The nuclear pore protein Nup88 binds specifically lamin A, but not B-type lamins, in vitro and in vivo. Expression of green fluorescent protein–lamin A in cells causes a masking of binding sites for Nup88 antibodies, which disappears in cells expressing mutants of lamin A that are associated with laminopathic diseases.
Abstract
Nuclear pore complexes (NPCs) are embedded in the nuclear envelope (NE) and mediate bidirectional nucleocytoplasmic transport. Their spatial distribution in the NE is organized by the nuclear lamina, a meshwork of nuclear intermediate filament proteins. Major constituents of the nuclear lamina are A- and B-type lamins. In this work we show that the nuclear pore protein Nup88 binds lamin A in vitro and in vivo. The interaction is mediated by the N-terminus of Nup88, and Nup88 specifically binds the tail domain of lamin A but not of lamins B1 and B2. Expression of green fluorescent protein–tagged lamin A in cells causes a masking of binding sites for Nup88 antibodies in immunofluorescence assays, supporting the interaction of lamin A with Nup88 in a cellular context. The epitope masking disappears in cells expressing mutants of lamin A that are associated with laminopathic diseases. Consistently, an interaction of Nup88 with these mutants is disrupted in vitro. Immunoelectron microscopy using Xenopus laevis oocyte nuclei further revealed that Nup88 localizes to the cytoplasmic and nuclear face of the NPC. Together our data suggest that a pool of Nup88 on the nuclear side of the NPC provides a novel, unexpected binding site for nuclear lamin A.
INTRODUCTION
The nuclear and cytoplasmic compartments of eukaryotic cells are spatially separated by the nuclear envelope (NE). The NE is composed of an outer nuclear membrane (ONM) and an inner nuclear membrane (INM), the nuclear lamina, and nuclear pore complexes (NPCs). NPCs mediate all molecular exchange between the nucleus and the cytoplasm of interphase cells and, in vertebrates, they consist of ∼30 different nucleoporins (or Nups) (Cronshaw et al., 2002). Nups are typically organized in distinct subcomplexes that form the major building blocks of the NPC. The octagonal core of the NPC is formed by the central framework that is sandwiched between cytoplasmic and nuclear ring moieties (Hinshaw et al., 1992; Akey and Radermacher, 1993; Stoffler et al., 2003; Beck et al., 2004). Filamentous structures decorate the cytoplasmic and nuclear face of the NPC, known as cytoplasmic filaments and nuclear basket, respectively (Lim and Fahrenkrog, 2006; Lim et al., 2008).
The nucleoporin Nup88 resides on the cytoplasmic face of the NPC and is found in a biochemically defined complex with the nucleoporins Nup214 and Nup358 as well as the nuclear export factor CRM1 (Bastos et al., 1997; Fornerod et al., 1997; Roth et al., 2003; Bernad et al., 2004, 2006). In Drosophila, Nup88 was also found intranuclearly in association with chromatin (Capelson et al., 2010). Nup88 is composed of two structural domains: the N-terminal two-thirds of the protein (residues 1–584 in humans) are folded into a β-propeller based on secondary structure prediction, and the C-terminal third (residues 585–741) is predicted to be largely α-helical in structure, thereby forming a coiled-coil (residues 585–651) (Fornerod et al., 1997; Schwartz, 2005). The coiled-coil domain is mediating the interaction with Nup214 (Bastos et al., 1997; Fornerod et al., 1997; Bernad et al., 2004), whereas the β-propeller domain of Nup88 interacts with the NPC-targeting domain of the nucleoporin Nup98, which is thought to localize to both the nuclear and cytoplasmic side of the NPC (Griffis et al., 2003). The Nup88/Nup214 complex does not seem to be directly required for nuclear protein import, but it is crucial for CRM1-mediated nuclear export (Roth et al., 2003; Bernad et al., 2006). Consequently, Nup88 depletion in Drosophila and human cells is causing nuclear accumulation of NF-κB transcription factors, which are CRM1 targets (Uv et al., 2000; Roth et al., 2003; Takahashi et al., 2008). Interestingly, Nup88 is often found overexpressed in a variety of tumor tissues (Martinez et al., 1999; Gould et al., 2002; Agudo et al., 2004; Zhang et al., 2007), although the exact link between Nup88 expression and cancer has remained elusive.
The nuclear lamina is a network of lamins and lamin-binding proteins that are embedded in the INM (Gruenbaum et al., 2005; Dechat et al., 2008; Furukawa et al., 2009). Lamins are type-V intermediate filament (IF) proteins, which have a short N-terminal head domain, a long α-helical coiled-coil domain (the rod domain), and a C-terminal tail domain (Stuurman et al., 1998). This tail domain contains a globular region, which adopts an immunoglobulin (Ig)-like fold (Dhe-Paganon et al., 2002; Krimm et al., 2002a, 2002b). Based on their biochemical properties and their behavior during mitosis, lamins are grouped into A and B types (Gruenbaum et al., 2005). B-type lamins arise from distinct genes and are constitutively expressed in cells throughout development, whereas A-type lamins arise from a single gene (LMNA) by alternative splicing and are only expressed in later stages of development and in differentiated cells (Hutchison et al., 2001; Broers et al., 2006). Studies in Caenorhabditis elegans, Drosophila melanogaster, and cultured human cells suggested that B-type lamins are essential for viability (Lenz-Bohme et al., 1997; Liu et al., 2000; Harborth et al., 2001), whereas mice disrupted in LMNA exhibited normal embryonic development with postnatal growth retardation (Sullivan et al., 1999). Mutations in LMNA as well as several INM proteins, such as emerin or MAN1, are associated with a diverse array of human diseases called laminopathies. The diseases are often tissue-specific and range from muscular dystrophy and lipodystrophy to premature aging syndromes (Broers et al., 2006; Worman and Bonne, 2007). Disease-related mutations are found all over LMNA, but a hot spot for mutations is the Ig fold within the tail domain, which mediates the interaction with lamin-binding proteins (Shumaker et al., 2008).
Lamins and the nuclear lamina are major determinants of nuclear architecture and as such they are involved, for example, in maintaining the structural integrity of the NE, chromatin organization, and spacing of NPCs (Lenz-Bohme et al., 1997; Sullivan et al., 1999; Liu et al., 2000; Broers et al., 2006). An interaction between nucleoporins and B-type lamins has been described for Nup153 and Nup53 (Smythe et al., 2000; Hawryluk-Gara et al., 2005). Here we demonstrate binding of Nup88 to lamin A, both in vitro and in vivo. The interaction sites lie within the N-terminal domain of Nup88 and the tail of lamin A. Our study provides new insights into the relationship between NPCs and lamins and suggests an implication in laminopathic diseases.
RESULTS
Nup88 binds to lamin A
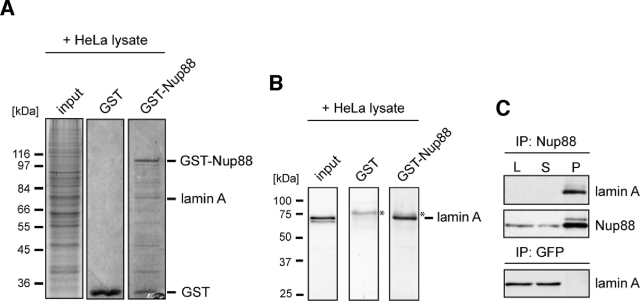
To identify new interaction partners of Nup88, we performed glutathione S-transferase (GST)-pull-down assays using purified, recombinant GST-tagged Nup88 fusion protein immobilized on glutathione sepharose beads. The beads were incubated with a nuclear extract of HeLa S3 cells, and proteins that bound GST-Nup88 or GST were separated by SDS–PAGE and detected using colloidal blue staining. Protein bands that were visible in the GST-Nup88 pull-down, but not in the GST control (Figure 1A), were identified by mass spectrometry. This analysis identified the polypeptide of ∼74 kDa protein as lamin A (Figure 1A). To further confirm that the ∼74 kDa polypeptide bound to GST-Nup88 is in fact lamin A, we immunoblotted the GST pull-down assays with a polyclonal antibody specific against lamin A (not recognizing lamin C). As shown in Figure 1B, lamin A copurified with GST-Nup88 but not with GST. A protein with slightly higher molecular weight than lamin A (indicated by *), however, is recognized unspecifically by the lamin A antibody in both pull-down assays (Figure 1B).
