Abstract
Africa is inferred to be the continent of origin for all modern human populations, but the details of human prehistory and evolution in Africa remain largely obscure owing to the complex histories of hundreds of distinct populations. We present data for more than 580,000 SNPs for several hunter-gatherer populations: the Hadza and Sandawe of Tanzania, and the ≠Khomani Bushmen of South Africa, including speakers of the nearly extinct N|u language. We find that African hunter-gatherer populations today remain highly differentiated, encompassing major components of variation that are not found in other African populations. Hunter-gatherer populations also tend to have the lowest levels of genome-wide linkage disequilibrium among 27 African populations. We analyzed geographic patterns of linkage disequilibrium and population differentiation, as measured by FST, in Africa. The observed patterns are consistent with an origin of modern humans in southern Africa rather than eastern Africa, as is generally assumed. Additionally, genetic variation in African hunter-gatherer populations has been significantly affected by interaction with farmers and herders over the past 5,000 y, through both severe population bottlenecks and sex-biased migration. However, African hunter-gatherer populations continue to maintain the highest levels of genetic diversity in the world.
Keywords: human evolution, population genetics, Khoisan
African human populations are the most genetically diverse in the world (1–4), but inference about African demographic history, evolution, and disease associations has been limited by relatively few genetic samples and scarce archaeological remains in many regions (5, 6). Modern humans are generally thought to have originated in eastern Africa, where the earliest anatomically modern skulls have been found (5), and because populations outside of Africa carry a subset of the genetic diversity found in eastern Africa (1, 7). Before 5,000 y ago, most of sub-Saharan Africa was sparsely occupied by a collection of linguistically and culturally diverse hunter-gatherer (HG) populations (5). Within the past 5,000 y the majority of HG populations in Africa have disappeared, either through assimilation into expanding agropastoral (farmer and herder) groups or by extinction. The remaining HG groups include the forest Pygmy populations of central Africa, isolated click-speaking populations of Tanzania, and the “Bushmen” of the Kalahari Desert region of southern Africa. Even some of these groups, however, have been experiencing a transition to agricultural subsistence over the past 100 y (8).
The expansion of agropastoralist populations has likely had a major impact on the distribution of genetic variation within Africa (3, 9–12). However, the extent to which these groups interacted with local HG is poorly understood because HG variation has been poorly characterized. Do the current HG populations represent agriculturalists who recently reverted to hunting-gathering or mixed subsistence strategies, as has been seen in other parts of the world (13), or do sub-Saharan HG represent geographically isolated remnants of populations from which other African populations diverged early in human prehistory? A recent study of noncoding autosomal sequences suggests that central African Pygmies and their agriculturalist neighbors diverged approximately 60,000 y ago, but inference of divergence was based on fewer than 500 SNPs (14). Further, a recently published Kalahari “Bushman” genome identified more than 700,000 unique SNPs, suggestive of high and ancient phylogenetic divergence of southern African KhoeSan (15). However, population genetic inference of African demographic history requires both the characterization of intrapopulation variation and the comparison of samples from the numerous geographically dispersed and highly structured populations of this continent. To explore questions about HG demographic history, we present the largest dataset to date of genomic SNPs for the remaining click-speaking (or Khoisan-speaking) HG populations in eastern and southern Africa, analyzed jointly with three other HG and 21 agriculturalist populations, including newly generated data from seven northern African populations.
The Khoisan languages of Africa are unique in incorporating a large variety of click consonants. The Hadza and Sandawe of Tanzania are the only Khoisan-speaking populations to reside outside of southern Africa. However, inclusion of the Hadza and Sandawe in the Khoisan language family is highly contentious among linguists (16, 17). Despite their small population sizes (the Hadza number only ≈1,000 individuals), the Khoisan-speakers exhibit a large amount of Y-chromosomal and mitochondrial genetic diversity. Many of their Y- and mtDNA lineages are quite rare, but diversity within the lineages may date to more than 50,000 y ago. The southern KhoeSan or “Bushmen” populations (see Materials and Methods for population nomenclature) today occupy the Kalahari Desert region of Botswana, Namibia, and northern South Africa. Haploid (i.e., Y and mtDNA) genetic studies routinely recognize Khoisan-speaking Ju Bushmen groups as the most divergent population in the world (11, 18, 19). The deepest basal splits in Y-chromosome and mitochondrial phylogenetic trees are between southern Ju Bushmen lineages and eastern African lineages (2). Only a handful of KhoeSan individuals have been characterized in detail for multiple genetic loci (15, 19, 20): these individuals originated from the northern Kalahari/Okavango region. Thus, only a very small amount of the potentially great genetic variation in southern Africa has been examined previously.
Results
We investigated the demographic history and population structure of African HG populations using primarily whole-genome Illumina 650K, 550K, and Affymetrix 500K and 6.0 SNP arrays (SI Appendix, Table S1). We also present 550K custom array data for 90 Khoisan-speaking individuals from the Hadza and Sandawe populations of Tanzania and the ≠Khomani Bushmen of South Africa, merged with corresponding data from an additional 24 African populations (SI Appendix, SI Appendix, Table S1). We use a variety of unique analyses to identify the ancestry of recently admixed genomic segments, severity of population bottlenecks, and the location of ancestral origins within Africa.
Population Structure Within Africa.
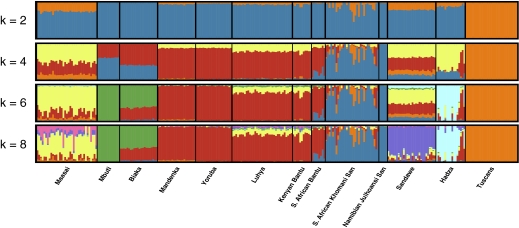
We identify putative ancestral clusters within sub-Saharan Africa by applying an unsupervised, maximum-likelihood clustering analysis (21) to 12 African and one European population for k possible ancestral populations (k = 2 through 8; Fig. 1). At k = 4, we see a western African/Bantu-speaking cluster, an eastern African cluster, a cluster representing Europeans that likely also signifies ancestral variation maintained in eastern Africa (e.g., Maasai and Sandawe populations), and finally, a cluster that links all our HG populations. Populations in eastern African have the highest proportion of the European ancestral cluster (represented by the Italian Tuscans, HapMap3), supporting prior models of the Out of Africa (OOA) migration originating from a population of eastern African ancestry (1). Higher k values (6 through 8) extract each of the HG populations as a distinct ancestral group [although eastern and western forest Pygmies do not differentiate at these low k (2 through 8) values, they do differentiate at higher k values (3)] (Fig. 1). The Sandawe and Hadza populations contain relatively high amounts of eastern African ancestry (>40%), which at k = 8 splits into Sandawe-specific and Hadza-specific clusters. The high fraction of eastern African ancestry indicates that these groups have likely been part of a greater eastern African population pool for a substantial number of generations. Additionally, at k = 4 through 8 it seems that the South African ≠Khomani Bushmen and the Hadza have both absorbed recent migrants from other clusters; individuals with partial Bantu ancestry in the Hadza, and Bantu and European ancestry in the ≠Khomani Bushmen (Fig. 1). Log-likelihood estimates increased as k was increased, indicating a better fit between the data and model at higher k values (SI Appendix, Fig. S1).
