2008;135:216–226. doi: 10.1016/j.cell.2008.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Antonarakis SE, McKusick VA. OMIM passes the 1,000-disease-gene mark. Nat. Genet. 2000;25:11. doi: 10.1038/75497. [DOI] [PubMed] [Google Scholar]
- 4.Hamosh A, Scott AF, Amberger J, Valle D, McKusick VA. Online Mendelian Inheritance in Man (OMIM) Hum. Mutat. 2000;15:57–61. doi: 10.1002/(SICI)1098-1004(200001)15:1<57::AID-HUMU12>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 5.Goh KI, Cusick ME, Valle D, Childs B, Vidal M, Barabási AL. The human disease network. Proc. Natl. Acad. Sci. U.S.A. 2007;104:8685–8690. doi: 10.1073/pnas.0701361104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barabási AL, Oltvai ZN. Network biology: Understanding the cell’s functional organization. Nat. Rev. Genet. 2004;5:101–113. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- 7.Feldman I, Rzhetsky A, Vitkup D. Network properties of genes harboring inherited disease mutations. Proc. Natl. Acad. Sci. U.S.A. 2008;105:4323–4328. doi: 10.1073/pnas.0701722105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Albert R. Scale-free networks in cell biology. J. Cell Sci. 2005;118:4947–4957. doi: 10.1242/jcs.02714. [DOI] [PubMed] [Google Scholar]
- 9.Alon U. Network motifs: Theory and experimental approaches. Nat. Rev. Genet. 2007;8:450–461. doi: 10.1038/nrg2102. [DOI] [PubMed] [Google Scholar]
- 10.Spirin V, Mirny LA. Protein complexes and functional modules in molecular networks. Proc. Natl. Acad. Sci. U.S.A. 2003;100:12123–12128. doi: 10.1073/pnas.2032324100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aerssens J, Paulussen AD. Pharmacogenomics and acquired long QT syndrome. Pharmacogenomics. 2005;6:259–270. doi: 10.1517/14622416.6.3.259. [DOI] [PubMed] [Google Scholar]
- 12.Miller-Jensen K, Janes KA, Brugge JS, Lauffenburger DA. Common effector processing mediates cell-specific responses to stimuli. Nature. 2007;448:604–608. doi: 10.1038/nature06001. [DOI] [PubMed] [Google Scholar]
- 13.Jordan JD, Landau EM, Iyengar R. Signaling networks: The origins of cellular multitasking. Cell. 2000;103:193–200. doi: 10.1016/s0092-8674(00)00112-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roden DM. Clinical practice. Long-QT syndrome. N. Engl. J. Med. 2008;358:169–176. doi: 10.1056/NEJMcp0706513. [DOI] [PubMed] [Google Scholar]
- 15.Roden DM. Human genomics and its impact on arrhythmias. Trends Cardiovasc. Med. 2004;14:112–116. doi: 10.1016/j.tcm.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 16.Roden DM. Long QT syndrome: Reduced repolarization reserve and the genetic link. J. Intern. Med. 2006;259:59–69. doi: 10.1111/j.1365-2796.2005.01589.x. [DOI] [PubMed] [Google Scholar]
- 17.Wang D, Patel VV, Ricciotti E, Zhou R, Levin MD, Gao E, Yu Z, Ferrari VA, Lu MM, Xu J, Zhang H, Hui Y, Cheng Y, Petrenko N, Yu Y, FitzGerald GA. Cardiomyocyte cyclooxygenase-2 influences cardiac rhythm and function. Proc. Natl. Acad. Sci. U.S.A. 2009;106:7548–7552. doi: 10.1073/pnas.0805806106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kilborn MJ, Woosley RL. Registry for torsades de pointes with drug treatment exists. BMJ. 2001;322:672–673. [PMC free article] [PubMed] [Google Scholar]
- 19.Layton D, Key C, Shakir SA. Prolongation of the QT interval and cardiac arrhythmias associated with cisapride: Limitations of the pharmacoepidemiological studies conducted and proposals for the future. Pharmacoepidemiol. Drug Saf. 2003;12:31–40. doi: 10.1002/pds.781. [DOI] [PubMed] [Google Scholar]
