Abstract
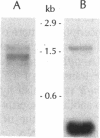
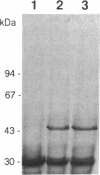
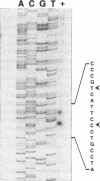
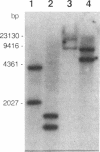
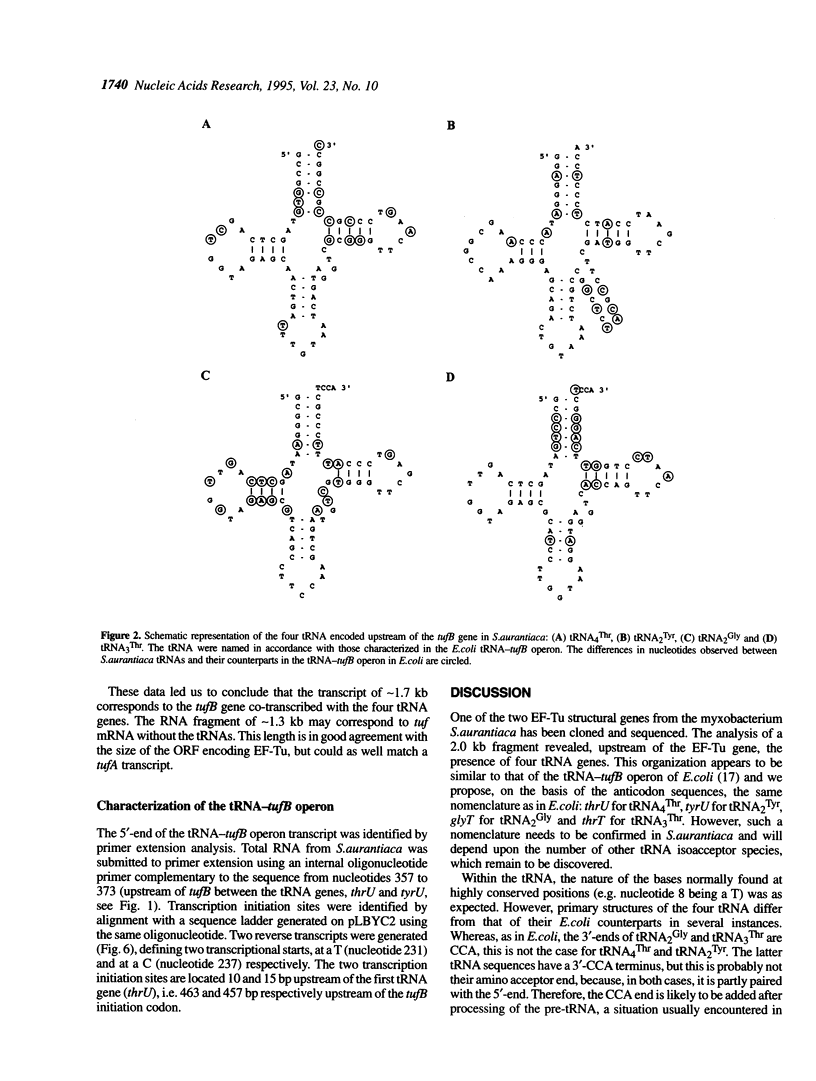
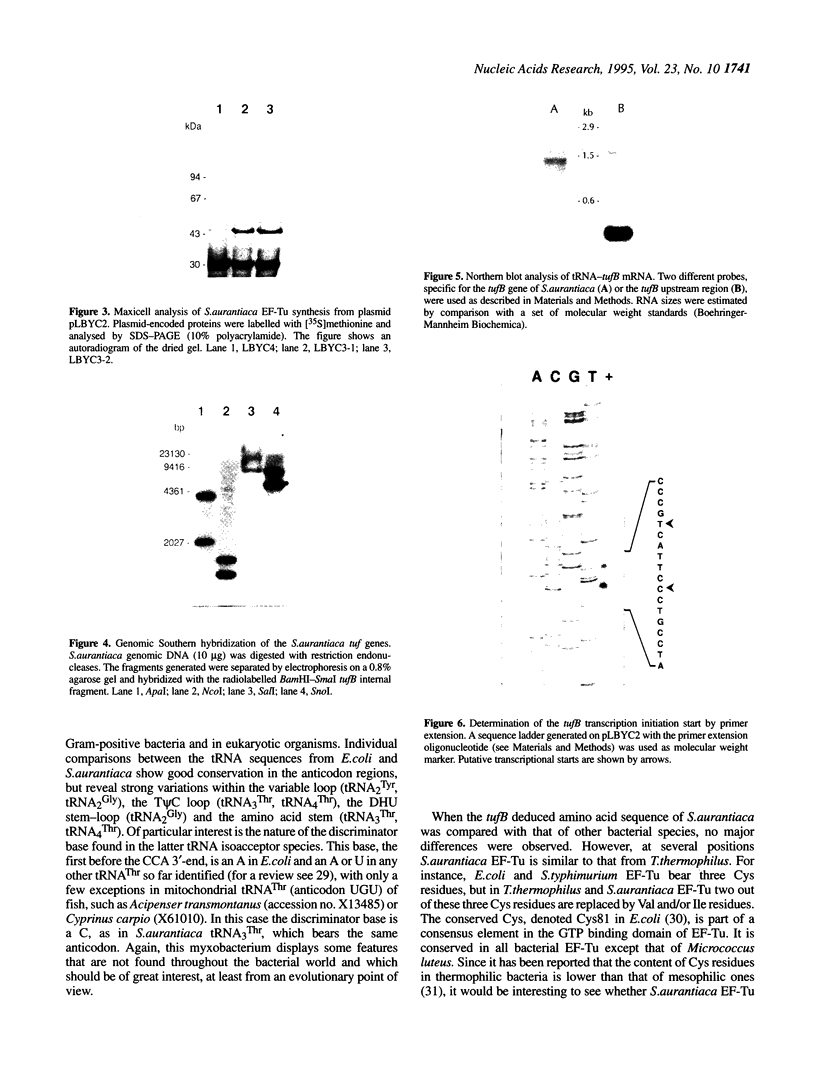
The tufB gene, encoding elongation factor Tu (EF-Tu), from the myxobacterium Stigmatella aurantiaca was cloned and sequenced. It is preceded by four tRNA genes, the first ever described in myxobacteria. The tRNA synthesized from these genes and the general organization of the locus seem identical to that of Escherichia coli, but differences of potential importance were found in the tRNA sequences and in the intergenic regions. The primary structure of EF-Tu was deduced from the tufB DNA sequence. The factor is composed of 396 amino acids, with a predicted molecular mass of 43.4 kDa, which was confirmed by expression of tufB in maxicells. Sequence comparisons between S.aurantiaca EF-Tu and other bacterial homologues from E.coli, Salmonella typhimurium and Thermus thermophilus displayed extensive homologies (75.9%). Among the variable positions, two Cys residues probably involved in the temperature sensitivity of E.coli and S.typhimurium EF-Tu are replaced in T.thermophilus and S.aurantiaca EF-Tu. Since two or even three tuf genes have been described in other bacterial species, the presence of multiple tuf genes was sought for. Southern and Northern analysis are consistent with two tuf genes in the genome of S.aurantiaca. Primer extension experiments indicate that the four tRNA genes and tufB are organized in a single operon.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- An G., Friesen J. D. The nucleotide sequence of tufB and four nearby tRNA structural genes of Escherichia coli. Gene. 1980 Dec;12(1-2):33–39. doi: 10.1016/0378-1119(80)90013-x. [DOI] [PubMed] [Google Scholar]