Fig. 1.
Ancestral population clusters in sub-Saharan Africa. An unsupervised clustering algorithm, ADMIXTURE (21), was used to analyze population structure among 12 sub-Saharan African populations using ≈461K autosomal SNP loci. We plot k: 2, 4, 6, 8 ancestral populations. European Tuscans were included to allow for potential recent European admixture in South Africans. We randomly chose a subset of 30 unrelated Maasai and Luhya for representation in this figure. At k = 4, all HG retain shared ancestry (in blue), and South African Bantu-speakers are likely to have recently absorbed 10–20% KhoeSan ancestry. At k = 8, HG populations emerge with four distinct, ancestral population clusters.
We also calculated Fst between clusters using the cluster-specific allele frequencies generated from ADMIXTURE (Table 1). Because the Fst is estimated here from the cluster-based allele frequencies rather than the population-based frequencies, much of the influence of recent migration on our estimates has been removed. Ancient migration and mis-estimation of cluster-based frequencies are potential sources of bias in this approach, but admixture between highly diverged populations, such as the Bushmen and Europeans, is likely to be detected and removed. The southern Bushmen, central forest Pygmies, and the Hadza, compared with Europeans, have Fst estimates in excess of 0.23 (Table 1), approximately twice the average Fst between other global populations (1). Pairwise comparisons with the Hadza tend to be exceptionally elevated, likely owing to the affect of genetic drift during a recent, severe bottleneck in that population (Fig. 4 B and C).
Table 1.
Fst estimates for k = 8 clusters assigned with ADMIXTURE
| Cluster* | European | Sandawe | Hadza | Eastern Africa | Maasai† | Western African | Forest Pygmies |
| European | |||||||
| Sandawe | 0.135 | ||||||
| Hadza | 0.256 | 0.158 | |||||
| Eastern Africa | 0.117 | 0.054 | 0.154 | ||||
| Maasai† | 0.172 | 0.108 | 0.218 | 0.104 | |||
| Western Africa | 0.169 | 0.053 | 0.16 | 0.046 | 0.103 | ||
| Forest Pygmies | 0.23 | 0.102 | 0.158 | 0.105 | 0.167 | 0.084 | |
| Southern KhoeSan | 0.25 | 0.122 | 0.222 | 0.131 | 0.194 | 0.115 | 0.107 |
*Cluster names correspond to the population or geographic affinities identified in Fig. 1, assuming k = 8 ancestral populations. Fst was calculated only on the basis of the allele frequencies estimated within each cluster to exclude recent admixture (e.g., Fst between the forest Pygmies and other clusters corresponds to the “green” cluster in Fig. 1).
†Although the majority of Maasai ancestry is assigned to the eastern African (or “yellow”) cluster, a second ancestry is also found primarily in the Maasai, indicated in Fig. 1 by the “pink” cluster. Preliminary results suggest that this ancestry is likely of North African origin.
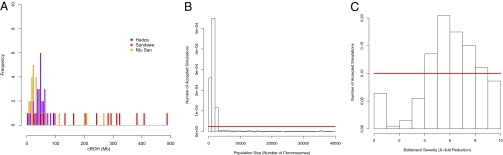
Fig. 4.
Runs of homozygosity among Khoisan-speakers. (A) Long runs of homozygosity were calculated for individuals in the Hadza, Sandawe, and ≠Khomani Bushmen populations. Runs were constrained to a minimum of 1 Mb, and two missing genotypes were allowed per run. cROH are plotted for all individuals; the y-axis represents counts of individuals. The Hadza distribution differs markedly from the other two populations, with 65% (n = 11/17) of individuals having cROH >100 Mb. This distribution is consistent with a severe, recent bottleneck in the Hadza. (B) Simulated posterior distribution of effective population size in the Hadza, generated by sampling from a uniform distribution of Ne and keeping simulated parameters within 20% of the observed fROH with REJECTOR (29). (C) Simulated posterior distribution of bottleneck severity in the Hadza, as modeled above.
Genetic Diversity Within Africa.
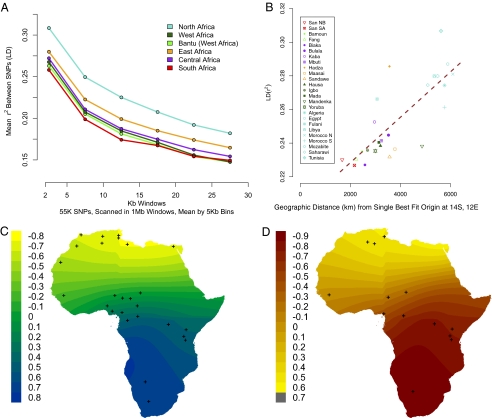
We merged SNP data for 27 populations across multiple genomic array platforms (Materials and Methods and SI Appendix, Table S1) and created a set of 55,000 autosomal SNPs common to all platforms. Linkage disequilibrium (LD) was measured using r2 between pairs of SNPs in sliding 1-Mb windows and estimated from equal sample sizes for each population. Within Africa, the HG populations tend to have the lowest levels of LD; the five populations with the lowest mean LD are South African ≠Khomani Bushman, Biaka Pygmies, Namibian Bushmen, Fang (not HG), and Tanzanian Sandawe (SI Appendix, Fig. S13). LD values are generally indistinguishable between agropastoralist African populations (e.g., Yoruba and Maasai); all northern African populations are elevated relative to sub-Saharan Africa (Fig. 2A). LD measured by r2 is a function of both recombination rate (c) and effective population size (Ne) such that E(r2) = 1/(4Nec) (22). Lower values of r2 in the South African KhoeSan, Biaka, and Sandawe HGs are either the result of larger effective population sizes or higher recombination rates in these populations (23, 24). Population substructure or recent admixture is expected to increase LD (23, 25), and thus our r2 estimates for African HGs may even be conservatively high given historical gene flow from Bantu-speaking and Europeans into these populations (Fig. 1). Assuming genome-wide recombination rates do not vary greatly between human populations, the LD results are consistent with several African HG groups (e.g., Bushmen, Biaka, and Sandawe) having greater Ne than almost all other African populations. Elevated LD in our samples of the HG Hadza and Mbuti is consistent with recent bottlenecks in these populations [see below and Patin et al. (14)].