FIGURE 1:
Lamin A is a novel interaction partner of Nup88. Bacterially expressed GST-Nup88 and GST were bound to glutathione sepharose beads and incubated with HeLa S3 nuclear extract. Bound fractions were analyzed by SDS–PAGE, colloidal blue staining, and mass spectroscopy (A) and immunoblotting using anti–lamin A antibodies (B). Lamin A was bound to GST-Nup88, but not to GST. (C) HeLa cell lysates were immunoprecipitated with anti-Nup88 antibodies, and the lysate (L), immune supernatant (S), and immune precipitate (P) were analyzed by SDS–PAGE and immunoblotted with anti-Nup88 and anti–lamin A antibodies. Lamin A coimmunoprecipitated with Nup88.
Next, we carried out immunoprecipitation assays with antibodies directed against Nup88 to confirm the interaction between Nup88 and lamin A in a cellular context. Lysates from whole HeLa cells were incubated with monoclonal anti-Nup88 antibodies directed against the N-terminal domain, precipitated and the lysate (L), supernatant (S), and pellet (P) fractions were separated by SDS–PAGE. After transfer to a polyvinylidene difluoride (PVDF) membrane, the membrane was probed with antibodies against Nup88 and lamin A, respectively. As shown in Figure 1C, lamin A was readily coprecipitated with Nup88 from HeLa whole-cell lysates but not with anti-GFP (green fluorescent protein) antibodies. Together our data therefore indicated that Nup88 associates with lamin A in vitro and in vivo.
The N-terminal domain of Nup88 binds the tail domain of lamin A
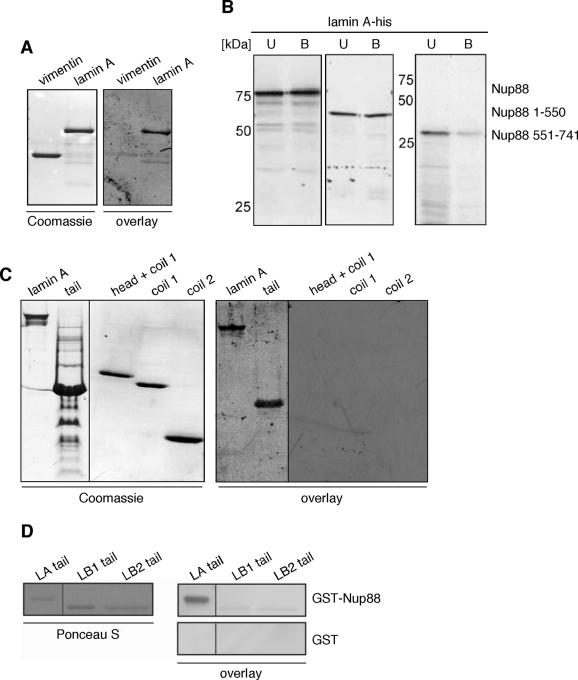
To confirm that Nup88 and lamin A are interacting, we carried out blot-overlay assays using recombinantly expressed lamin A and in vitro transcribed/translated 35S-labeled Nup88 (Figure 2A). Nup88 and lamin A both harbor coiled-coil domains and, to rule out unspecific coiled-coil interactions between the two proteins, we used bacterially expressed vimentin as control. Vimentin is a cytoplasmic IF protein with a domain organization similar to that of lamin A (Herrmann et al., 2007). Similar amounts of vimentin and lamin A were subjected to SDS–PAGE (Figure 2A, Coomassie), transferred to a PVDF membrane, and overlaid with 35S-labeled Nup88. As shown in Figure 2A (overlay), Nup88 associated with lamin A, but not vimentin, indicating that Nup88 binds lamin A specifically.
FIGURE 2:
The N-terminal domain of Nup88 is interacting with the tail domain of lamin A. (A) Blot-overlay assay employing bacterially expressed and purified vimentin and lamin A and in vitro expressed and 35S-labeled Nup88. Samples of purified vimentin and lamin A were separated by SDS–PAGE, stained with Coomassie blue (left panel), or transferred to a PVDF membrane and overlaid with 35S-labeled Nup88 (right panel). 35S-labeled Nup88 was visualized by autoradiography. (B) Bacterially expressed his-tagged lamin A was bound to nickel beads and incubated with in vitro synthesized 35S-labeled, full-length, N-terminal (residues 1–550) or C-terminal (residues 551–741) Nup88. Unbound and bound fractions were analyzed by SDS–PAGE and autoradiography. 35S-labeled, full-length Nup88 and N-terminal 35S-Nup88 were binding to lamin A-his, whereas C-terminal 35S-Nup88 was not able to bind lamin A-his. (C) Blot-overlay assay using bacterially expressed and purified lamin A and lamin A fragments and in vitro expressed and 35S-labeled Nup88. Samples of purified lamin A were separated by SDS–PAGE, stained with Coomassie blue (left panel), or transferred to a PVDF membrane and overlaid with 35S-labeled Nup88 (right panel). 35S-labeled Nup88 was visualized by autoradiography. (D) Blot-overlay assay using bacterially expressed and purified tail domains of lamin A, lamin B1 and B2, and bacterially expressed and purified GST-Nup88 as well as GST alone. Samples of purified tail domain of lamin A (LA tail), lamin B1 (LB1 tail), and B2 (LB2 tail) were separated by SDS–PAGE, transferred to a PVDF membrane, stained with Ponceau S (left panel), and overlaid with purified GST-Nup88 and GST, respectively (right panel). Bound Nup88 was visualized by Western blot analysis with anti-Nup88 antibody. No binding of GST to the lamin tail domains was observed by immunoblotting with an anti-GST antibody.
Next we aimed to determine the domains of Nup88 and lamin A that are required for the interaction between the two proteins. We therefore performed in vitro binding assays using purified recombinant his-tagged lamin A that was immobilized on Ni-sepharose beads and incubated with distinct in vitro synthesized 35S-labeled Nup88 fragments, in other words, full-length Nup88, the N-terminal domain (residues 1–550), and the C-terminal domain (residues 551–741), respectively (Figure 2B). Bound (B) and unbound (U) fractions were separated by SDS–PAGE, transferred to a PVDF membrane, and analyzed by autoradiography. As shown in Figure 2B, Nup88 (left panel) and the N-terminal domain (middle panel) both interacted with lamin A, but not so the C terminus (right panel).
To define which domain of lamin A interacts with Nup88, we next carried out blot-overlay assays using recombinantly expressed lamin A and fragments thereof and in vitro transcribed/translated 35S-labeled Nup88. We used full-length lamin A, the tail domain of lamin A (tail, residues 243–664), the head domain with coil 1 (head + coil 1, residues 1–263), the coil 1 (residues 24–263), and the coil 2 (residues 264–402) of lamin A, respectively. Similar amounts of proteins were subjected to SDS–PAGE (Figure 2C, Coomassie) and overlaid with 35S-labeled Nup88 after transfer to a PVDF membrane. We found that Nup88 associated with lamin A and the lamin A tail but not with fragments of the head or the rod domain of lamin A (Figure 2C, overlay).
To further confirm the specific binding of Nup88 to lamin A, we carried out blot-overlay assays using recombinantly expressed lamin-A tail (residues 243–664) as well as lamin B1 (residues 245–568) and lamin B2 tail (residues 238–600) and purified, recombinantly expressed GST-Nup88. Similar amounts of the tail domains were subjected to SDS–PAGE (Figure 2D, Ponceau S), transferred to a PVDF membrane, overlaid with GST-Nup88, and probed with a monoclonal antibody (mAb) against the N terminus of Nup88. This revealed that Nup88 is specifically binding to the lamin-A tail, but not to the lamin-B1 and lamin-B2 tails (Figure 2D, overlay). No binding of either tail domain to GST was observed. Together our data indicate that Nup88 is specifically binding to A-type, but not to B-type lamins and that the interaction involves the N-terminal domain of Nup88 as well as the tail domain of lamin A.
