Abstract
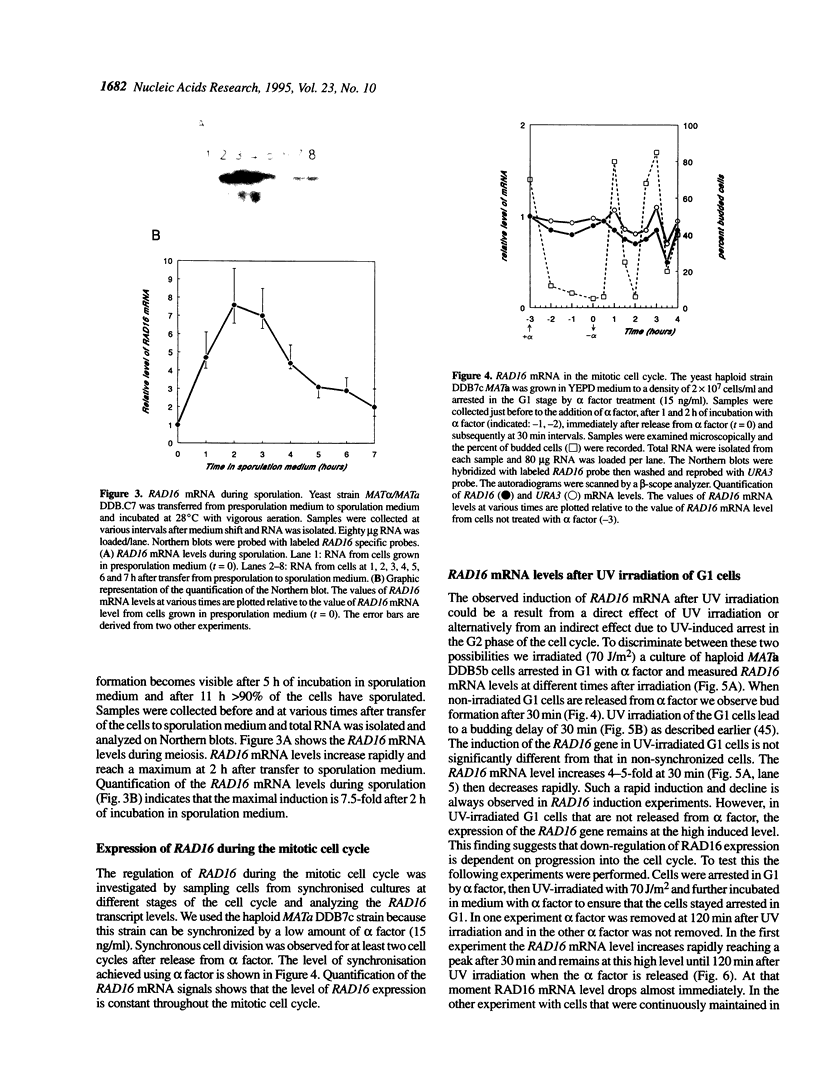
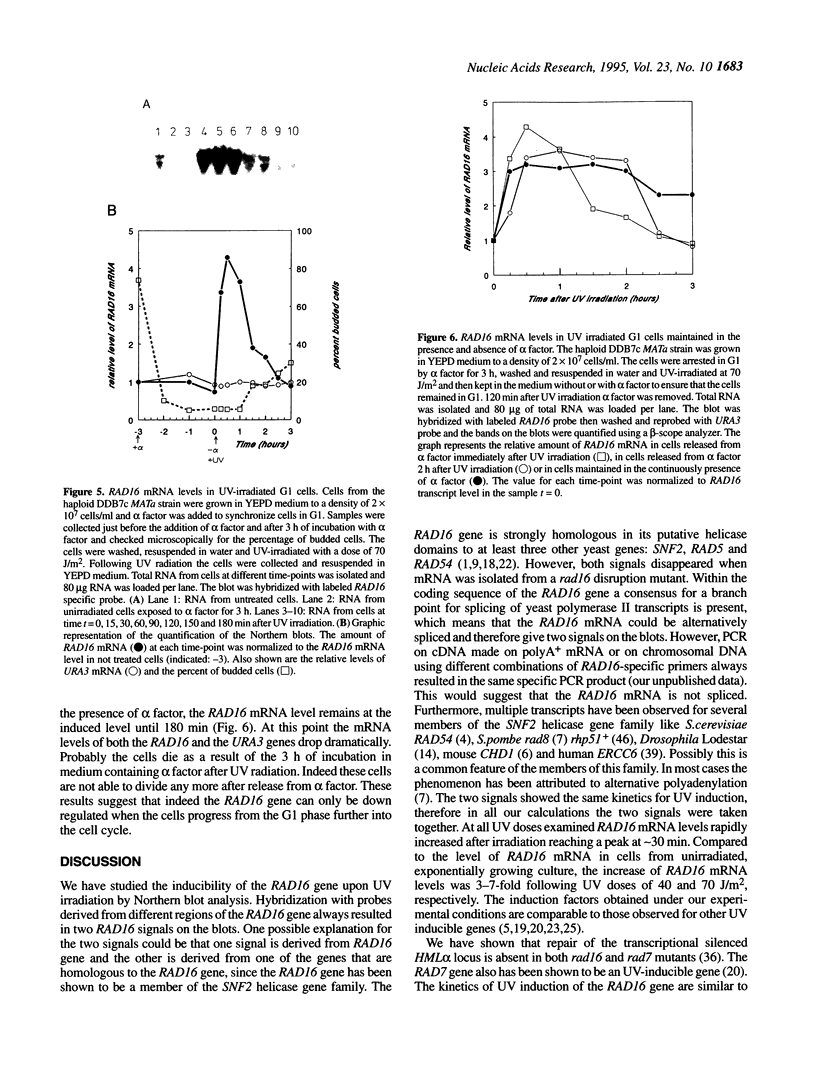
The RAD16 gene product has been shown to be essential for the repair of the silenced mating type loci [Bang et al. (1992) Nucleic Acids Res. 20, 3925-3931]. More recently we demonstrated that the RAD16 and RAD7 proteins are also required for repair of non-transcribed strands of active genes in Saccharomyces cerevisiae [Waters et al. (1993) Mol. Gen. Genet. 239, 28-32]. We have studied the regulation of the RAD16 gene and found that the RAD16 transcript levels increased up to 7-fold upon UV irradiation. Heat shock at 42 degrees C also results in elevated levels of RAD16 mRNA. In sporulating MAT alpha/MATa diploid cells RAD16 mRNA is also induced. The basal level of the RAD16 transcript is constant during the mitotic cell cycle. G1-arrested cells show normal induction of RAD16 mRNA upon UV irradiation demonstrating that the induction is not a secondary consequence of G2 cell cycle arrest following UV irradiation. However, in cells arrested in G1 the induction of RAD16 mRNA after UV irradiation is not followed by a rapid decline as occurs in normal growing cells suggesting that the down regulation of RAD16 transcription is dependent on progression into the cell cycle.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- BURNS V. W. X-ray-induced division delay of individual yeast cells. Radiat Res. 1956 May;4(5):394–412. [PubMed] [Google Scholar]
- Bang D. D., Verhage R., Goosen N., Brouwer J., van de Putte P. Molecular cloning of RAD16, a gene involved in differential repair in Saccharomyces cerevisiae. Nucleic Acids Res. 1992 Aug 11;20(15):3925–3931. doi: 10.1093/nar/20.15.3925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole G. M., Mortimer R. K. Failure to induce a DNA repair gene, RAD54, in Saccharomyces cerevisiae does not affect DNA repair or recombination phenotypes. Mol Cell Biol. 1989 Aug;9(8):3314–3322. doi: 10.1128/mcb.9.8.3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole G. M., Schild D., Lovett S. T., Mortimer R. K. Regulation of RAD54- and RAD52-lacZ gene fusions in Saccharomyces cerevisiae in response to DNA damage. Mol Cell Biol. 1987 Mar;7(3):1078–1084. doi: 10.1128/mcb.7.3.1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole G. M., Schild D., Mortimer R. K. Two DNA repair and recombination genes in Saccharomyces cerevisiae, RAD52 and RAD54, are induced during meiosis. Mol Cell Biol. 1989 Jul;9(7):3101–3104. doi: 10.1128/mcb.9.7.3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doe C. L., Murray J. M., Shayeghi M., Hoskins M., Lehmann A. R., Carr A. M., Watts F. Z. Cloning and characterisation of the Schizosaccharomyces pombe rad8 gene, a member of the SNF2 helicase family. Nucleic Acids Res. 1993 Dec 25;21(25):5964–5971. doi: 10.1093/nar/21.25.5964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elledge S. J., Davis R. W. Identification and isolation of the gene encoding the small subunit of ribonucleotide reductase from Saccharomyces cerevisiae: DNA damage-inducible gene required for mitotic viability. Mol Cell Biol. 1987 Aug;7(8):2783–2793. doi: 10.1128/mcb.7.8.2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery H. S., Schild D., Kellogg D. E., Mortimer R. K. Sequence of RAD54, a Saccharomyces cerevisiae gene involved in recombination and repair. Gene. 1991 Jul 31;104(1):103–106. doi: 10.1016/0378-1119(91)90473-o. [DOI] [PubMed] [Google Scholar]