Fig. 2.
Genome-wide LD and Fst in African populations. (A) Each line represents the LD decay averaged across populations within each of six geographic regions; regions are described in SI Appendix, Table S1. LD (r2) between SNPs was calculated in sliding 1-Mb windows. The r2 estimates were binned by the genetic distance between SNPs in 5-Kb bins. HG populations have the lowest LD curves (SI Appendix, Fig. S13 shows population-specific LD decay curves). (B) We assessed a possible point of origin by regressing LD onto geographic distance. Regression for the single best fit for geographic origin is shown, with a correlation coefficient of 0.78, to the point 14°S, 12°E. (C) Map is shown using mean LD within 0–5 Kb. The highest correlation coefficient in blue indicates the best fit with a potential geographic origin. Crosses indicate the geographic coordinates of the sampled populations. (D) We assessed a possible point of origin by regressing Fst between African populations and Europe onto geographic distance, using a waypoint in the Near East. Populations were grouped for Fst analysis; crosses indicate the geographic mean for each population grouping.
To find the best-fit location for an origin of modern humans within Africa, we regressed LD statistics by distance in kilometers from different points on the continent (Fig. 2B), in a manner similar to the heterozygosity regressions of Ramanchandran et al. (1). Under a serial founder model, populations further from the origin will have smaller estimated Ne and higher LD on average. Therefore, in this model, positive correlations indicate a good fit with the serial founder model. The highest positive correlations occur when the point of origin is in southwestern Africa (using mean LD at 5 Kb; SI Appendix, Fig. S3). Bantu-speaking populations in eastern and southern Africa were removed for this analysis, because they are considered recent migrants and would not reflect the ancient patterns of migration (5, 9, 26), but exclusion of these samples made little difference to the LD maps. Regressions of LD on distance from southwestern Africa were highly statistically significant (at 5-Kb windows, P ≈ 4.9 × 10−6) (Fig. 2C). Best-fit (Materials and Methods) locations based on LD are consistent with a common origin in southern Africa. A point of origin in southwestern Africa was approximately 300–1,000 times more likely than in eastern Africa; detailed likelihood statistics are available in SI Appendix, Table S6. The best-fit regression originates at 14°S latitude and 12°E longitude (Fig. 2 B and C) with an r of 0.78. The geographic pattern in Fig. 2 is driven by exceptionally low LD in the southern Africa KhoeSan and central African Biaka pygmies.
Because LD estimates can be affected by recent admixture, we used a second statistic, Fst, to infer the location of origin within Africa. Fst was calculated from cluster-specific allele frequencies using the ADMIXTURE software at k = 14. Similar to Tishkoff et al. (3), we still detect discrete population membership at k = 14. We used the inferred genetic clusters, rather than the population-based allele frequency estimates, to remove the effect of recent migration on Fst. This procedure will, for example, minimize the bias of European gene flow on allele frequency estimates from the South African and Namibian Bushmen samples, and the cluster-based Fst estimate will better reflect the divergence of the Bushman population before arrival of recent migrants. This procedure does, however, decrease the number of independent points from 27 in the LD analysis to 13 within Africa. Using Fst between each African cluster and Europe (Italy) as a sample OOA population, we performed the regression and permutation analysis of geographic origin as described above for LD. The best-fit regression originates at 20°S latitude and 22°E longitude (Fig. 2D), with an r of −0.84. With fewer observations the P value is not as low as in the LD analysis; however, we still find a P value of 0.06 after correcting for multiple tests via 1,000 permutations. Given that the Fst estimates are not collinear with the LD decay estimates (because the ADMIXTURE model does not take LD into account), this provides an additional line of evidence for a southern origin for modern humans.
To provide an array-independent assessment of heterozygosity we typed the ≠Khomani Bushmen and Hadza at high resolution for HLA-A, B, and C, arguably the most polymorphic of human genes. In comparison with other African populations the Hadza have low HLA-A, B, and C diversity but unusually high frequency (0.375; SI Appendix, Table S5) of HLA-B*44:03, features often characteristic of small populations that have been subject to pressure from infectious disease (27). In contrast, the ≠Khomani Bushmen have the greatest HLA-A heterozygosity (30 alleles, H = 0.95; SI Appendix, Table S4), not only among African populations but throughout the world. ≠Khomani Bushmen remain among the most heterozygous populations through the HLA-B and C loci (H = 0.94 and 0.89, respectively; SI Appendix, Table S4). Further attesting to the extraordinary HLA diversity and low genome-wide LD of the ≠Khomani Bushmen, we estimated 82 different HLA-A, B, and C haplotypes in the panel studied, compared with a mean of 25 haplotypes per population worldwide (SI Appendix).
Demographic History of the Khoisan HG.
All HG populations in our study segregated into unique population clusters, separate from neighboring agriculturalist populations, indicating that the HG groups derive from distinct ancestral populations (Fig. 1 and SI Appendix, Fig. S2). We asked whether there was evidence of recent gene flow between HG and agriculturalists by examining the population affiliation of both the haploid systems and the autosomes. Y-chromosome and mtDNA haplogroups were assigned on the basis of approximately 2,000 SNPs per haploid system. Virtually every individual in the ≠Khomani Bushmen sample carried a highly diverged mtDNA haplogroup that is indigenous to southern Africa, 93% L0d1; two individuals carried basal pre-L0a'f lineages that have an unknown geographic distribution, but given that KhoeSan individuals retain many of the deep divergence mtDNA haplogroups, it is likely that this lineage is also endemic to the KhoeSan (18) (SI Appendix, Table S3). However, ≈35% of the paternal lineages were of either European or Bantu origin. Across all three click-speaking populations the contribution of maternal haplogroups from putative agriculturalists varied from 0 to 28%, whereas the corresponding paternal contribution of agriculturalists varied from 30% to 42% (SI Appendix, Table S3).