- 20.González MA, Estes KS. Pharmacokinetic overview of oral second-generation H1 antihistamines. Int. J. Clin. Pharmacol. Ther. 1998;36:292–300. [PubMed] [Google Scholar]
- 21.Hoffmann P, Warner B. Are hERG channel inhibition and QT interval prolongation all there is in drug-induced torsadogenesis? A review of emerging trends. J. Pharmacol. Toxicol. Methods. 2006;53:87–105. doi: 10.1016/j.vascn.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 22.Berger SI, Posner JM, Ma’ayan A. Genes2Networks: Connecting lists of gene symbols using mammalian protein interactions databases. BMC Bioinformatics. 2007;8:372. doi: 10.1186/1471-2105-8-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Camargo LM, Collura V, Rain JC, Mizuguchi K, Hermjakob H, Kerrien S, Bonnert TP, Whiting PJ, Brandon NJ. Disrupted in Schizophrenia 1 Interactome: Evidence for the close connectivity of risk genes and a potential synaptic basis for schizophrenia. Mol. Psychiatry. 2007;12:74–86. doi: 10.1038/sj.mp.4001880. [DOI] [PubMed] [Google Scholar]
- 24.Hyduke DR, Jarboe LR, Tran LM, Chou KJ, Liao JC. Integrated network analysis identifies nitric oxide response networks and dihydroxyacid dehydratase as a crucial target in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 2007;104:8484–8489. doi: 10.1073/pnas.0610888104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lim J, Hao T, Shaw C, Patel AJ, Szabó G, Rual JF, Fisk CJ, Li N, Smolyar A, Hill DE, Barabási AL, Vidal M, Zoghbi HY. A protein-protein interaction network for human inherited ataxias and disorders of Purkinje cell degeneration. Cell. 2006;125:801–814. doi: 10.1016/j.cell.2006.03.032. [DOI] [PubMed] [Google Scholar]
- 26.Chen J, Aronow BJ, Jegga AG. Disease candidate gene identification and prioritization using protein interaction networks. BMC Bioinformatics. 2009;10:73. doi: 10.1186/1471-2105-10-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen JY, Shen C, Sivachenko AY. Mining Alzheimer disease relevant proteins from integrated protein interactome data. Pac. Symp. Biocomput. 2006:367–378. [PubMed] [Google Scholar]
- 28.Petersen CI, McFarland TR, Stepanovic SZ, Yang P, Reiner DJ, Hayashi K, George AL, Roden DM, Thomas JH, Balser JR. In vivo identification of genes that modify ether-a-go-go-related gene activity in Caenorhabditis elegans may also affect human cardiac arrhythmia. Proc. Natl. Acad. Sci. U.S.A. 2004;101:11773–11778. doi: 10.1073/pnas.0306005101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Köhler S, Bauer S, Horn D, Robinson PN. Walking the interactome for prioritization of candidate disease genes. Am. J. Hum. Genet. 2008;82:949–958. doi: 10.1016/j.ajhg.2008.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kemeny JG, Snell JL. Finite Markov Chains. New York: Springer-Verlag; 1983. [Google Scholar]
- 31.Freudenberg J, Propping P. A similarity-based method for genome-wide prediction of disease-relevant human genes. Bioinformatics. 2002;18 Suppl. 2:S110–S115. doi: 10.1093/bioinformatics/18.suppl_2.s110. [DOI] [PubMed] [Google Scholar]