- Anborgh P. H., Parmeggiani A., Jonák J. Site-directed mutagenesis of elongation factor Tu. The functional and structural role of residue Cys81. Eur J Biochem. 1992 Sep 1;208(2):251–257. doi: 10.1111/j.1432-1033.1992.tb17180.x. [DOI] [PubMed] [Google Scholar]
- Arai K., Clark B. F., Duffy L., Jones M. D., Kaziro Y., Laursen R. A., L'Italien J., Miller D. L., Nagarkatti S., Nakamura S. Primary structure of elongation factor Tu from Escherichia coli. Proc Natl Acad Sci U S A. 1980 Mar;77(3):1326–1330. doi: 10.1073/pnas.77.3.1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bremaud L., Derijard B., Cenatiempo Y. Selective amplification of DNA fragments coding for the G domain of factors IF2 and EF-Tu, two G proteins from the myxobacterium Stigmatella aurantiaca. PCR Methods Appl. 1993 Dec;3(3):195–199. doi: 10.1101/gr.3.3.195. [DOI] [PubMed] [Google Scholar]
- Cheng Y. L., Kalman L. V., Kaiser D. The dsg gene of Myxococcus xanthus encodes a protein similar to translation initiation factor IF3. J Bacteriol. 1994 Mar;176(5):1427–1433. doi: 10.1128/jb.176.5.1427-1433.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Furano A. V. The elongation factor Tu coded by the tufA gene of Escherichia coli K-12 is almost identical to that coded by the tufB gene. J Biol Chem. 1977 Mar 25;252(6):2154–2157. [PubMed] [Google Scholar]
- Giegé R., Puglisi J. D., Florentz C. tRNA structure and aminoacylation efficiency. Prog Nucleic Acid Res Mol Biol. 1993;45:129–206. doi: 10.1016/s0079-6603(08)60869-7. [DOI] [PubMed] [Google Scholar]