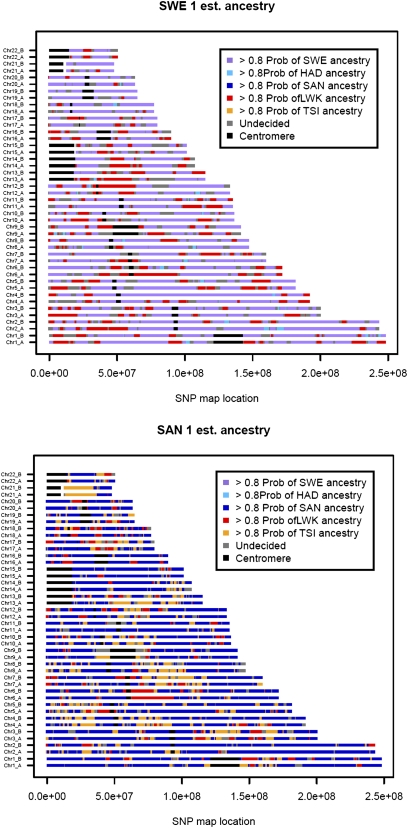
We used phased data from admixed individuals to visualize the distribution of ancestry along each chromosome, assuming the number of ancestral populations was equal to three. A principal components (PCA)-based analysis of genomic local ancestry (28) in 40 SNP windows was implemented for the Khoisan-speaking populations (Materials and Methods). Motivated by the ADMIXTURE results in Fig. 1, ancestral populations for the potentially admixed Sandawe individuals were represented by the Sandawe, Hadza, and Kenyan Luhya (Bantu) populations. As illustrated in Fig. 3A, for one Sandawe genome, segments of probable Bantu ancestry tended to be long, often spanning more than 10 Mb, and very little Hadza ancestry could be detected. In the South African ≠Khomani Bushmen, we assumed three possible source populations: ≠Khomani Bushmen, Kenyan Luhya, or European Tuscans. One example, individual SA1 (Fig. 3B), did not report European ancestry among parents or grandparents, yet displays a fairly high amount of apparent European ancestry.
Fig. 3.
Local ancestry assignment along phased chromosomes. Two individuals with potential admixture (Fig. 1) were projected onto the principal component space of three putative ancestral populations. The three ancestral populations differed for Sandawe individuals (SWE, Sandawe; HAD, Hadza; LWK, Luhya Bantu) and South African ≠Khomani Bushmen (SAN, KhoeSan; TSI, European Tuscan; LWK, Luhya Bantu). Ancestry was assigned in 40-SNP windows along phased chromosomes (haplotypes A and B) by calculating the minimal distance to an ancestral population (28). Ancestry from Bantu-speaking agriculturalists seems to have occurred relatively recently, as indicated by many ≥10-Mb segments. Switch errors in the phasing could potentially shorten the length of these migrant tracts, but with low levels of admixture, phase switch errors are less likely to lengthen inferred migrant tracts.
Distributions of the cumulative amount of runs of homozygosity (cROH, where we define a run of homozygosity to be >500 Kb) were calculated for all individuals in the three click-speaking populations (Fig. 4) (Materials and Methods). The Tanzanian Hadza population differed strikingly from the other two groups; approximately 65% of Hadza individuals have a cROH >100 Mb, and on average the fraction of the genome in runs of homozygosity (fROH) is 8% in the Hadza. We found no strong correlation between the number of missing genotypes and cROH (SI Appendix, Fig. S5). In comparison with the 52 Human Genome Diversity Project (HGDP) populations, higher fROH values are found in four Native American populations, which have been subject to serial founder events.
The elevated mean and variance of cROH in the Hadza population is indicative of a severe and possibly recent population bottleneck. To test this hypothesis we performed a number of demographic simulations using a rejection-based approximate Bayesian algorithm (29) (REJECTOR; Materials and Methods), focusing on the summary statistic fROH. On the basis of an observed fROHHadza of 8%, the most likely estimate of effective population size (Ne) of the Hadza was 2,590 [95% confidence interval (CI) 475–21,222]; current census size of the Hadza is thought to be only 1,000 (30) (Fig. 4B). Single population estimates of Ne vary widely (19, 31), but among a global sample of HGDP populations only Native Americans and the Kalash have an Ne <3,000. We estimate the mean severity of a bottleneck in the Hadza to be sixfold (95% CI 0.4–9.67) (Fig. 4C). The fROH statistic did not seem to be sensitive to the time of the bottleneck. The corresponding estimates of mean Ne in the neighboring Sandawe are 7,000 (95% CI 3,340–23,200) and 11,670 in the ≠Khomani Bushmen (95% CI 5,760–28,650). Posterior distributions of bottleneck severity were flat for the Sandawe and Bushmen (SI Appendix, Fig. S6).
Loci Under Selection.
We scanned five African HG populations for ongoing selective sweeps using the iHS statistic (32) (SI Appendix, Fig. S8). We identified regions of the genome characterized by extreme iHS values in each population using the procedures described by Pickrell et al. (33). The most extreme 1% regions generally do not overlap between HG groups. We find eight regions that are in the most extreme 10 loci from a single population and are also in the top 5% for at least two additional African HG populations. These regions include genes such as POT1 and TUSC3, associated with carcinogenesis, a section on chromosome 6 (30.2–30.6, Build Hg36) of the HLA region that contains several tripartite motif-containing genes that have been implicated in pathogen response, and a region on chromosome 9 that is in the top 10% in all five African HG populations and contains nine genes, including IKBKAP mutations, which result in familial dysautonomia (SI Appendix, Fig. S8).
We also surveyed a subset of known loci with functions related to infectious disease and that may have been under recent selection owing to increased population density and contact between groups. One SNP, rs2395029, is a missense Val-to-Gly mutation in the HLA complex P5 (HCP5); the function of this protein is not well understood. The G allele, likely ancestral, has been shown to be associated with reduced viral load in HIV-positive individuals, explaining 10% of the variance in HIV viral load among infected Europeans (34). A second genome-wide association study also found that in European individuals the G allele is associated with HIV nonprogression (35) (odds ratio 3.47). We found our sample of the Sandawe to carry rs2395029 at an elevated 17% frequency (excluding close relatives), substantially higher than in other populations from HGDP– Centre d'Etude du Polymorphisme Humain (CEPH) (SI Appendix, Fig. S7). The SNP is absent in the Hadza and ≠Khomani Bushmen. The iHS scan, meant to measure ongoing or incomplete selective sweeps, gave a score of 2.79 (absolute value) to the rs2395029 allele in the Sandawe population; values >2 are generally indicative of selection (32) (although this window is not within the top 1% of the iHS scores). The cross-population extended haplotype homozygosity XP-EHH and composite likelihood ratio selection statistics also find the HCP5 locus significant at an empirical P < 0.01 level in the Sandawe (but not in other HG populations), with Europeans used for the cross-population comparison.
Discussion
Population Structure.