Nup88 localizes to both sides of the NPC
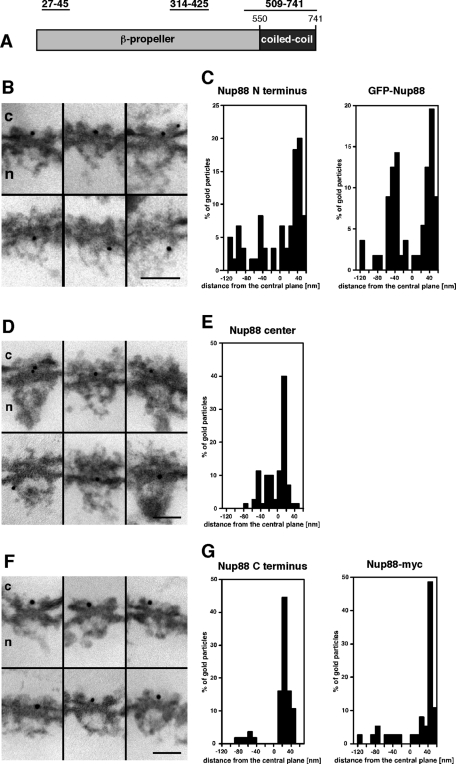
Nup88 was previously mapped to the cytoplasmic face of the NPC in Xenopus oocyte nuclei using an antibody recognizing the C terminus of Nup88 (Bernad et al., 2004). The interaction between Nup88 and lamin A suggested that an additional pool of Nup88 might reside on the nuclear side of the NPC. In an effort to more precisely determine the position of Nup88 within the NPC, we performed immuno-electron microscopy (immuno-EM) using Xenopus oocyte nuclei and domain-specific antibodies against human Nup88. Xenopus oocyte nuclei were isolated manually and incubated with antibodies directed against the N terminus (residues 27–45), the central region (residues 314–425), and the C terminus of Nup88 (residues 509–741), respectively (Figure 3A), that were directly conjugated to 8-nm colloidal gold and processed for thin-sectioning EM. As shown in Figure 3B, antibodies against the N terminus of Nup88 were recognizing epitopes on both the nuclear and the cytoplasmic side of the NPC. Quantification of the gold particle distribution with respect to the central plane of the NE revealed that ∼40% of the gold particles were associated with the nuclear face of the NPC at a mean distance of -53.8 nm ± 22.6 nm from the central plane (Figure 3C). Together with corresponding mean radial distance of 37.1 nm ± 13.9 nm, this distance corresponds to an epitope near the nuclear ring moiety of the NPC. The remaining 60% of the gold particles were found on the cytoplasmic side of the NPC, with a mean distance of 34.7 nm ± 15.1 nm from the central plane and a mean radial distance of 22.2 nm ± 15.1 nm.
FIGURE 3:
Domain topology of endogenous and ectopically expressed Nup88 within the NPC. Nuclei were isolated manually and labeled with antibodies directly conjugated to 8-nm colloidal gold. (A) Schematic representation of Nup88 domain organization and antibody epitope. (B and D) Nup88 localizes to both the cytoplasmic and the nuclear side of the NPC with a polyclonal Nup88 antibody directed against the N terminus of Nup88 and a mAb against its central region. Shown are selected examples of gold-labeled NPCs in cross-section. c, cytoplasm; n, nucleus. (C and E) Quantitative analysis of the gold particle distribution associated with the NPC using an antibody against the N terminus of Nup88 and the central region. Sixty (N terminus) and 70 (center) gold particles were scored. (C) Quantitative analysis of the gold particle distribution associated with the NPC using an antibody against the N-terminal GFP-tag of Nup88. Fifty-five gold particles were scored. (F) Nup88 is found mainly on the cytoplasmic side of the NPC with a monoclonal Nup88 antibody directed against an epitope in the C terminus of Nup88. Shown are selected examples of gold-labeled NPCs in cross-section. c, cytoplasm; n, nucleus. (G) Left: Quantitative analysis of the gold particle distribution associated with the NPC using antibodies against the Nup88 C terminus. Fifty-seven gold particles were scored. Right: Quantitative analysis of the gold particle distribution with anti-myc antibodies. Thirty-eight gold particles were scored. Scale bars, 100 nm.
To confirm the localization of the N terminus of Nup88, we expressed N-terminally GFP-tagged Nup88 in Xenopus oocytes (GFP-Nup88). Plasmids were microinjected into the oocytes, and the localization of the incorporated proteins was determined by using a monoclonal anti-GFP antibody directly coupled to 8-nm colloidal gold. The anti-GFP antibody recognized epitopes both on the cytoplasmic and the nuclear side of the NPC. As shown in Figure 3C, quantification of the labeling pattern relative to the central plane of the NPC revealed that 51% of the particles were detected on the cytoplasmic side at a mean distance of 38.1 nm ± 12.2 nm and a mean radial distance of 27.3 nm ± 15.4 nm and that 49% of the gold particles were found on the nucleoplasmic side with a mean distance of –49.8 nm ± 26.1 nm from the central plane and a mean radial distance of 24.2 nm ± 15.4 nm, consistent with the localization data of the untagged N terminus of Nup88. These data suggest that indeed a pool of Nup88 is localizing to the nucleoplasmic side of the NPC.
Next, we determined the position of the central region of Nup88 within the NPC. As shown in Figure 3D, these antibodies against the central region of Nup88 were recognizing epitopes on both the nuclear and the cytoplasmic side of the NPC. Quantification of the gold particle distribution revealed that ∼40% of the gold particles were associated with the nuclear face of the NPC at a mean distance of –31.9 nm ± 16.7 nm from the central plane (Figure 3E). Together with corresponding mean radial distance of 25.7 nm ± 19.6 nm, this distance corresponds to an epitope near the nuclear ring moiety of the NPC. The remaining 60% of the gold particles were found on the cytoplasmic side of the NPC, with a mean distance of 16.8 nm ± 8.6 nm from the central plane and a mean radial distance of 8.2 nm ± 8.7 nm.
To revise the localization of Nup88’s C terminus by immuno-EM, we next used antibodies against the C terminus of Nup88 conjugated to 8-nm colloidal gold. These antibodies recognized epitopes almost exclusively on the cytoplasmic side of the NPC (Figure 3F). Quantification of the labeling pattern (Figure 3G) relative to the central plane revealed that 87.7% of the gold particles were detected at a mean distance of 30.7 nm ± 9.2 nm from the central plane with a radial distance of 16.5 nm ± 12.8 nm, which is consistent with a previous ultrastructural localization study of the C-terminus of Xenopus laevis Nup88 (Bernad et al., 2004).
We further localized the position of recombinant Nup88 with a C-terminal myc-tag (Nup88-myc) using anti-myc antibodies directly coupled to 8-nm colloidal gold. Quantification of the labeling distribution (Figure 3G) revealed that 76.3% of the gold particles were associated with the cytoplasmic side of the NPC (mean distance from the central plane of the NPC of 41.6 nm ± 12.2 nm, with a corresponding radial distance of 33.2 nm ± 22 nm). Together our data indicate that the C terminus of Nup88 is more accessible on the cytoplasmic as compared with the nuclear side of the NPC, whereas the N terminus of Nup88 and its central region are roughly equally accessible on both sides. We therefore conclude that Nup88 is residing on both sides of the NPC.
Expression of GFP-lamin A is masking epitopes for Nup88 antibodies
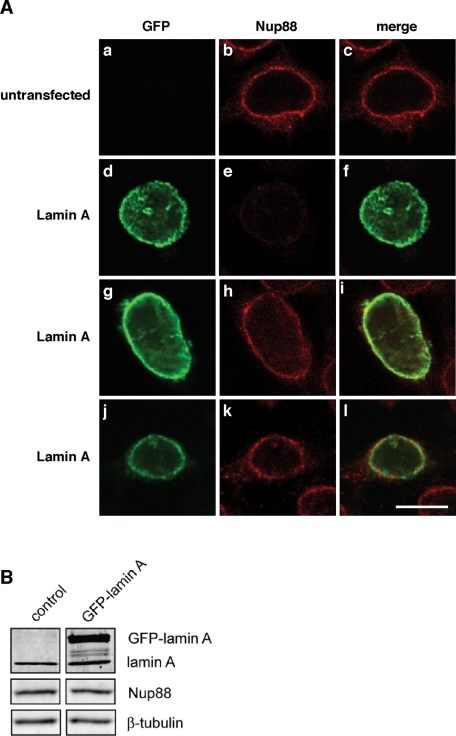
After having established that Nup88 interacts with lamin A and that Nup88 localizes to both sides of the NE, we next examined whether ectopic expression of lamin A affected Nup88 in HEK 293 cells. HEK cells were transfected with GFP-tagged lamin A, and the effect on Nup88 was analyzed by indirect immunofluorescence using antibodies against the center of Nup88 (residues 314–425). GFP-lamin A was readily expressed and localized to the NE and the nucleoplasm 24 h posttransfection (Figure 4A, d, g, and j). Nup88 in untransfected HEK 293 cells was found predominantly at NPCs as indicated by a nucleoporin-typical rim staining of the NE (Figure 4A, b), whereas expression of GFP-lamin A resulted in barely detectable Nup88 (Figure 4A, e). In contrast, GFP-lamin A expression had no influence on the accessibility of epitopes recognized by the mAb 414, which is directed against phenylalanine–glycine (FG)-repeat nucleoporins (Supplemental Figure S1, second row). Moreover, expression of GFP-lamin B1 did not affect the accessibility of Nup88 epitopes (Figure S1, first row), indicating that GFP-lamin A expression specifically constrained epitopes recognized by the antibodies against residues 314–425 of Nup88. Masking of the epitopes was observed in 57.8% of the transfected cells (680 cells counted). Masking occurred with different intensity (Figure 4A, e, and Figure S1, third and fourth rows), and quantification of the fluorescence signal revealed a reduction of 35% in cells transfected with GFP-lamin A as compared with nontransfected cells.