- Finley D., Ozkaynak E., Varshavsky A. The yeast polyubiquitin gene is essential for resistance to high temperatures, starvation, and other stresses. Cell. 1987 Mar 27;48(6):1035–1046. doi: 10.1016/0092-8674(87)90711-2. [DOI] [PubMed] [Google Scholar]
- Friedberg E. C. Deoxyribonucleic acid repair in the yeast Saccharomyces cerevisiae. Microbiol Rev. 1988 Mar;52(1):70–102. doi: 10.1128/mr.52.1.70-102.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girdham C. H., Glover D. M. Chromosome tangling and breakage at anaphase result from mutations in lodestar, a Drosophila gene encoding a putative nucleoside triphosphate-binding protein. Genes Dev. 1991 Oct;5(10):1786–1799. doi: 10.1101/gad.5.10.1786. [DOI] [PubMed] [Google Scholar]
- Hurd H. K., Roberts C. W., Roberts J. W. Identification of the gene for the yeast ribonucleotide reductase small subunit and its inducibility by methyl methanesulfonate. Mol Cell Biol. 1987 Oct;7(10):3673–3677. doi: 10.1128/mcb.7.10.3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R. E., Henderson S. T., Petes T. D., Prakash S., Bankmann M., Prakash L. Saccharomyces cerevisiae RAD5-encoded DNA repair protein contains DNA helicase and zinc-binding sequence motifs and affects the stability of simple repetitive sequences in the genome. Mol Cell Biol. 1992 Sep;12(9):3807–3818. doi: 10.1128/mcb.12.9.3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston L. H., White J. H., Johnson A. L., Lucchini G., Plevani P. The yeast DNA polymerase I transcript is regulated in both the mitotic cell cycle and in meiosis and is also induced after DNA damage. Nucleic Acids Res. 1987 Jul 10;15(13):5017–5030. doi: 10.1093/nar/15.13.5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones J. S., Prakash L., Prakash S. Regulated expression of the Saccharomyces cerevisiae DNA repair gene RAD7 in response to DNA damage and during sporulation. Nucleic Acids Res. 1990 Jun 11;18(11):3281–3285. doi: 10.1093/nar/18.11.3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones J. S., Prakash L. Transcript levels of the Saccharomyces cerevisiae DNA repair gene RAD18 increase in UV irradiated cells and during meiosis but not during the mitotic cell cycle. Nucleic Acids Res. 1991 Feb 25;19(4):893–898. doi: 10.1093/nar/19.4.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurent B. C., Treitel M. A., Carlson M. Functional interdependence of the yeast SNF2, SNF5, and SNF6 proteins in transcriptional activation. Proc Natl Acad Sci U S A. 1991 Apr 1;88(7):2687–2691. doi: 10.1073/pnas.88.7.2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little J. W., Mount D. W. The SOS regulatory system of Escherichia coli. Cell. 1982 May;29(1):11–22. doi: 10.1016/0092-8674(82)90085-x. [DOI] [PubMed] [Google Scholar]