It is unclear to what extent the ancestral African population was structured at the time of the OOA migration by modern humans, ≈60,000 y ago (36, 37). Deep ancestral population structure within Africa would affect inference of divergence time among African and non-African populations, extent of admixture between expanding modern humans and Neanderthals (38), and the causes of divergence during the origin of modern humans (i.e., demographic growth, unique mutations affecting behavior, etc.). By examining genomic data from six HG populations from southern, central, and eastern Africa (i.e., Ju and ≠Khomani Bushmen, Mbuti and Biaka, Hadza and Sandwe) in conjunction with 21 other African populations, we characterized the structure and relationship between these populations. Frequentist population structure analyses (Fig. 1 and SI Appendix, Fig. S2) demonstrate that the HG populations are highly differentiated, both among themselves and compared with other African populations (19, 39). Thus, they do not seem to derive from agriculturalist populations and instead represent geographically distinct populations isolated from other groups for thousands of years.
However, all African HG populations do share an ancestral cluster at small k levels (Fig. 1). A similar phenomenon was observed in the Tishkoff et al. (3) microsatellite structure analysis and suggests that central African Pygmies, southern KhoeSan, and Tanzanian click-speakers share common ancestry distinct from other African populations, either through a more recent common ancestor or through long-distance migration. Population divergence between the eastern and western Pygmy populations, here represented by the Mbuti and Biaka, respectively, is estimated to have occurred approximately 15,000–27,000 y ago (14, 40, 41). Our Y-chromosome microsatellite data from the highly diverged, basal KhoeSan A3b1-M51 clade, found exclusively in southern Africa, has a most recent common ancestor estimated at 40,000 y ago (SI Appendix, Fig. S11) (42). If the time to the most recent common ancestor (TMRCA) within the KhoeSan population is 40,000 y ago, then this is among the first genetic evidence that the KhoeSan have continuously occupied southern Africa for at least the entire span of the Upper Paleolithic (43). This conclusion assumes that any past geographic movement of the KhoeSan ancestral population would have spread the distribution of A3b1 lineages, and these lineages would survive in contemporary populations. African HG populations seem to have been highly structured at the time of the OOA migration, but further sophisticated modeling of autosomal data are necessary to confirm this.
Geographic Origins and Population Size.
LD decays more rapidly between SNPs in African populations than in non-African populations, and single-gene studies indicate there is variation in levels of LD among African populations (44, 45) (Fig. 2A). We present LD decay plots based on genome-wide data for 27 African populations. Of the five populations with the lowest levels of LD, four are HG populations: the ≠Khomani and Namibian Bushmen, Sandawe and Biaka Pygmies; the Fang from Cameroon also have very low LD. This suggests that the largest human effective population sizes have been in African HG populations. We estimate Ne in the ≠Khomani Bushmen to be high, namely 11,600, on the basis of fROH simulations. We note, however, that an estimate of Ne using the fraction of the genome in runs of homozygosity is an indicator of relatively recent effective population size, not of the ancestral population living tens of thousands of years ago.
Recently Tishkoff et al. (3) suggested a potential origin for modern humans in southern Africa, on the basis of heterozygosity estimates from microsatellite data. However, their sample of KhoeSan was small, and the directionality of a southwestern origin of humans based on heterozygosity could have been driven by the inclusion of a highly admixed (and thus highly heterozygous) “Coloured” population. To account for these concerns, we used a large sample from two KhoeSan populations (n = 47) and different statistics: Fst and mean genomic LD. At levels of mean LD between 0 and 30 Kb, the best-fit geographic origin is found in southwestern Africa, likely driven by the exceptionally low LD in the Bushmen (South Africa and Namibia) and Biaka Pygmies (Central African Republic). Fst comparisons also support a southern African origin. Geographic dispersal of modern humans from southern Africa is consistent with the earliest archaeological evidence for worked bone awls, inscribed ostrich eggshell, and climatic evidence indicating a more hospitable climate in southern Africa than eastern Africa until 60,000–70,000 y ago (46–48). The large number of unique SNPs not present in dbSNP but identified in the Namibian KB1 (15) “Bushman” genome is also suggestive of the greater diversity present in these KhoeSan populations, but resequencing of larger sample sizes from many geographically separated African populations is required to confirm this conclusion.
Extremely elevated LD, increased runs of homozygosity, and very low HLA and haplotype heterozygosity estimates in comparison with other sub-Saharan African populations (Figs. 2 and 4 and SI Appendix, Tables S2 and S4) all indicate a severe population bottleneck in the Hadza. We estimate their Ne to be only 2,500 individuals, which corresponds to an ancestral population size of 15,000 (given the estimated sixfold bottleneck). Strikingly, our Ne estimate is more than twice the current census size of the Hadza, suggesting that the bottleneck is ongoing and that historically the Hadza had a much larger population size. Either our current population sample reflects a substantial founder effect from a larger eastern African HG population, or the entire population has experienced a bottleneck that narrowed its geographic distribution to present day Lake Eyasi. The geographic extent of the historic Hadza population is unknown and is complicated by the lack of evidence for substantial gene flow between the Hadza and neighboring Sandawe, who live only 150 km away (Fig. 3A).
Selection in HG.
We find little overlap between HG populations in ongoing or incomplete selective sweeps (SI Appendix, Fig. S8), even between populations in similar environments such as the central African Pygmies or the Tanzanian Hadza and Sandawe. This is consistent with the ancient and continuing separation of these groups (Figs. 1 and 2) and the global observation in Pickrell et al. (33) that ongoing selective sweeps tend to be regionally specific. Direct comparison of haplotype-based statistics, such as iHS, between samples can be confounded by differences in SNP selection and phasing (SI Appendix). The five HG populations examined here (SI Appendix, Fig. S8) were jointly phased for the same 461K SNPs with a diverse set of trio-based haplotype seeds from Yoruba, Maasai, and Europeans and duo-seeds from the Sandawe and ≠Khomani Bushmen.
We identified evidence for positive selection on the HCP5 locus in the Tanzanian Sandawe together with an increased frequency of the rs2395029 [G] allele. There was no evidence of a recent demographic bottleneck in the Sandawe that could explain such a high allele frequency by drift (SI Appendix). The rs2395029 [G] allele has been associated with protection from HIV by virtue of LD with HLA-B*57:01, where putative causal amino acid variants have been identified (34). We found that the G allele is not in LD with HLA-B*57:01 in the Sandawe; instead it tends to be on the background of HLA-B*49:01, which encodes none of the HIV-protective amino acids of B*57. Additionally, in Europeans rs2395029 [G] is in nearly complete LD with HLA-B*57:01 and is used as a diagnostic marker for hypersensitivity to abacavir, a drug prescribed during HIV therapy (49). Our results suggest both that the HPC5 locus is under strong selection in the Sandawe independently of B*57:01 and that rs2395029 [G] is not a diagnostic marker for B*57:01 in individuals with African ancestry, among whom this allele has recombined onto a variety of haplotypes. It remains to be determined whether the selection has been due to an increase in HIV prevalence in Tanzania or another infectious disease.