FIGURE 4:
Expression of GFP-lamin A is masking an antibody epitope within the N terminus of Nup88. (A) HEK cells transfected with GFP-lamin A (d–f, g–i, and j–l) were prepared for immunofluorescence 24 h after transfection using mAbs directed against residues 314–425 of Nup88 (e and f). Cells transfected with GFP-lamin A showed a decreased signal for Nup88 staining as compared with untransfected control cells (a–c). The masking can be circumvented by the use of a polyclonal antibody directed against residues 27–45 in the N terminus of Nup88 (h and i) or methanol fixation in combination with the monoclonal Nup88 antibody against residues 314–425 (k and l). Scale bars, 10 μm. (B) Protein levels of Nup88 were analyzed in HEK cells transfected with GFP-lamin A or mock transfected control cells. No changes in protein levels of Nup88 were observed when compared with β-tubulin.
To confirm that the loss of Nup88 labeling was in fact due to epitope masking and not due to reduced expression of Nup88, we performed Western blot analysis to determine Nup88 levels in HEK 293 cells transfected with GFP-lamin A. As shown in Figure 4B, Nup88 and endogenous lamin A expression were similar in control and GFP-lamin A–transfected cells. Furthermore, epitope masking of Nup88 was not observed when we used polyclonal antibodies against residues 27–45 in the N terminus of Nup88 (Figure 4A, h) or in methanol-fixed cells that were stained with antibodies against residues 314–425 (Figure 4A, k). Together, these results demonstrate that lamin A binds to Nup88 in vivo and that the binding site is within residues 314–425 of Nup88, which is in good agreement with our in vitro data that revealed the tail of lamin A to interact with the N-terminal domain of the Nup88 (Figure 2B).
Mutations in the Ig fold of lamin A abolish the interaction with Nup88 in cells
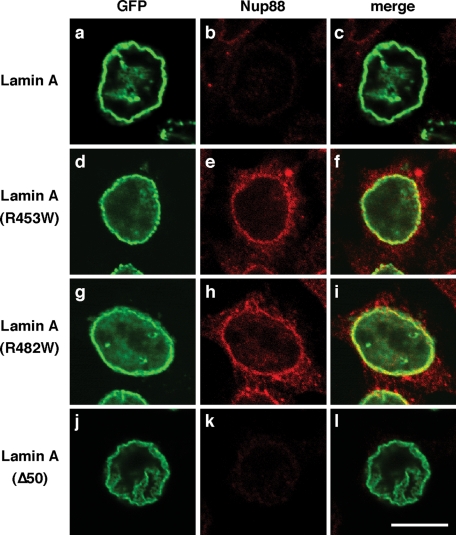
To study the effect of disease-related mutations in lamin A on epitope masking of Nup88, we next expressed GFP-lamin A and GFP-lamin A mutants in HEK 293 cells. The Ig fold in the lamin A tail is a hot spot for mutations associated with laminopathies (Broers et al., 2006; Worman and Bonne, 2007), and we analyzed two mutants in this domain: an arginine-to-tryptophan mutation in residue 453 (R453W) that causes Emery–Dreifuss muscular dystrophy (EDMD), and an arginine 482 mutated to tryptophan (R482W) that is associated with Dunnigan-type familial partial lipodystrophy (FPLD). Furthermore, we used a lamin-A mutant with a deletion of 50 amino acids in the tail domain of lamin A downstream of the Ig fold (lamin AΔ50), a mutation that is associated with Hutchinson–Gilford progeria syndrome (HGPS). HEK 293 cells were transiently transfected with GFP-lamin A(R453W), GFP-lamin A(R482W), and GFP-lamin AΔ50, respectively, and were immunolabeled with our mAbs against residues 314–425 of Nup88. We found that expression of GFP-lamin A(R453W) (Figure 5, d–f) and GFP-lamin A(R482W) (Figure 5, g–i) did not coincided with epitope masking of Nup88 antibodies, in contrast to the effect of wild-type GFP-lamin A (Figure 5, a–c) and GFP-lamin AΔ50 (Figure 5, j–l), expression, respectively. These data indicate that the R453W and the R482W mutations in lamin A disrupt the association with Nup88, whereas GFP-lamin AΔ50 is still able to bind Nup88, and the Ig fold of lamin A (residues 436–544) at least in part is contributing to the binding of Nup88. Consistent with this notion, we observe a weaker binding of the lamin A Ig fold as compared with the lamin A tail and no binding of the R453W and R482W Ig fold mutants in vitro (Supplemental Figure S2).
FIGURE 5:
EDMD-associated lamin A(R453W) and FPLD-associated lamin A(R482W) mutants do not interact with Nup88. HEK cells transfected with GFP-lamin A(R453W) (d–f), GFP-lamin A(R482W) (g–i), GFP-lamin AΔ50 (j–l), or wild-type GFP-lamin A (a–c) were prepared for immunofluorescence 24 h posttransfection using mAbs directed against residues 314–425. Epitope masking was observed in cells transfected with wild-type GFP-lamin A (b) and GFP-lamin AΔ50 (k), respectively, but not in cells expressing the GFP-tagged lamin A(R453W) (e) and lamin A(R482W) (h) mutants.
DISCUSSION
NPCs and the nuclear lamina are major constituents of the NE. We report here a new link between NPCs and lamins and show that the nucleoporin Nup88 binds to the nuclear IF protein lamin A, both in vitro and in vivo. The interaction between Nup88 and lamin A involves the N terminus of Nup88 and the tail domain of lamin A. Importantly, Nup88 binds specifically to lamin A but not to lamin B1 and B2 or to the cytoplasmic IF protein vimentin. The interaction of Nup88 with lamin A appears disrupted in the presence of two mutations in the Ig fold that are found in patients with EDMD and FPLD but not by an HGPS-related mutation. This new information about a novel interaction between NPCs and lamins will be an important consideration when deciphering underlying molecular mechanisms of laminopathic diseases.
Lamin A is a novel interaction partner of Nup88
Our biochemical search for novel Nup88 interacting proteins has led to the identification of the nuclear IF protein lamin A as its binding partner (Figures 1 and 2, A–C). The observed interaction seems to be specific for nuclear lamin A, because Nup88 is not interacting with the cytoplasmic IF protein vimentin in a blot-overlay assay (Figure 2A) and with nuclear lamins B1 and B2 (Figure 2D). Nup88, however, has been implicated in CRM1-mediated nuclear protein export, and lamin A is imported into the nucleus in an importin α/β-dependent manner (Loewinger and McKeon, 1988; Fornerod et al., 1997; Adam et al., 2008), suggesting that the Nup88–lamin A complex might somewhat resemble a nucleocytoplasmic transport–related complex. We could exclude this transport-related complex, however, as CRM1, importin α, and β, despite their presence in the rabbit reticulocyte lysate used to produce 35S-labeled lamin A, did not copurify with the Nup88–lamin A complex in solution-binding assays (Supplemental Figure S3).