- 32.Chen L, Marquardt ML, Tester DJ, Sampson KJ, Ackerman MJ, Kass RS. Mutation of an A-kinase-anchoring protein causes long-QT syndrome. Proc. Natl. Acad. Sci. U.S.A. 2007;104:20990–20995. doi: 10.1073/pnas.0710527105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ueda K, Valdivia C, Medeiros-Domingo A, Tester DJ, Vatta M, Farrugia G, Ackerman MJ, Makielski JC. Syntrophin mutation associated with long QT syndrome through activation of the nNOS-SCN5A macromolecular complex. Proc. Natl. Acad. Sci. U.S.A. 2008;105:9355–9360. doi: 10.1073/pnas.0801294105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arking DE, Pfeufer A, Post W, Kao WH, Newton-Cheh C, Ikeda M, West K, Kashuk C, Akyol M, Perz S, Jalilzadeh S, Illig T, Gieger C, Guo CY, Larson MG, Wichmann HE, Marban E, O’Donnell CJ, Hirschhorn JN, Kääb S, Spooner PM, Meitinger T, Chakravarti A. A common genetic variant in the NOS1 regulator NOS1AP modulates cardiac repolarization. Nat. Genet. 2006;38:644–651. doi: 10.1038/ng1790. [DOI] [PubMed] [Google Scholar]
- 35.Nielsen NH, Winkel BG, Kanters JK, Schmitt N, Hofman-Bang J, Jensen HS, Bentzen BH, Sigurd B, Larsen LA, Andersen PS, Haunsø S, Kjeldsen K, Grunnet M, Christiansen M, Olesen SP. Mutations in the Kv1.5 channel gene KCNA5 in cardiac arrest patients. Biochem. Biophys. Res. Commun. 2007;354:776–782. doi: 10.1016/j.bbrc.2007.01.048. [DOI] [PubMed] [Google Scholar]
- 36.The Gene Ontology Consortium. The Gene Ontology project in 2008. Nucleic Acids Res. 2008;36:D440–D444. doi: 10.1093/nar/gkm883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hamosh A, Scott AF, Amberger JS, Bocchini CA, McKusick VA. Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res. 2005;33:D514–D517. doi: 10.1093/nar/gki033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hedley PL, Jørgensen P, Schlamowitz S, Wangari R, Moolman-Smook J, Brink PA, Kanters JK, Corfield VA, Christiansen M. The genetic basis of long QT and short QT syndromes: A mutation update. Hum. Mutat. 2009;30:1486–1511. doi: 10.1002/humu.21106. [DOI] [PubMed] [Google Scholar]
- 39.Patel U, Pavri BB. Short QT syndrome: A review. Cardiol. Rev. 2009;17:300–303. doi: 10.1097/CRD.0b013e3181c07592. [DOI] [PubMed] [Google Scholar]
- 40.Schimpf R, Veltmann C, Wolpert C, Borggrefe M. Channelopathies: Brugada syndrome, long QT syndrome, short QT syndrome, and CPVT. Herz. 2009;34:281–288. doi: 10.1007/s00059-009-3238-1. [DOI] [PubMed] [Google Scholar]
- 41.Zellerhoff S, Pistulli R, Mönnig G, Hinterseer M, Beckmann BM, Köbe J, Steinbeck G, Kääb S, Haverkamp W, Fabritz L, Gradaus R, Breithardt G, Schulze-Bahr E, Bocker D, Kirchhof P. Atrial arrhythmias in long-QT syndrome under daily life conditions: A nested case control study. J. Cardiovasc. Electrophysiol. 2009;20:401–407. doi: 10.1111/j.1540-8167.2008.01339.x. [DOI] [PubMed] [Google Scholar]
- 42.Barrett CF, Tsien RW. The Timothy syndrome mutation differentially affects voltage- and calcium-dependent inactivation of CaV1.2 L-type calcium channels. Proc. Natl. Acad. Sci. U.S.A. 2008;105:2157–2162. doi: 10.1073/pnas.0710501105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weir RA, Petrie CJ, Murday V, Findlay IN. Andersen-Tawil syndrome. Int. J. Cardiol. doi: 10.1016/j.ijcard.2009.01.057. in press. [DOI] [PubMed] [Google Scholar]
- 44.Bär KJ, Koschke M, Boettger MK, Berger S, Kabisch A, Sauer H, Voss A, Yeragani VK. Acute psychosis leads to increased QT variability in patients suffering from schizophrenia. Schizophr. Res. 2007;95:115–123. doi: 10.1016/j.schres.2007.05.034. [DOI] [PubMed] [Google Scholar]
- 45.Glassman AH, Bigger JT., Jr Antipsychotic drugs: Prolonged QTc interval, torsade de pointes, and sudden death. Am. J. Psychiatry. 2001;158:1774–1782. doi: 10.1176/appi.ajp.158.11.1774. [DOI] [PubMed] [Google Scholar]