Conclusions
We present a targeted analysis of African HG genomes, including genomic SNP array data for the Tanzanian Hadza, Sandawe, and the South African ≠Khomani Bushmen. We find that the six sub-Saharan HG populations share common ancestry distinct from agriculturalists (Fig. 1 and SI Appendix, Fig. S2). Analyses of LD and heterozygosity suggest that HGs, especially the click-speaking ≠Khomani and Namibian Bushmen, are among the most diverse of all human populations. LD and Fst patterns support the hypothesis that all human populations originated from southern Africa (Fig. 2 and Table 1), although we caution that sampling in Africa remains sparse; our sample size of Bushmen numbers just 47 individuals, and other highly variable populations may be discovered. Additionally, the interaction between agropastoralist populations and HG over the past 5,000 y has markedly affected HG populations in a variety of ways, including sex-biased gene flow from agropastoralists into HG groups, demographic bottlenecks [in the Hadza and central Pygmies (41)], and possibly unique selection pressures, for example those associated with infectious disease. We suggest that HG groups in Africa are currently experiencing rapid evolutionary change, as reflected in their patterns of genomic diversity.
Materials and Methods
Samples.
Sampling of the ≠Khomani Bushmen in Upington, South Africa and neighboring villages occurred in 2006. Institution review board approval was obtained from Stanford University. ≠Khomani N|u-speaking individuals, local community leaders, traditional leaders, nonprofit organizations, and a legal counselor were all consulted regarding the aims of this research before collection of DNA. All individuals consented orally to participation. DNA via saliva (Orogene kits) and ethnographic information regarding ancestry, language, and parental place of birth were collected for all participants. Hadza individuals were collected in the Arusha district of Tanzania; the Commission for Science and Technology and the National Institute for Medical Research of Tanzania approved the sample collection (50). Sandawe individuals were collected in 2004 [informed consent described in Li et al. and Donnelly et al. (51, 52)]. Samples from the ≠Khomani Bushmen, Hadza, and Sandawe were typed on the Illumina Beadchip 550K custom v2 chip designed by 23andMe, Inc. and processed through 23andMe’s partner laboratory. Genomic data for comparable loci were also obtained from HGDP-CEPH (19) and HapMap3 (http://hapmap.ncbi.nlm.nih.gov/), publicly available resources. These genotype data were merged with samples typed on the Affymetrix 6.0, Affymetrix 500K, Illumina 1M for a total of 27 African populations (using PLINK v1.07; SI Appendix, Table S1). Data for western African populations genotyped on the Affymetrix 500K were obtained from Bryc et al. (28) (available in the database of Genotypes and Phenotypes), and North African population data generated for this manuscript, shown in SI Appendix. A total of 55,000 SNPs were identified as being common to all platforms (available at www-evo.stanford.edu/pubs.html).
Nomenclature.
We use the term “Khoisan” to refer to the linguistic family consisting of click-speaking populations as defined by Greenberg (53). The term “KhoeSan” refers to both the pastoralist and HG Khoisan-speakers of southern Africa, including the Nama, Ju|hoansi, and N|u. “San” is indicative of a historic HG subsistence lifestyle, regardless of the specific Khoisan southern African subfamily language affiliation and has been often used in the literature interchangeably with “Bushmen.” Many individuals in our sample preferred to be referred to as “Bushmen.”
Population Structure.
An unsupervised clustering algorithm, ADMIXTURE (21), was run on our three unique Khoisan-speaking populations, HGDP-CEPH sub-Saharan Africans, HapMap3 Kenyan Luhya, Maasai, and Italian Tuscans. Eight ancestral clusters (k = 1 through 14) in total were tested successively. Log-likelihoods for each k clusters are shown in SI Appendix. Replicate runs of k ancestral populations did not result in substantially different individual ancestral frequencies. Fst based on allele frequencies was calculated in ADMIXTURE for each identified cluster at k = 8 and k = 14.
Phasing.
Parent-offspring (PO) pairs were first identified using segments sharing identity by descent; PO pairs in the Sandawe and ≠Khomani San (absent in the Hadza) were phased separately using BEAGLE software (54). The phased Khoisan PO pairs were then combined with HapMap3 trio-phased Yoruba, Maasai, and European population samples (n = 30) for a larger seed ancestral haplotype pool. HGDP African populations, Tanzanian Hadza and Sandawe, and South African ≠Khomani San were then phased together in BEAGLE.
Runs of Homozygosity and Rejection Algorithm.
Long runs of homozygosity were calculated with PLINK v1.07 software (55) (http://pngu.mgh.harvard.edu/~purcell/plink/download.shtml) under the following parameters: SNP density of 50 SNPs per window, minimum ROH length of 1 Mb, two missing genotypes per window allowed, one heterozygote per window allowed, sliding window of 1 Mb, minor allele frequency 5%. These parameters were similar to those chosen by Nalls et al. (56) and Auton et al. (57) for their calculations of the accumulated ROH. Estimates of the fROH for rejection algorithm simulations used the same parameters as above, except that all missing genotypes had been imputed in BEAGLE. We investigated sensitivity to the number of missing genotypes and correlation between cROH and the number of ROH segments (SI Appendix, Fig. S5). Using a rejection algorithm program we inferred demographic history by comparing simulated cROH estimates with observed cROH estimates. Extensive documentation of the rejection algorithm method, accuracy, and the software REJECTOR (29) package are available online (SI Appendix and www.rejector.org).
Linkage Disequilbrium.
LD, r2, was calculated between all SNPs in sliding 1-Mb windows. Pairs of SNPs were then binned by their genomic distance in 5-Kb bins. Each population was randomly subsampled 10 times for 12 individuals, LD decay was calculated independently, and mean LD for each bin was averaged across the 10 subsamples for the final population estimate (Fig. 2). To infer goodness-of-fit of a serial founder effect model across the continent of Africa using LD, we first created a 1° × 1° grid of latitude/longitude points across the continent. For each of these, we calculated great circle distances from our potential point-of-origin to every population. For distances between western and southern Africa we used a waypoint in Cameroon to reflect realistic land-based migration. We then calculated the regression of LD on distance, estimated via smoothing the empirical LD decay measurements with exponential decay functions. Best-fit was identified by the explained variance “r.” This method was similarly applied to the pairwise Fst estimates between African populations and Europe, using nonadmixed allele frequencies estimated from ADMIXTURE, k = 14, and with an additional waypoint in the Near East for the OOA populations. For clusters consisting of multiple populations, such as western African Bantu-speakers, the geographic location of the cluster Fst was calculated as the average distance to the centroid for the longitude and the sine average distance to the centroid for the latitude. We used MapViewer (http://www.goldensoftware.com) to create the Kriging interpolation plot of the correlation coefficients.