The previous mapping of Nup88 to the cytoplasmic face of the NPC (Bernad et al., 2004) seems at first glance at odds with a potentially nuclear association with lamin A. We therefore performed a systematic immuno-EM analysis using a combination of domain-specific antibodies and the expression of epitope-tagged versions of Nup88 in Xenopus oocytes. By a similar strategy we have previously successfully unraveled the complex organization of several nucleoporins within the three-dimensional architecture of the NPC (Fahrenkrog et al., 2002; Paulillo et al., 2005; Schwarz-Herion et al., 2007). Here we revealed that Nup88 localizes to both sides of the NPC (Figure 3). Whereas the N terminus and the central region of Nup88 are roughly equally accessible on the cytoplasmic and the nuclear face of the NPC (Figure 3, B–E), the C terminus of Nup88 appears to be largely masked on the nuclear side of the NPC (Figure 3, F and G), which is consistent with a previous study (Bernad et al., 2004). The unequal accessibility of the C terminus of Nup88 would be best explained by the existence of different Nup88 complexes on the cytoplasmic and nuclear side of the NPC, respectively. Nup88 is known to form a complex with Nup214 and Nup358 (Bastos et al., 1997; Fornerod et al., 1997; Roth et al., 2003; Bernad et al., 2004, 2006), and Nup214 and Nup358 are found exclusively anchored on the cytoplasmic side of the NPC (Walther et al., 2002; Paulillo et al., 2005), suggesting that these are the cytoplasmic binding partners of Nup88. Our data shown here suggest lamin A as nuclear binding partner of Nup88. Another putative nuclear binding partner might be Nup98, as Nup98 localizes to both sides of the NPC and was shown to interact with Nup88 (Griffis et al., 2003). It has not been studied, however, on which side of the NPC the interaction between Nup88 and Nup98 occurs. It will also be interesting to see if other nucleoplasmic nucleoporins, such as Nup153 and Tpr (Walther et al., 2001; Fahrenkrog et al., 2002; Frosst et al., 2002; Krull et al., 2004), exhibit binding sites for Nup88.
A further characterization of the interaction between Nup88 and lamin A showed that Nup88’s N-terminal two thirds are binding the tail of lamin A (Figure 2). Within the tail of lamin A, the Ig fold region is frequently mutated in laminopathies (Worman and Bonne, 2007). The R453W and R482W mutations in lamin A are associated with EDMD and FPLD, respectively, and the interaction of Nup88 with these two lamin A mutants is abrogated in cells (Figure 5) and in vitro (Supplemental Figure S2). Together these data indicate that, within the tail domain of lamin A, the Ig fold region is implicated in the binding between Nup88 and lamin A. Whether the Ig fold is the exclusive binding site for Nup88 necessitates further investigations. For future studies, it will be interesting to see whether Nup88 plays a role in the molecular mechanisms underlying EDMD and FPLD.
The interaction between Nup88 and lamin A appears not affected in the presence of lamin AΔ50 (Figure 5), which is found in patients suffering from HGPS (De Sandre-Giovannoli et al., 2003; Eriksson et al., 2003) as well as in normal aged cells (Scaffidi and Misteli, 2006). Interestingly, the expression of Nup88 was found to be down-regulated in both HGPS and normal aged cells (Ly et al., 2000), which might be due to the presence of lamin AΔ50 or due to reduced levels of wild-type lamin A. Along this line, we found that depletion of lamin A from HeLa cells by RNA interference coincided with a decrease in Nup88 protein levels (Supplemental Figure S3B). Nup88 protein levels were reduced by 40% in cells treated with siRNA against lamin A as compared with cells treated with control siRNA (cyclophilin B) or untreated cells (Supplemental Figure S3C). A likely explanation for this observation is that reduced lamin A levels lead to unstable free Nup88 that is more rapidly degraded, as seen for other nucleoporins (Bernad et al., 2004). Alternatively, the observed reduction of Nup88 protein levels might be due to changes in gene expression due to the lack of lamin A, as it is involved in transcriptional regulation (Andres and Gonzalez, 2009).
Expression of GFP-lamin A in HEK 293 cells caused reduced Nup88 staining with antibodies against the residues 314–425 (Figures 4 and 5). This reduced staining is likely due to epitope masking rather than a decrease in Nup88 expression (Figure 4B). Antigen masking can occur as a result of conformational changes in the antigen, posttranslational modifications, or interaction with other macromolecules that physically block the epitope. Our data suggest that excess lamin A physically blocks Nup88 epitopes. A high-affinity antibody is typically capable of displacing interacting proteins from an antigen by mass action (Tunnah et al., 2005). Chemical cross-linking can prevent such displacement and might explain why the masking is observed in formaldehyde-fixed samples but not in methanol-fixed samples (Supplemental Figure S1). Interestingly, the masking phenotype is lost upon expression of lamin A(R453W) and lamin A(R482W) (Figure 5), suggesting that the mutant lamin A variants are not recruited to Nup88.
Future studies are required to elucidate the function of the Nup88–lamin A interaction. In our immunoprecipitation assays we used conditions that seem to extract the soluble pool of lamin A rather than its pool in the nuclear lamina (Moir et al., 2000; Muralikrishna et al., 2004). It will therefore be interesting to see whether Nup88 in fact interacts with the soluble and the lamina pool of lamin A and if LAP2α, which binds the nucleoplasmic lamin A, is involved in the Nup88–lamin A complex.
MATERIALS AND METHODS
Constructs
Plasmids pEGF (epidermal growth factor)P-lamin A and pEGFP (enhanced green fluorescent protein)-lamin B1 were provided by Robert D. Goldman (Northwestern University, Chicago, IL), and plasmid pEGFP-lamin AΔ50 by Harald Herrmann (German Cancer Research Center, DKFZ, Heidelberg). N-terminally tagged GST-Nup88 was cloned into NcoI/KpnI-cut pGEX-CS vector (Amersham-Pharmacia, Little Chalfont, UK) and N-terminally tagged GFP-Nup88 into KpnI-cut pEGFP-C1 (BD Bioscience, Clontech, Allschwil, Switzerland). C-terminally myc-his–tagged Nup88 was cloned into KpnI/XbaI-cut pcDNA3.1 myc-His A (Invitrogen, Paisley, UK). The pET-lamin A Ig was constructed with HindIII and XhoI restriction sites into pET24a. pET-lamin A Ig(R453W) and pEGFP-lamin A(R453W) as well as pET-lamin A Ig(R482W) and pEGFP-lamin A(R482W) constructs were provided by Teiba Al-Haboubi (Biozentrum, Basel, Switzerland). They were generated by site–directed mutagenesis with primers (5′-GAGGGCAAGTTTGTCTGGCTGCGCAACAACTCC) and (3′-GGAGTTGTTGCGCAGCCAGACAAACTTGCCCTC) as well as primers (5′-CCCTTGCTGACTTACTGGTTCCCACCAAAGTTC) and (3′-GAACTTTGGTGGGAACCAGTAAGTCAGCAAGGG) on pET-lamin A Ig and pEGFP-lamin A, respectively.
Antibodies
A peptide-specific antibody against residues 27–45 (LREGLKNQSPTEAEKPASS-C) of human Nup88 (1:1000 [vol/vol] for immunofluorescence) was generated in rabbits and affinity-purified. The polyclonal anti–lamin A antibody (1:500 [vol/vol]) was provided by Robert D. Goldman (Northwestern University, Chicago, IL) (Dechat et al., 2007). Further primary antibodies were the mAbs against amino acids 314–425 of human Nup88 (1:500 [vol/vol]; BD Bioscience, Pharmingen, Allschwil, Switzerland), amino acids 509–741 of human Nup88 (1:25 [vol/vol]; Novocastra, Newcastle, UK), mAB414 (1:2000 [vol/vol]; Covance, Berkeley, CA), anti–β-tubulin (1:2000 [vol/vol]; Chemicon, Billerica, MA), anti–importin α (1:1000 [vol/vol]; Sigma Aldrich, St. Louis, MO), anti–importin β (1:1000 [vol/vol]; BD Biosciences, PharMingen, Allschwil, Switzerland), as well as the polyclonal anti-CRM1 (1:1000 [vol/vol]; Abcam, Cambridge, UK), anti-GFP (Santa Cruz Biotechnology, Santa Cruz, CA), and anti-GST (Abcam, Cambridge, UK) antibodies. Secondary antibodies include anti–mouse IgG-Alexa 568 and anti–rabbit IgG-Alexa 568 from Molecular Probes (Paisley, UK) and used at 1:1000 [vol/vol].
Cell culture and transfections
HEK cells were grown in DMEM supplemented with 10% fetal bovine serum plus penicillin and streptomycin. Cells were transfected using TransIt-293 transfection reagent (Mirus Bio LLC, Madison, WI) following the instructions of the manufacturer.