- 46.Vetter VL, Elia J, Erickson C, Berger S, Blum N, Uzark K, Webb CL. Cardiovascular monitoring of children and adolescents with heart disease receiving medications for attention deficit/hyperactivity disorder: A scientific statement from the American Heart Association Council on Cardiovascular Disease in the Young Congenital Cardiac Defects Committee and the Council on Cardiovascular Nursing. Circulation. 2008;117:2407–2423. doi: 10.1161/CIRCULATIONAHA.107.189473. [DOI] [PubMed] [Google Scholar]
- 47.Johnson JN, Hofman N, Haglund CM, Cascino GD, Wilde AA, Ackerman MJ. Identification of a possible pathogenic link between congenital long QT syndrome and epilepsy. Neurology. 2009;72:224–231. doi: 10.1212/01.wnl.0000335760.02995.ca. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Minoretti P, Politi P, Martinelli V, Emanuele E, Bertona M, Falcone C, Geroldi D. QT interval duration in apparently healthy men is associated with depression-related personality trait neuroticism. J. Psychosom. Res. 2006;61:19–23. doi: 10.1016/j.jpsychores.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 49.Li C, Hu D, Shang L, Ma S, Liu W, Li Y, Ma Z, Tang C, Mei Y, Wang L. Surgical left cardiac sympathetic denervation for long QT syndrome: Effects on QT interval and heart rate. Heart Vessels. 2005;20:137–141. doi: 10.1007/s00380-005-0820-1. [DOI] [PubMed] [Google Scholar]
- 50.Shamsuzzaman AS, Ackerman MJ, Kara T, Lanfranchi P, Somers VK. Sympathetic nerve activity in the congenital long-QT syndrome. Circulation. 2003;107:1844–1847. doi: 10.1161/01.CIR.0000066284.34258.59. [DOI] [PubMed] [Google Scholar]
- 51.Omichi C, Momose Y, Kitahara S. Congenital long QT syndrome presenting with a history of epilepsy: Misdiagnosis or relationship between channelopathies of the heart and brain? Epilepsia. 2010;51:289–292. doi: 10.1111/j.1528-1167.2009.02267.x. [DOI] [PubMed] [Google Scholar]
- 52.Pfeufer A, Sanna S, Arking DE, Müller M, Gateva V, Fuchsberger C, Ehret GB, Orrú M, Pattaro C, Köttgen A, Perz S, Usala G, Barbalic M, Li M, Pütz B, Scuteri A, Prineas RJ, Sinner MF, Gieger C, Najjar SS, Kao WH, Muhleisen TW, Dei M, Happle C, Möhlenkamp S, Crisponi L, Erbel R, Jöckel KH, Naitza S, Steinbeck G, Marroni F, Hicks AA, Lakatta E, Müller-Myhsok B, Pramstaller PP, Wichmann HE, Schlessinger D, Boerwinkle E, Meitinger T, Uda M, Coresh J, Kääb S, Abecasis GR, Chakravarti A. Common variants at ten loci modulate the QT interval duration in the QTSCD study. Nat. Genet. 2009;41:407–414. doi: 10.1038/ng.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Newton-Cheh C, Eijgelsheim M, Rice KM, de Bakker PI, Yin X, Estrada K, Bis JC, Marciante K, Rivadeneira F, Noseworthy PA, Sotoodehnia N, Smith NL, Rotter JI, Kors JA, Witteman JC, Hofman A, Heckbert SR, O’Donnell CJ, Uitterlinden AG, Psaty BM, Lumley T, Larson MG, Stricker BH. Common variants at ten loci influence QT interval duration in the QTGEN Study. Nat. Genet. 2009;41:399–406. doi: 10.1038/ng.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Milan DJ, Kim AM, Winterfield JR, Jones IL, Pfeufer A, Sanna S, Arking DE, Amsterdam AH, Sabeh KM, Mably JD, Rosenbaum DS, Peterson RT, Chakravarti A, Kääb S, Roden DM, MacRae CA. Drug-sensitized zebrafish screen identifies multiple genes, including GINS3, as regulators of myocardial repolarization. Circulation. 2009;120:553–559. doi: 10.1161/CIRCULATIONAHA.108.821082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Poluzzi E, Raschi E, Moretti U, De Ponti F. Drug-induced torsades de pointes: Data mining of the public version of the FDA Adverse Event Reporting System (AERS) Pharmacoepidemiol. Drug Saf. 2009;18:512–518. doi: 10.1002/pds.1746. [DOI] [PubMed] [Google Scholar]