Supplementary Material
Acknowledgments
We thank Andy Reynolds and Mike Palmer for technical assistance; Dara Torgerson, Lindsey Roth, and Saunak Sen for discussion; the University of California, San Francisco (UCSF) Biostatistics High Performance Computing System for computing resources; and the Tanzanian and South African participants who generously contributed DNA. This work was funded by the Center for Human Origins and Evolution, the Morrison Institute for Population and Resource Studies at Stanford University, a UCSF Chancellor's Graduate Research Fellowship (to C.R.G.), and National Institute of Health Grants 3R01HG003229 and 3R01GM028016.
Footnotes
This Feature Article is part of a series identified by the Editorial Board as reporting findings of exceptional significance.
Conflict of interest statement: The authors from 23andMe, Inc. (C.R.G., J.M.M., L.H., and J.L.M.) declare competing financial interests as employees at and stock holders of 23andMe, Inc. SNP arrays designed by 23andMe were used to generate a unique dataset reported in this article. To our knowledge, affiliation with 23andMe, Inc. did not bias the results or discussion of results reported in this article.
This article is a PNAS Direct Submission.
Data deposition: Genotype data will be hosted on a Stanford University website: http://www-evo.stanford.edu/pubs.html.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1017511108/-/DCSupplemental.
References
- 1.Ramachandran S, et al. Support from the relationship of genetic and geographic distance in human populations for a serial founder effect originating in Africa. Proc Natl Acad Sci USA. 2005;102:15942–15947. doi: 10.1073/pnas.0507611102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Semino O, Santachiara-Benerecetti AS, Falaschi F, Cavalli-Sforza LL, Underhill PA. Ethiopians and Khoisan share the deepest clades of the human Y-chromosome phylogeny. Am J Hum Genet. 2002;70:265–268. doi: 10.1086/338306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tishkoff SA, et al. The genetic structure and history of Africans and African Americans. Science. 2009;324:1035–1044. doi: 10.1126/science.1172257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vigilant L, Stoneking M, Harpending H, Hawkes K, Wilson AC. African populations and the evolution of human mitochondrial DNA. Science. 1991;253:1503–1507. doi: 10.1126/science.1840702. [DOI] [PubMed] [Google Scholar]
- 5.Phillipson DW. African Archaeology. Cambridge, UK: Cambridge Univ Press; 2005. [Google Scholar]
- 6.Mitchell P. Genetics and southern African prehistory: An archaeological view. J Anthropol Sci. 2010;88:73–92. [PubMed] [Google Scholar]
- 7.Gonder MK, Mortensen HM, Reed FA, de Sousa A, Tishkoff SA. Whole-mtDNA genome sequence analysis of ancient African lineages. Mol Biol Evol. 2007;24:757–768. doi: 10.1093/molbev/msl209. [DOI] [PubMed] [Google Scholar]
- 8.Newman JL. The Ecological Basis for Subsistence Change Among the Sandawe of Tanzania. Washington, DC: National Academies; 1970. [Google Scholar]
- 9.Beleza S, Gusmão L, Amorim A, Carracedo A, Salas A. The genetic legacy of western Bantu migrations. Hum Genet. 2005;117:366–375. doi: 10.1007/s00439-005-1290-3. [DOI] [PubMed] [Google Scholar]
- 10.Destro-Bisol G, et al. Variation of female and male lineages in sub-Saharan populations: The importance of sociocultural factors. Mol Biol Evol. 2004;21:1673–1682. doi: 10.1093/molbev/msh186. [DOI] [PubMed] [Google Scholar]
- 11.Tishkoff SA, et al. History of click-speaking populations of Africa inferred from mtDNA and Y chromosome genetic variation. Mol Biol Evol. 2007;24:2180–2195. doi: 10.1093/molbev/msm155. [DOI] [PubMed] [Google Scholar]
- 12.Sikora M, Laayouni H, Calafell F, Comas D, Bertranpetit J. A genomic analysis identifies a novel component in the genetic structure of sub-Saharan African populations. Eur J Hum Genet. 2011;19:84–88. doi: 10.1038/ejhg.2010.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Waters T. Comment on “Recent origin and cultural reversion of a hunter-gatherer group”. PLoS Biol. 2005;3 doi: 10.1371/journal.pbio.0030269. e269, author reply e270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Patin E, et al. Inferring the demographic history of African farmers and pygmy hunter-gatherers using a multilocus resequencing data set. PLoS Genet. 2009;5:e1000448. doi: 10.1371/journal.pgen.1000448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schuster SC, et al. Complete Khoisan and Bantu genomes from southern Africa. Nature. 2010;463:943–947. doi: 10.1038/nature08795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Güldemann T, Stoneking M. A historical appraisal of clicks: A linguistic and genetic population perspective. Annu Rev Anthropol. 2008;37:93–109. [Google Scholar]
- 17.Sands B, Güldemann T. What click languages can and can't tell us about language origins. In: Botha R, Knight C, editors. The Cradle of Language. Oxford: Oxford University Press; 2009. p. 204. [Google Scholar]
- 18.Behar DM, et al. Genographic Consortium. The dawn of human matrilineal diversity. Am J Hum Genet. 2008;82:1130–1140. doi: 10.1016/j.ajhg.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li JZ, et al. Worldwide human relationships inferred from genome-wide patterns of variation. Science. 2008;319:1100–1104. doi: 10.1126/science.1153717. [DOI] [PubMed] [Google Scholar]
- 20.Jakobsson M, et al. Genotype, haplotype and copy-number variation in worldwide human populations. Nature. 2008;451:998–1003. doi: 10.1038/nature06742. [DOI] [PubMed] [Google Scholar]
- 21.Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19:1655–1664. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hill W, Robertson A. Linkage disequilibrium in finite populations. Theor Appl Genet. 1968;38:226–231. doi: 10.1007/BF01245622. [DOI] [PubMed] [Google Scholar]
- 23.Pritchard JK, Przeworski M. Linkage disequilibrium in humans: Models and data. Am J Hum Genet. 2001;69:1–14. doi: 10.1086/321275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Serre D, Nadon R, Hudson TJ. Large-scale recombination rate patterns are conserved among human populations. Genome Res. 2005;15:1547–1552. doi: 10.1101/gr.4211905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wilson JF, Goldstein DB. Consistent long-range linkage disequilibrium generated by admixture in a Bantu-Semitic hybrid population. Am J Hum Genet. 2000;67:926–935. doi: 10.1086/303083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berniell-Lee G, et al. Genetic and demographic implications of the Bantu expansion: Insights from human paternal lineages. Mol Biol Evol. 2009;26:1581–1589. doi: 10.1093/molbev/msp069. [DOI] [PubMed] [Google Scholar]