Preparation of nuclei from HeLa cell suspension cultures by osmotic swelling
HeLa S3 cells were grown in suspension and harvested by centrifugation at 600 × g for 5 min. The pelleted cells were washed in 5 volumes of prechilled Earle’s balanced salt solution, resuspended in 10 volumes of RSB buffer (0.01 M NaCl, 1.5 mM MgCl2, 0.01 M Tris-HCl, pH 7.4) and incubated for 10 min on ice. Cells with swollen cytoplasm were homogenized in a prechilled glass Dounce homogenizer and examined in the phase contrast microscope. The homogenized cell suspension with a free nuclei to intact cell ratio of 9 or greater was centrifuged at 1000 × g for 3 min to pellet the nuclei and washed in 10 volumes of RSB buffer.
Preparation of HeLa S3 nuclear extract
HeLa S3 nuclei (3 × 106) were dissolved in 300 μl of lysis buffer (1× phosphate-buffered saline [PBS], 150 mM KAc, 5 mM MgAc, 5 mM CaCl2, DNase at 50 μg/ml), homogenized in a prechilled Dounce homogenizer, and incubated with GST and GST-Nup88 bound to glutathione sepharose beads, respectively.
Immunofluorescence
HEK 293 cells were grown on poly-lysine–coated glass coverslips and fixed either in 2% formaldehyde for 15 min or –20°C methanol for 5 min, washed three times for 10 min with PBS, and permeabilized with PBS containing 1% bovine serum albumin (BSA) and 0.2% Triton X-100 for 5 min on ice. Next the cells were washed three times for 10 min in PBS containing 1% BSA, incubated with the appropriate primary antibodies for 1 h, washed three times in PBS containing 1% BSA, incubated with the appropriate secondary antibodies for 1 h, washed four times for 10 min with PBS, mounted with a drop of Mowiol, and stored at 4°C until viewed. Cells were imaged using a confocal laser-scanning microscope (Leica TCS NT/SP5; Vienna, Austria). Images were recorded using the microscope system software and processed using Adobe Photoshop (Adobe Systems, Mountain View, CA).
Gel electrophoresis and immunoblotting
Cells were resuspended in lysis buffer containing 50 mM Tris-HCl, pH 7.8, 150 mM NaCl, 1% Nonidet P-40, and protease inhibitor cocktail tablets (Roche, Basel, Switzerland). Proteins were separated by SDS–PAGE (10%) and transferred onto a PVDF membrane. The membrane was incubated in I-Block solution (Tropix, Bedford, MA) containing 0.1% Tween-20 (blocking solution) overnight at 4°C, incubated in blocking solution containing a primary antibody directed against either Nup88 (1:800 [vol/vol]) or anti–β-tubulin (1:1000 [vol/vol]; Chemicon, Billerica, MA) for 1 h followed by washing three times with PBS containing 0.1% Tween-20. Next, the membrane was incubated in the dark with anti–mouse IRDye 800 (1:10,000 [vol/vol]; LI-COR Biosciences, Lincoln, NE) in blocking solution. Images were recorded using the Odyssey infrared imaging system and analyzed by Odyssey Application Software v2 (LI-COR Biosciences).
Immunoprecipitation
Subconfluent HeLa cells (3 × 106) were trypsinized, washed with PBS, resuspended in 160 μl of lysis buffer (50 mM Tris-HCl, pH 7.4, 250 mM NaCl, 0.1% Triton X-100, 2 mM EDTA-Na2, 10% glycerol, and protease inhibitor;), vortexed, and incubated for 10 min at 37°C. The cells were pelleted at 16,000 × g for 10 min, and the supernatant was transferred to a fresh tube. The supernatant was cleared with 20 μl of protein G-agarose beads (Santa Cruz Biotechnology) and incubated for 30 min at 4°C, centrifuged at 1000 × g for 30 s at 4°C, and transferred to a fresh tube. Mouse monoclonal anti-Nup88 antibody (1 μg; BD Bioscience, PharMingen) was added and incubated for 2 h on ice. Prewashed protein G-agarose slurry (40 μl) was added to the lysate and incubated at 4°C on a rocker platform for 1 h. The immunoprecipitate was collected by centrifugation at 1000 × g for 30 s at 4°C, and the supernatant was carefully removed and kept to analyze as unbound fraction. The pellet was washed three times with lysis buffer, resuspended in electrophoresis sample buffer, boiled at 95°C for 5 min, and subjected to electrophoresis and Western blotting.
In vitro transcription and translation
Nup88, lamin A, and fragments of both proteins were obtained by in vitro transcription and translation using the TNT-coupled reticulocyte lysate system (Promega, Madison, WI) in the presence of residue l-[35S]methionine-cysteine (Amersham Bioscience, Uppsala, Sweden) following the instructions of the manufacturer.
Expression of recombinant Nup88 and lamin A
GST, GST-Nup88, and lamin A-his were expressed in Escherichia coli BL21 (DE3) cells. Protein expression was induced with 0.5 mM isopropyl-beta-d-thiogalactopyranoside (IPTG) for 5 h at 25°C. Cells were lysed by sonication (Branson Digital Sonifier; Branson Ultrasonics Corporation, Danbury, CT) in 2× PBS containing 1% Triton X-100 and protease inhibitor (Thermo Scientific, Waltham, MA). Lysed cells were spun at 60,000 × g for 1 h, and the cleared lysates were stored at –80°C.
In vitro binding assays
The in vitro interaction between Nup88 and lamin A was tested by binding GST and GST-Nup88 to glutathione sepharose beads (Amersham Bioscience) or lamin A-his to Ni-sepharose 6 fast flow beads (GE Healthcare, formerly Amersham Biosciences) for 1 h at 4°C. Beads were washed twice with 2× PBS containing 1% Triton X-100 and protease inhibitor (Thermo Scientific). In vitro produced Nup88, Nup88 (1–550), Nup88 (551–741), lamin A tail (residues 243–664), lamin A Ig (residues 436–545), lamin A Ig(R453W), and lamin A Ig(R482W) were allowed to bind for 16–20 h at 4°C. After binding, beads were washed twice in 2× PBS containing 1% Triton X-100 and protease inhibitor followed by two washes in 2× PBS. Samples were eluted in 50 μl of SDS sample buffer, analyzed on a 10% SDS–polyacrylamide gel, and detected by autoradiography.
Blot-overlay assay
Purified vimentin (provided by Harald Herrmann, DKFZ, Heidelberg, Germany) and lamin A (provided by Dale K. Shumaker, Northwestern University, Chicago, IL) were resolved on SDS–PAGE, transferred to a PVDF membrane, blocked with blocking solution over night at 4°C and incubated with in vitro transcribed/translated 35S-labeled Nup88. After washing, binding of Nup88 was visualized by autoradiography. Similarly, purified lamin A tail (provided by Dale K. Shumaker) and rod fragments (provided by Larisa Kapinos, Biozentrum, University of Basel, Switzerland) were resolved and incubated with in vitro transcribed/translated 35S-labeled Nup88.
Purified tail domains of lamin A, lamin B1 and lamin B2 (provided by Dale K. Shumaker) were resolved on SDS–PAGE, transferred to a PVDF membrane, blocked with blocking solution overnight at 4°C, and incubated with bacterially expressed and purified GST-Nup88. After washing, binding of Nup88 was visualized by Western blot analysis with anti-Nup88 antibody.
Immuno-EM
Mature (stage 6) oocytes were surgically removed from female Xenopus laevis, and their nuclei were isolated and labeled with antibodies directly conjugated to 8-nm colloidal gold as described previously (Fahrenkrog et al., 2002; Paulillo et al., 2005). Expression of GFP-Nup88 and Nup88-myc was as described previously (Fahrenkrog et al., 2002; Paulillo et al., 2005). EM micrographs were recorded on a Phillips CM-100 transmission electron microscope equipped with a CCD camera.
Supplementary Material
Acknowledgments
We thank Teiba Al-Haboubi, Robert D. Goldman, Harald Herrmann, and Dale K. Shumaker for plasmids and antibodies as well as Dale K. Shumaker and Larisa Kapinos for purified lamin proteins. This work is supported by research grants from the Swiss National Science Foundation (SNF grant 31003A-125488 to BF and 31003A-118053 to UK), the Fonds de la Recherche Scientifique Belgium (to BF), as well as the Kanton Basel Stadt and the M.E. Müller Foundation of Switzerland (to BF).
Abbreviations used:
- EDMD
Emery–Dreifuss muscular dystrophy
- FPLD
familial partial lipodystrophy
- GFP
green fluorescent protein
- GST
glutathione S-transferase
- HGPS
Hutchinson–Gilford progeria syndrome
- IF
intermediate filament
- INM
inner nuclear membrane
- NE
nuclear envelope
- NPC
nuclear pore complex
- Nup
nucleoporin
- ONM
outer nuclear membrane
- PVDF
polyvinylidene difluoride
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E10-05-0463) on February 2, 2011.