- Madura K., Prakash S. Nucleotide sequence, transcript mapping, and regulation of the RAD2 gene of Saccharomyces cerevisiae. J Bacteriol. 1986 Jun;166(3):914–923. doi: 10.1128/jb.166.3.914-923.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madura K., Prakash S., Prakash L. Expression of the Saccharomyces cerevisiae DNA repair gene RAD6 that encodes a ubiquitin conjugating enzyme, increases in response to DNA damage and in meiosis but remains constant during the mitotic cell cycle. Nucleic Acids Res. 1990 Feb 25;18(4):771–778. doi: 10.1093/nar/18.4.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madura K., Prakash S. The Saccharomyces cerevisiae DNA repair gene RAD2 is regulated in meiosis but not during the mitotic cell cycle. Mol Cell Biol. 1990 Jun;10(6):3256–3257. doi: 10.1128/mcb.10.6.3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madura K., Prakash S. Transcript levels of the Saccharomyes cerevisiae DNA repair gene RAD23 increase in response to UV light and in meiosis but remain constant in the mitotic cell cycle. Nucleic Acids Res. 1990 Aug 25;18(16):4737–4742. doi: 10.1093/nar/18.16.4737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClanahan T., McEntee K. DNA damage and heat shock dually regulate genes in Saccharomyces cerevisiae. Mol Cell Biol. 1986 Jan;6(1):90–96. doi: 10.1128/mcb.6.1.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClanahan T., McEntee K. Specific transcripts are elevated in Saccharomyces cerevisiae in response to DNA damage. Mol Cell Biol. 1984 Nov;4(11):2356–2363. doi: 10.1128/mcb.4.11.2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muris D. F., Vreeken K., Carr A. M., Broughton B. C., Lehmann A. R., Lohman P. H., Pastink A. Cloning the RAD51 homologue of Schizosaccharomyces pombe. Nucleic Acids Res. 1993 Sep 25;21(19):4586–4591. doi: 10.1093/nar/21.19.4586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson T. A., Prakash L., Prakash S., Osley M. A., Reed S. I. Regulation of CDC9, the Saccharomyces cerevisiae gene that encodes DNA ligase. Mol Cell Biol. 1985 Jan;5(1):226–235. doi: 10.1128/mcb.5.1.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby S. W., Szostak J. W. Specific Saccharomyces cerevisiae genes are expressed in response to DNA-damaging agents. Mol Cell Biol. 1985 Jan;5(1):75–84. doi: 10.1128/mcb.5.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiestl R. H., Reynolds P., Prakash S., Prakash L. Cloning and sequence analysis of the Saccharomyces cerevisiae RAD9 gene and further evidence that its product is required for cell cycle arrest induced by DNA damage. Mol Cell Biol. 1989 May;9(5):1882–1896. doi: 10.1128/mcb.9.5.1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt M. E., Brown T. A., Trumpower B. L. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 1990 May 25;18(10):3091–3092. doi: 10.1093/nar/18.10.