- 27.Gendzekhadze K, et al. Co-evolution of KIR2DL3 with HLA-C in a human population retaining minimal essential diversity of KIR and HLA class I ligands. Proc Natl Acad Sci USA. 2009;106:18692–18697. doi: 10.1073/pnas.0906051106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bryc K, et al. Genome-wide patterns of population structure and admixture in West Africans and African Americans. Proc Natl Acad Sci USA. 2010;107:786–791. doi: 10.1073/pnas.0909559107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jobin MJ, Mountain JL. REJECTOR: Software for population history inference from genetic data via a rejection algorithm. Bioinformatics. 2008;24:2936–2937. doi: 10.1093/bioinformatics/btn540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blurton Jones NG, Smith LC, O'Connell JF, Hawkes K, Kamuzora CL. Demography of the Hadza, an increasing and high density population of Savanna foragers. Am J Phys Anthropol. 1992;89:159–181. doi: 10.1002/ajpa.1330890204. [DOI] [PubMed] [Google Scholar]
- 31.Conrad DF, et al. A worldwide survey of haplotype variation and linkage disequilibrium in the human genome. Nat Genet. 2006;38:1251–1260. doi: 10.1038/ng1911. [DOI] [PubMed] [Google Scholar]
- 32.Voight BF, Kudaravalli S, Wen X, Pritchard JK. A map of recent positive selection in the human genome. PLoS Biol. 2006;4:e72. doi: 10.1371/journal.pbio.0040072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pickrell JK, et al. Signals of recent positive selection in a worldwide sample of human populations. Genome Res. 2009;19:826–837. doi: 10.1101/gr.087577.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fellay J, et al. A whole-genome association study of major determinants for host control of HIV-1. Science. 2007;317:944–947. doi: 10.1126/science.1143767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Limou S, et al. ANRS Genomic Group. Genomewide association study of an AIDS-nonprogression cohort emphasizes the role played by HLA genes (ANRS Genomewide Association Study 02) J Infect Dis. 2009;199:419–426. doi: 10.1086/596067. [DOI] [PubMed] [Google Scholar]
- 36.Harding RM, McVean G. A structured ancestral population for the evolution of modern humans. Curr Opin Genet Dev. 2004;14:667–674. doi: 10.1016/j.gde.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 37.Plagnol V, Wall JD. Possible ancestral structure in human populations. PLoS Genet. 2006;2:e105. doi: 10.1371/journal.pgen.0020105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Green RE, et al. A draft sequence of the Neandertal genome. Science. 2010;328:710–722. doi: 10.1126/science.1188021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhivotovsky LA, Rosenberg NA, Feldman MW. Features of evolution and expansion of modern humans, inferred from genomewide microsatellite markers. Am J Hum Genet. 2003;72:1171–1186. doi: 10.1086/375120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Quintana-Murci L, et al. Maternal traces of deep common ancestry and asymmetric gene flow between Pygmy hunter-gatherers and Bantu-speaking farmers. Proc Natl Acad Sci USA. 2008;105:1596–1601. doi: 10.1073/pnas.0711467105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Batini C, et al. Insights into the demographic history of african pygmies from complete mitochondrial genomes. Mol Biol Evol. 2011;28:1099–1110. doi: 10.1093/molbev/msq294. [DOI] [PubMed] [Google Scholar]
- 42.Cruciani F, et al. A back migration from Asia to sub-Saharan Africa is supported by high-resolution analysis of human Y-chromosome haplotypes. Am J Hum Genet. 2002;70:1197–1214. doi: 10.1086/340257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mitchell P. The Archaeology of Southern Africa. Cambridge, UK: Cambridge Univ Press; 2002. [Google Scholar]
- 44.Tarazona-Santos E, Tishkoff SA. Divergent patterns of linkage disequilibrium and haplotype structure across global populations at the interleukin-13 (IL13) locus. Genes Immun. 2005;6:53–65. doi: 10.1038/sj.gene.6364149. [DOI] [PubMed] [Google Scholar]
- 45.Tishkoff SA, et al. Global patterns of linkage disequilibrium at the CD4 locus and modern human origins. Science. 1996;271:1380–1387. doi: 10.1126/science.271.5254.1380. [DOI] [PubMed] [Google Scholar]
- 46.Henshilwood CS, d'Errico F, Marean CW, Milo RG, Yates R. An early bone tool industry from the Middle Stone Age at Blombos Cave, South Africa: implications for the origins of modern human behaviour, symbolism and language. J Hum Evol. 2001;41:631–678. doi: 10.1006/jhev.2001.0515. [DOI] [PubMed] [Google Scholar]
- 47.Scholz CA, et al. East African megadroughts between 135 and 75 thousand years ago and bearing on early-modern human origins. Proc Natl Acad Sci USA. 2007;104:16416–16421. doi: 10.1073/pnas.0703874104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Texier PJ, et al. From the Cover: A Howiesons Poort tradition of engraving ostrich eggshell containers dated to 60,000 years ago at Diepkloof Rock Shelter, South Africa. Proc Natl Acad Sci USA. 2010;107:6180–6185. doi: 10.1073/pnas.0913047107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Colombo S, et al. Swiss HIV Cohort Study. The HCP5 single-nucleotide polymorphism: A simple screening tool for prediction of hypersensitivity reaction to abacavir. J Infect Dis. 2008;198:864–867. doi: 10.1086/591184. [DOI] [PubMed] [Google Scholar]
- 50.Knight A, et al. African Y chromosome and mtDNA divergence provides insight into the history of click languages. Curr Biol. 2003;13:464–473. doi: 10.1016/s0960-9822(03)00130-1. [DOI] [PubMed] [Google Scholar]
- 51.Li H, et al. Geographically separate increases in the frequency of the derived ADH1B*47His allele in eastern and western Asia. Am J Hum Genet. 2007;81:842–846. doi: 10.1086/521201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Donnelly MP, et al. The distribution and most recent common ancestor of the 17q21 inversion in humans. Am J Hum Genet. 2010;86:161–171. doi: 10.1016/j.ajhg.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Greenberg JH. The Languages of Africa. Bloomington, IN: Indiana University; 1963. [Google Scholar]
- 54.Browning SR, Browning BL. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am J Hum Genet. 2007;81:1084–1097. doi: 10.1086/521987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Purcell S, et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nalls MA, et al. Measures of autozygosity in decline: Globalization, urbanization, and its implications for medical genetics. PLoS Genet. 2009;5:e1000415. doi: 10.1371/journal.pgen.1000415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Auton A, et al. Global distribution of genomic diversity underscores rich complex history of continental human populations. Genome Res. 2009;19:795–803. doi: 10.1101/gr.088898.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.