REFERENCES
- Adam SA, Sengupta K, Goldman RD. Regulation of nuclear lamin polymerization by importin alpha. J Biol Chem. 2008;283:8462–8468. doi: 10.1074/jbc.M709572200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agudo D, Gomez-Esquer F, Martinez-Arribas F, Nunez-Villar MJ, Pollan M, Schneider J. Nup88 mRNA overexpression is associated with high aggressiveness of breast cancer. Int J Cancer. 2004;109:717–720. doi: 10.1002/ijc.20034. [DOI] [PubMed] [Google Scholar]
- Akey CW, Radermacher M. Architecture of the Xenopus nuclear pore complex revealed by three-dimensional cryo-electron microscopy. J Cell Biol. 1993;122:1–19. doi: 10.1083/jcb.122.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andres V, Gonzalez JM. Role of A-type lamins in signaling, transcription, and chromatin organization. J Cell Biol. 2009;187:945–957. doi: 10.1083/jcb.200904124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastos R, Ribas de Pouplana L, Enarson M, Bodoor K, Burke B. Nup84, a novel nucleoporin that is associated with CAN/Nup214 on the cytoplasmic face of the nuclear pore complex. J Cell Biol. 1997;137:989–1000. doi: 10.1083/jcb.137.5.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck M, Forster F, Ecke M, Plitzko JM, Melchior F, Gerisch G, Baumeister W, Medalia O. Nuclear pore complex structure and dynamics revealed by cryoelectron tomography. Science. 2004;306:1387–1390. doi: 10.1126/science.1104808. [DOI] [PubMed] [Google Scholar]
- Bernad R, Engelsma D, Sanderson H, Pickersgill H, Fornerod M. Nup214-Nup88 nucleoporin subcomplex is required for CRM1-mediated 60 S preribosomal nuclear export. J Biol Chem. 2006;281:19378–19386. doi: 10.1074/jbc.M512585200. [DOI] [PubMed] [Google Scholar]
- Bernad R, Van Der Velde H, Fornerod M, Pickersgill H. Nup358/RanBP2 attaches to the nuclear pore complex via association with Nup88 and Nup214/CAN and plays a supporting role in CRM1-mediated nuclear protein export. Mol Cell Biol. 2004;24:2373–2384. doi: 10.1128/MCB.24.6.2373-2384.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broers JL, Ramaekers FC, Bonne G, Yaou RB, Hutchison CJ. Nuclear lamins: laminopathies and their role in premature ageing. Physiol Rev. 2006;86:967–1008. doi: 10.1152/physrev.00047.2005. [DOI] [PubMed] [Google Scholar]
- Capelson M, Liang Y, Schulte R, Mair W, Wagner U, Hetzer MW. Chromatin-bound nuclear pore components regulate gene expression in higher eukaryotes. Cell. 2010;140:372–383. doi: 10.1016/j.cell.2009.12.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronshaw JM, Krutchinsky AN, Zhang W, Chait BT, Matunis MJ. Proteomic analysis of the mammalian nuclear pore complex. J Cell Biol. 2002;158:915–927. doi: 10.1083/jcb.200206106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Sandre-Giovannoli A, et al. Lamin a truncation in Hutchinson-Gilford progeria. Science. 2003;300:2055. doi: 10.1126/science.1084125. [DOI] [PubMed] [Google Scholar]
- Dechat T, Pfleghaar K, Sengupta K, Shimi T, Shumaker DK, Solimando L, Goldman RD. Nuclear lamins: major factors in the structural organization and function of the nucleus and chromatin. Genes Dev. 2008;22:832–853. doi: 10.1101/gad.1652708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dechat T, Shimi T, Adam SA, Rusinol AE, Andres DA, Spielmann HP, Sinensky MS, Goldman RD. Alterations in mitosis and cell cycle progression caused by a mutant lamin A known to accelerate human aging. Proc Natl Acad Sci USA. 2007;104:4955–4960. doi: 10.1073/pnas.0700854104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhe-Paganon S, Werner ED, Chi YI, Shoelson SE. Structure of the globular tail of nuclear lamin. J Biol Chem. 2002;277:17381–17384. doi: 10.1074/jbc.C200038200. [DOI] [PubMed] [Google Scholar]
- Eriksson M, et al. Recurrent de novo point mutations in lamin A cause Hutchinson-Gilford progeria syndrome. Nature. 2003;423:293–298. doi: 10.1038/nature01629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahrenkrog B, Maco B, Fager AM, Koser J, Sauder U, Ullman KS, Aebi U. Domain-specific antibodies reveal multiple-site topology of Nup153 within the nuclear pore complex. J Struct Biol. 2002;140:254–267. doi: 10.1016/s1047-8477(02)00524-5. [DOI] [PubMed] [Google Scholar]
- Fornerod M, van Deursen J, van Baal S, Reynolds A, Davis D, Murti KG, Fransen J, Grosveld G. The human homologue of yeast CRM1 is in a dynamic subcomplex with CAN/Nup214 and a novel nuclear pore component Nup88. EMBO J. 1997;16:807–816. doi: 10.1093/emboj/16.4.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frosst P, Guan T, Subauste C, Hahn K, Gerace L. Tpr is localized within the nuclear basket of the pore complex and has a role in nuclear protein export. J Cell Biol. 2002;156:617–630. doi: 10.1083/jcb.200106046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa K, Ishida K, Tsunoyama TA, Toda S, Osoda S, Horigome T, Fisher PA, Sugiyama S. A-type and B-type lamins initiate layer assembly at distinct areas of the nuclear envelope in living cells. Exp Cell Res. 2009;315:1181–1189. doi: 10.1016/j.yexcr.2008.12.024. [DOI] [PubMed] [Google Scholar]
- Gould VE, Orucevic A, Zentgraf H, Gattuso P, Martinez N, Alonso A. Nup88 (karyoporin) in human malignant neoplasms and dysplasias: correlations of immunostaining of tissue sections, cytologic smears, and immunoblot analysis. Hum Pathol. 2002;33:536–544. doi: 10.1053/hupa.2002.124785. [DOI] [PubMed] [Google Scholar]
- Griffis ER, Xu S, Powers MA. Nup98 localizes to both nuclear and cytoplasmic sides of the nuclear pore and binds to two distinct nucleoporin subcomplexes. Mol Biol Cell. 2003;14:600–610. doi: 10.1091/mbc.E02-09-0582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruenbaum Y, Margalit A, Goldman RD, Shumaker DK, Wilson KL. The nuclear lamina comes of age. Nat Rev Mol Cell Biol. 2005;6:21–31. doi: 10.1038/nrm1550. [DOI] [PubMed] [Google Scholar]
- Harborth J, Elbashir SM, Bechert K, Tuschl T, Weber K. Identification of essential genes in cultured mammalian cells using small interfering RNAs. J Cell Sci. 2001;114:4557–4565. doi: 10.1242/jcs.114.24.4557. [DOI] [PubMed] [Google Scholar]
- Hawryluk-Gara LA, Shibuya EK, Wozniak RW. Vertebrate Nup53 interacts with the nuclear lamina and is required for the assembly of a Nup93-containing complex. Mol Biol Cell. 2005;16:2382–2394. doi: 10.1091/mbc.E04-10-0857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann H, Bar H, Kreplak L, Strelkov SV, Aebi U. Intermediate filaments: from cell architecture to nanomechanics. Nat Rev Mol Cell Biol. 2007;8:562–573. doi: 10.1038/nrm2197. [DOI] [PubMed] [Google Scholar]
- Hinshaw JE, Carragher BO, Milligan RA. Architecture and design of the nuclear pore complex. Cell. 1992;69:1133–1141. doi: 10.1016/0092-8674(92)90635-p. [DOI] [PubMed] [Google Scholar]
- Hutchison CJ, Alvarez-Reyes M, Vaughan OA. Lamins in disease: why do ubiquitously expressed nuclear envelope proteins give rise to tissue-specific disease phenotypes? J Cell Sci. 2001;114:9–19. doi: 10.1242/jcs.114.1.9. [DOI] [PubMed] [Google Scholar]
- Krimm I, Couprie J, Ostlund C, Worman HJ, Zinn-Justin S. 1H, 13C and 15N resonance assignments of the C-terminal domain of human lamin A/C. J Biomol NMR. 2002a;22:371–372. doi: 10.1023/a:1014943122068. [DOI] [PubMed] [Google Scholar]