- 56.Wishart DS, Knox C, Guo AC, Cheng D, Shrivastava S, Tzur D, Gautam B, Hassanali M. DrugBank: A knowledgebase for drugs, drug actions and drug targets. Nucleic Acids Res. 2008;36:D901–D906. doi: 10.1093/nar/gkm958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wishart DS, Knox C, Guo AC, Shrivastava S, Hassanali M, Stothard P, Chang Z, Woolsey J. DrugBank: A comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res. 2006;34:D668–D672. doi: 10.1093/nar/gkj067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Woosley RL. Drugs That Prolong the QT Interval and/or Induce Torsades de Pointes. Tucson, AZ: Arizona CERT; 2007. http://qtdrugs.org/. [Google Scholar]
- 59.Matavel A, Medei E, Lopes CM. PKA and PKC partially rescue long QT type 1 phenotype by restoring channel-PIP2 interactions. Channels (Austin) 2010;4:3–11. doi: 10.4161/chan.4.1.10227. [DOI] [PubMed] [Google Scholar]
- 60.Zhang J, Yang PL, Gray NS. Targeting cancer with small molecule kinase inhibitors. Nat. Rev. Cancer. 2009;9:28–39. doi: 10.1038/nrc2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang DY, Wang Y, Lau CP, Tse HF, Li GR. Both EGFR kinase and Src-related tyrosine kinases regulate human ether-à-go-go-related gene potassium channels. Cell. Signal. 2008;20:1815–1821. doi: 10.1016/j.cellsig.2008.06.006. [DOI] [PubMed] [Google Scholar]
- 62.Keiser MJ, Setola V, Irwin JJ, Laggner C, Abbas AI, Hufeisen SJ, Jensen NH, Kuijer MB, Matos RC, Tran TB, Whaley R, Glennon RA, Hert J, Thomas KL, Edwards DD, Shoichet BK, Roth BL. Predicting new molecular targets for known drugs. Nature. 2009;462:175–181. doi: 10.1038/nature08506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Platt R, Wilson M, Chan KA, Benner JS, Marchibroda J, McClellan M. The new Sentinel Network—Improving the evidence of medical-product safety. N. Engl. J. Med. 2009;361:645–647. doi: 10.1056/NEJMp0905338. [DOI] [PubMed] [Google Scholar]
- 64.Yildirim H, Adanir T, Atay A, Katircioğlu K, Savaci S. The effects of sevoflurane, isoflurane and desflurane onQTinterval of theECG. Eur. J.Anaesthesiol. 2004;21:566–570. doi: 10.1017/s0265021504007112. [DOI] [PubMed] [Google Scholar]
- 65.Fauchier L, Babuty D, Poret P, Autret ML, Cosnay P, Fauchier JP. Effect of verapamil on QT interval dynamicity. Am. J. Cardiol. 1999;83:807–808. doi: 10.1016/s0002-9149(98)01001-7. A10–A11. [DOI] [PubMed] [Google Scholar]
- 66.Beuming T, Skrabanek L, Niv MY, Mukherjee P, Weinstein H. PDZBase: A protein–protein interaction database for PDZ-domains. Bioinformatics. 2005;21:827–828. doi: 10.1093/bioinformatics/bti098. [DOI] [PubMed] [Google Scholar]
- 67.Breitkreutz BJ, Stark C, Reguly T, Boucher L, Breitkreutz A, Livstone M, Oughtred R, Lackner DH, Bähler J, Wood V, Dolinski K, Tyers M. The BioGRID Interaction Database: 2008 update. Nucleic Acids Res. 2008;36:D637–D640. doi: 10.1093/nar/gkm1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chatr-aryamontri A, Ceol A, Palazzi LM, Nardelli G, Schneider MV, Castagnoli L, Cesareni G. MINT: The Molecular INTeraction database. Nucleic Acids Res. 2007;35:D572–D574. doi: 10.1093/nar/gkl950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Diella F, Gould CM, Chica C, Via A, Gibson TJ. Phospho.ELM: A database of phosphorylation sites—Update 2008. Nucleic Acids Res. 2008;36:D240–D244. doi: 10.1093/nar/gkm772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kerrien S, Alam-Faruque Y, Aranda B, Bancarz I, Bridge A, Derow C, Dimmer E, Feuermann M, Friedrichsen A, Huntley R, Kohler C, Khadake J, Leroy C, Liban A, Lieftink C, Montecchi-Palazzi L, Orchard S, Risse J, Robbe K, Roechert B, Thorneycroft