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebastian J., Sancar G. B. A damage-responsive DNA binding protein regulates transcription of the yeast DNA repair gene PHR1. Proc Natl Acad Sci U S A. 1991 Dec 15;88(24):11251–11255. doi: 10.1073/pnas.88.24.11251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siede W., Friedberg A. S., Friedberg E. C. RAD9-dependent G1 arrest defines a second checkpoint for damaged DNA in the cell cycle of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1993 Sep 1;90(17):7985–7989. doi: 10.1073/pnas.90.17.7985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumrada R., Cooper T. G. Control of vacuole permeability and protein degradation by the cell cycle arrest signal in Saccharomyces cerevisiae. J Bacteriol. 1978 Oct;136(1):234–246. doi: 10.1128/jb.136.1.234-246.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terleth C., Schenk P., Poot R., Brouwer J., van de Putte P. Differential repair of UV damage in rad mutants of Saccharomyces cerevisiae: a possible function of G2 arrest upon UV irradiation. Mol Cell Biol. 1990 Sep;10(9):4678–4684. doi: 10.1128/mcb.10.9.4678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terleth C., van Sluis C. A., van de Putte P. Differential repair of UV damage in Saccharomyces cerevisiae. Nucleic Acids Res. 1989 Jun 26;17(12):4433–4439. doi: 10.1093/nar/17.12.4433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treger J. M., Heichman K. A., McEntee K. Expression of the yeast UB14 gene increases in response to DNA-damaging agents and in meiosis. Mol Cell Biol. 1988 Mar;8(3):1132–1136. doi: 10.1128/mcb.8.3.1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troelstra C., van Gool A., de Wit J., Vermeulen W., Bootsma D., Hoeijmakers J. H. ERCC6, a member of a subfamily of putative helicases, is involved in Cockayne's syndrome and preferential repair of active genes. Cell. 1992 Dec 11;71(6):939–953. doi: 10.1016/0092-8674(92)90390-x. [DOI] [PubMed] [Google Scholar]
- Van Houten B. Nucleotide excision repair in Escherichia coli. Microbiol Rev. 1990 Mar;54(1):18–51. doi: 10.1128/mr.54.1.18-51.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhage R., Zeeman A. M., de Groot N., Gleig F., Bang D. D., van de Putte P., Brouwer J. The RAD7 and RAD16 genes, which are essential for pyrimidine dimer removal from the silent mating type loci, are also required for repair of the nontranscribed strand of an active gene in Saccharomyces cerevisiae. Mol Cell Biol. 1994 Sep;14(9):6135–6142. doi: 10.1128/mcb.14.9.6135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker G. C. Inducible DNA repair systems. Annu Rev Biochem. 1985;54:425–457. doi: 10.1146/annurev.bi.54.070185.002233. [DOI] [PubMed] [Google Scholar]
- Waters R., Zhang R., Jones N. J. Inducible removal of UV-induced pyrimidine dimers from transcriptionally active and inactive genes of Saccharomyces cerevisiae. Mol Gen Genet. 1993 May;239(1-2):28–32. doi: 10.1007/BF00281597. [DOI] [PubMed] [Google Scholar]
- Weinert T. A., Hartwell L. H. The RAD9 gene controls the cell cycle response to DNA damage in Saccharomyces cerevisiae. Science. 1988 Jul 15;241(4863):317–322. doi: 10.1126/science.3291120. [DOI] [PubMed] [Google Scholar]
- Witkin E. M. Ultraviolet mutagenesis and inducible DNA repair in Escherichia coli. Bacteriol Rev. 1976 Dec;40(4):869–907. doi: 10.1128/br.40.4.869-907.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]