- Krimm I, Ostlund C, Gilquin B, Couprie J, Hossenlopp P, Mornon JP, Bonne G, Courvalin JC, Worman HJ, Zinn-Justin S. The Ig-like structure of the C-terminal domain of lamin A/C, mutated in muscular dystrophies, cardiomyopathy, and partial lipodystrophy. Structure. 2002b;10:811–823. doi: 10.1016/s0969-2126(02)00777-3. [DOI] [PubMed] [Google Scholar]
- Krull S, Thyberg J, Bjorkroth B, Rackwitz HR, Cordes VC. Nucleoporins as components of the nuclear pore complex core structure and Tpr as the architectural element of the nuclear basket. Mol Biol Cell. 2004;15:4261–4277. doi: 10.1091/mbc.E04-03-0165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz-Bohme B, Wismar J, Fuchs S, Reifegerste R, Buchner E, Betz H, Schmitt B. Insertional mutation of the Drosophila nuclear lamin Dm0 gene results in defective nuclear envelopes, clustering of nuclear pore complexes, and accumulation of annulate lamellae. J Cell Biol. 1997;137:1001–1016. doi: 10.1083/jcb.137.5.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim RY, Aebi U, Fahrenkrog B. Towards reconciling structure and function in the nuclear pore complex. Histochem Cell Biol. 2008;129:105–116. doi: 10.1007/s00418-007-0371-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim RY, Fahrenkrog B. The nuclear pore complex up close. Curr Opin Cell Biol. 2006;18:342–347. doi: 10.1016/j.ceb.2006.03.006. [DOI] [PubMed] [Google Scholar]
- Liu J, Rolef Ben-Shahar T, Riemer D, Treinin M, Spann P, Weber K, Fire A, Gruenbaum Y. Essential roles for Caenorhabditis elegans lamin gene in nuclear organization, cell cycle progression, and spatial organization of nuclear pore complexes. Mol Biol Cell. 2000;11:3937–3947. doi: 10.1091/mbc.11.11.3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewinger L, McKeon F. Mutations in the nuclear lamin proteins resulting in their aberrant assembly in the cytoplasm. EMBO J. 1988;7:2301–2309. doi: 10.1002/j.1460-2075.1988.tb03073.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ly DH, Lockhart DJ, Lerner RA, Schultz PG. Mitotic misregulation and human aging. Science. 2000;287:2486–2492. doi: 10.1126/science.287.5462.2486. [DOI] [PubMed] [Google Scholar]
- Martinez N, Alonso A, Moragues MD, Ponton J, Schneider J. The nuclear pore complex protein Nup88 is overexpressed in tumor cells. Cancer Res. 1999;59:5408–5411. [PubMed] [Google Scholar]
- Moir RD, Yoon M, Khuon S, Goldman RD. Nuclear lamins A and B1: different pathways of assembly during nuclear envelope formation in living cells. J Cell Biol. 2000;151:1155–1168. doi: 10.1083/jcb.151.6.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muralikrishna B, Thanumalayan S, Jagatheesan G, Rangaraj N, Karande AA, Parnaik VK. Immunolocalization of detergent-susceptible nucleoplasmic lamin A/C foci by a novel monoclonal antibody. J Cell Biochem. 2004;91:730–739. doi: 10.1002/jcb.10765. [DOI] [PubMed] [Google Scholar]
- Paulillo SM, Phillips EM, Koser J, Sauder U, Ullman KS, Powers MA, Fahrenkrog B. Nucleoporin domain topology is linked to the transport status of the nuclear pore complex. J Mol Biol. 2005;351:784–798. doi: 10.1016/j.jmb.2005.06.034. [DOI] [PubMed] [Google Scholar]
- Roth P, Xylourgidis N, Sabri N, Uv A, Fornerod M, Samakovlis C. The Drosophila nucleoporin DNup88 localizes DNup214 and CRM1 on the nuclear envelope and attenuates NES-mediated nuclear export. J Cell Biol. 2003;163:701–706. doi: 10.1083/jcb.200304046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaffidi P, Misteli T. Lamin A-dependent nuclear defects in human aging. Science. 2006;312:1059–1063. doi: 10.1126/science.1127168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz TU. Modularity within the architecture of the nuclear pore complex. Curr Opin Struct Biol. 2005;15:221–226. doi: 10.1016/j.sbi.2005.03.003. [DOI] [PubMed] [Google Scholar]
- Schwarz-Herion K, Maco B, Sauder U, Fahrenkrog B. Domain topology of the p62 complex within the 3-D architecture of the nuclear pore complex. J Mol Biol. 2007;370:796–806. doi: 10.1016/j.jmb.2007.05.030. [DOI] [PubMed] [Google Scholar]
- Shumaker DK, Solimando L, Sengupta K, Shimi T, Adam SA, Grunwald A, Strelkov SV, Aebi U, Cardoso MC, Goldman RD. The highly conserved nuclear lamin Ig-fold binds to PCNA: its role in DNA replication. J Cell Biol. 2008;181:269–280. doi: 10.1083/jcb.200708155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smythe C, Jenkins HE, Hutchison CJ. Incorporation of the nuclear pore basket protein nup153 into nuclear pore structures is dependent upon lamina assembly: evidence from cell-free extracts of Xenopus eggs. EMBO J. 2000;19:3918–3931. doi: 10.1093/emboj/19.15.3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoffler D, Feja B, Fahrenkrog B, Walz J, Typke D, Aebi U. Cryo–electron tomography provides novel insights into nuclear pore architecture: implications for nucleocytoplasmic transport. J Mol Biol. 2003;328:119–130. doi: 10.1016/s0022-2836(03)00266-3. [DOI] [PubMed] [Google Scholar]
- Stuurman N, Heins S, Aebi U. Nuclear lamins: their structure, assembly, and interactions. J Struct Biol. 1998;122:42–66. doi: 10.1006/jsbi.1998.3987. [DOI] [PubMed] [Google Scholar]
- Sullivan T, Escalante-Alcalde D, Bhatt H, Anver M, Bhat N, Nagashima K, Stewart CL, Burke B. Loss of A-type lamin expression compromises nuclear envelope integrity leading to muscular dystrophy. J Cell Biol. 1999;147:913–920. doi: 10.1083/jcb.147.5.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi N, van Kilsdonk JW, Ostendorf B, Smeets R, Bruggeman SW, Alonso A, van de Loo F, Schneider M, Van Den Berg WB, Swart GW. Tumor marker nucleoporin 88kDa regulates nucleocytoplasmic transport of NF-kappaB. Biochem Biophys Res Commun. 2008;374:424–430. doi: 10.1016/j.bbrc.2008.06.128. [DOI] [PubMed] [Google Scholar]
- Tunnah D, Sewry CA, Vaux D, Schirmer EC, Morris GE. The apparent absence of lamin B1 and emerin in many tissue nuclei is due to epitope masking. J Mol Histol. 2005;36:337–344. doi: 10.1007/s10735-005-9004-7. [DOI] [PubMed] [Google Scholar]
- Uv AE, Roth P, Xylourgidis N, Wickberg A, Cantera R, Samakovlis C. members only encodes a Drosophila nucleoporin required for rel protein import and immune response activation. Genes Dev. 2000;14:1945–1957. [PMC free article] [PubMed] [Google Scholar]
- Walther TC, Fornerod M, Pickersgill H, Goldberg M, Allen TD, Mattaj IW. The nucleoporin Nup153 is required for nuclear pore basket formation, nuclear pore complex anchoring and import of a subset of nuclear proteins. EMBO J. 2001;20:5703–5714. doi: 10.1093/emboj/20.20.5703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther TC, Pickersgill HS, Cordes VC, Goldberg MW, Allen TD, Mattaj IW, Fornerod M. The cytoplasmic filaments of the nuclear pore complex are dispensable for selective nuclear protein import. J Cell Biol. 2002;158:63–77. doi: 10.1083/jcb.200202088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worman HJ, Bonne G. “Laminopathies”: a wide spectrum of human diseases. Exp Cell Res. 2007;313:2121–2133. doi: 10.1016/j.yexcr.2007.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang ZY, Zhao ZR, Jiang L, Li JC, Gao YM, Cui DS, Wang CJ, Schneider J, Wang MW, Sun XF. Nup88 expression in normal mucosa, adenoma, primary adenocarcinoma and lymph node metastasis in the colorectum. Tumour Biol. 2007;28:93–99. doi: 10.1159/000099154. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.