D, Zhang Y, Apweiler R, Hermjakob H. IntAct—Open source resource for molecular interaction data. Nucleic Acids Res. 2007;35:D561–D565. doi: 10.1093/nar/gkl958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Maglott D, Ostell J, Pruitt KD, Tatusova T. Entrez Gene: Gene-centered information at NCBI. Nucleic Acids Res. 2007;35:D26–D31. doi: 10.1093/nar/gkl993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mewes HW, Dietmann S, Frishman D, Gregory R, Mannhaupt G, Mayer KF, Münsterkötter M, Ruepp A, Spannagl M, Stumpflen V, Rattei T. MIPS: Analysis and annotation of genome information in 2007. Nucleic Acids Res. 2008;36:D196–D201. doi: 10.1093/nar/gkm980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mishra GR, Suresh M, Kumaran K, Kannabiran N, Suresh S, Bala P, Shivakumar K, Anuradha N, Reddy R, Raghavan TM, Menon S, Hanumanthu G, Gupta M, Upendran S, Gupta S, Mahesh M, Jacob B, Mathew P, Chatterjee P, Arun KS, Sharma S, Chandrika KN, Deshpande N, Palvankar K, Raghavnath R, Krishnakanth R, Karathia H, Rekha B, Nayak R, Vishnupriya G, Kumar HG, Nagini M, Kumar GS, Jose R, Deepthi P, Mohan SS, Gandhi TK, Harsha HC, Deshpande KS, Sarker M, Prasad TS, Pandey A. Human protein reference database—2006 update. Nucleic Acids Res. 2006;34:D411–D414. doi: 10.1093/nar/gkj141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Salwinski L, Miller CS, Smith AJ, Pettit FK, Bowie JU, Eisenberg D. The Database of Interacting Proteins: 2004 update. Nucleic Acids Res. 2004;32:D449–D451. doi: 10.1093/nar/gkh086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Vastrik I, D’Eustachio P, Schmidt E, Joshi-Tope G, Gopinath G, Croft D, de Bono B, Gillespie M, Jassal B, Lewis S, Matthews L, Wu G, Birney E, Stein L. Reactome: A knowledge base of biologic pathways and processes. Genome Biol. 2007;8:R39. doi: 10.1186/gb-2007-8-3-r39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bult CJ, Eppig JT, Kadin JA, Richardson JE, Blake JA. Mouse Genome Database Group, The Mouse Genome Database (MGD): Mouse biology and model systems. Nucleic Acids Res. 2008;36:D724–D728. doi: 10.1093/nar/gkm961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ma’ayan A, Jenkins SL, Goldfarb J, Iyengar R. Network analysis of FDA approved drugs and their targets. Mt. Sinai J. Med. 2007;74:27–32. doi: 10.1002/msj.20002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Woosley RL. Discovering adverse reactions:Why does it take so long? Clin. Pharmacol. Ther. 2004;76:287–289. doi: 10.1016/j.clpt.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 79.Acknowledgments: We thank D. Roden for providing the original LQTS seed gene list that started this project and for additional helpful conversations. We also thank A. Chakravarti and the QTSCD Consortium for providing the QTSCD GWAS data set. Funding: This work was supported by the NIH (grant DK038761) and the Systems Biology Center New York (grant P50GM07558). S.I.B. was supported by Pharmacological Sciences Training Grant GM062754. R.I. and A.M. were supported by a Clinical and Translational Science Award (UL1RR029887) from National Center for Research Resources. Author contributions: S.I.B., A.M., and R.I. developed the initial ideas and hypotheses for the analyses. S.I.B. and A.M. developed the database and networks for the analyses. S.I.B. conducted all of the computational analyses. All authors analyzed the results and participated in writing the paper. Competing interests: The authors have no competing interests.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.