Abstract
During DNA sequencing projects one of the most labour intensive and highly skilled tasks is to view the original trace descriptions of gels and to adjudicate between conflicting readings. Given the current methods of calculating a consensus, the majority of the time employed in viewing traces and editing readings is actually devoted to making the poorer data fit the good data. We propose new consensus calculation algorithms that employ numerical estimates of base calling accuracy and which when used in conjunction with an automatic detector of contradictory data should greatly reduce the time spent checking and editing readings and hence improve DNA sequencing productivity.
Full text
PDF

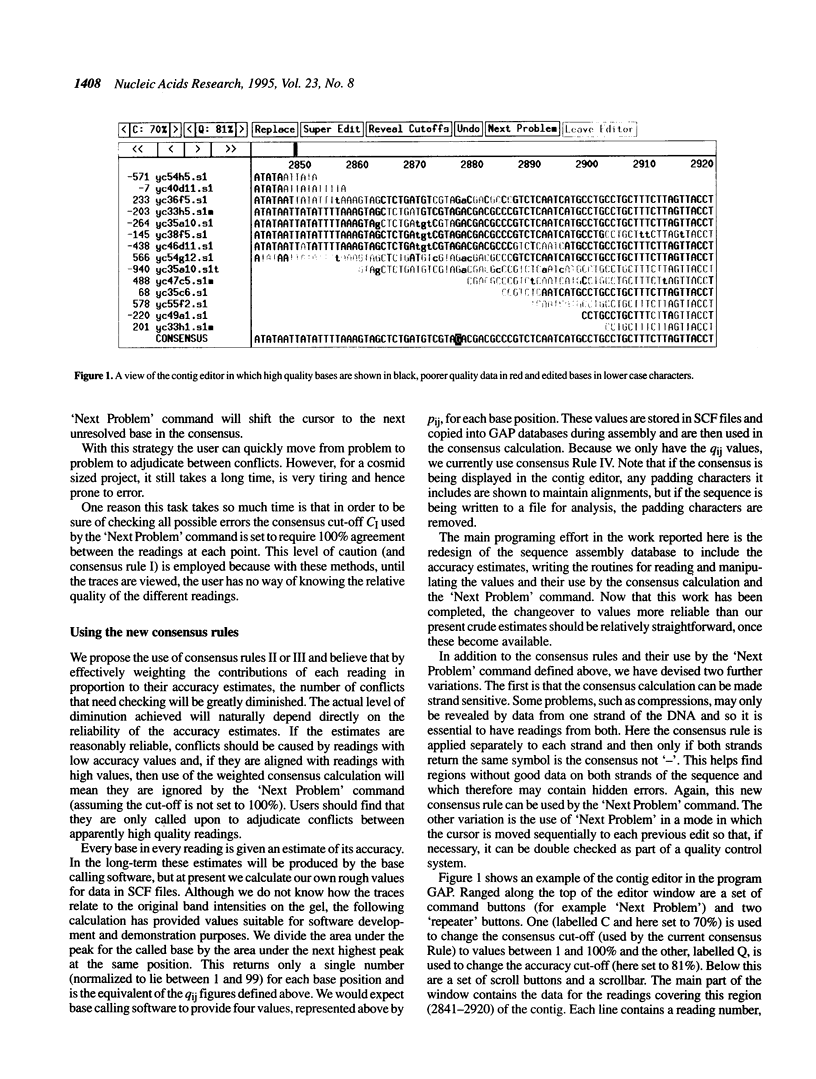


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Churchill G. A., Waterman M. S. The accuracy of DNA sequences: estimating sequence quality. Genomics. 1992 Sep;14(1):89–98. doi: 10.1016/s0888-7543(05)80288-5. [DOI] [PubMed] [Google Scholar]
- Dear S., Staden R. A sequence assembly and editing program for efficient management of large projects. Nucleic Acids Res. 1991 Jul 25;19(14):3907–3911. doi: 10.1093/nar/19.14.3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dear S., Staden R. A standard file format for data from DNA sequencing instruments. DNA Seq. 1992;3(2):107–110. doi: 10.3109/10425179209034003. [DOI] [PubMed] [Google Scholar]
- Giddings M. C., Brumley R. L., Jr, Haker M., Smith L. M. An adaptive, object oriented strategy for base calling in DNA sequence analysis. Nucleic Acids Res. 1993 Sep 25;21(19):4530–4540. doi: 10.1093/nar/21.19.4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleeson T., Hillier L. A trace display and editing program for data from fluorescence based sequencing machines. Nucleic Acids Res. 1991 Dec 11;19(23):6481–6483. doi: 10.1093/nar/19.23.6481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence C. B., Solovyev V. V. Assignment of position-specific error probability to primary DNA sequence data. Nucleic Acids Res. 1994 Apr 11;22(7):1272–1280. doi: 10.1093/nar/22.7.1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipshutz R. J., Taverner F., Hennessy K., Hartzell G., Davis R. DNA sequence confidence estimation. Genomics. 1994 Feb;19(3):417–424. doi: 10.1006/geno.1994.1089. [DOI] [PubMed] [Google Scholar]
- Smith S., Welch W., Jakimcius A., Dahlberg T., Preston E., Van Dyke D. High throughput DNA sequencing using an automated electrophoresis analysis system and a novel sequence assembly program. Biotechniques. 1993 Jun;14(6):1014–1018. [PubMed] [Google Scholar]
- Staden R. A new computer method for the storage and manipulation of DNA gel reading data. Nucleic Acids Res. 1980 Aug 25;8(16):3673–3694. doi: 10.1093/nar/8.16.3673. [DOI] [PMC free article] [PubMed] [Google Scholar]