- Gordon J. Regulation of the in vivo synthesis of the polypeptide chain elongation factors in Escherichia coli. Biochemistry. 1970 Feb 17;9(4):912–917. doi: 10.1021/bi00806a028. [DOI] [PubMed] [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983 Jun 5;166(4):557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Hudson L., Rossi J., Landy A. Dual function transcripts specifying tRNA and mRNA. Nature. 1981 Dec 3;294(5840):422–427. doi: 10.1038/294422a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes D. Both genes for EF-Tu in Salmonella typhimurium are individually dispensable for growth. J Mol Biol. 1990 Sep 5;215(1):41–51. doi: 10.1016/S0022-2836(05)80093-2. [DOI] [PubMed] [Google Scholar]
- Jaskunas S. R., Lindahl L., Nomura M. Identification of two copies of the gene for the elongation factor EF-Tu in E. coli. Nature. 1975 Oct 9;257(5526):458–462. doi: 10.1038/257458a0. [DOI] [PubMed] [Google Scholar]
- Jones M. D., Petersen T. E., Nielsen K. M., Magnusson S., Sottrup-Jensen L., Gausing K., Clark B. F. The complete amino-acid sequence of elongation factor Tu from Escherichia coli. Eur J Biochem. 1980 Jul;108(2):507–526. doi: 10.1111/j.1432-1033.1980.tb04748.x. [DOI] [PubMed] [Google Scholar]
- Jurnak F. Structure of the GDP domain of EF-Tu and location of the amino acids homologous to ras oncogene proteins. Science. 1985 Oct 4;230(4721):32–36. doi: 10.1126/science.3898365. [DOI] [PubMed] [Google Scholar]
- Kagawa Y., Nojima H., Nukiwa N., Ishizuka M., Nakajima T., Yasuhara T., Tanaka T., Oshima T. High guanine plus cytosine content in the third letter of codons of an extreme thermophile. DNA sequence of the isopropylmalate dehydrogenase of Thermus thermophilus. J Biol Chem. 1984 Mar 10;259(5):2956–2960. [PubMed] [Google Scholar]
- Kaiser D. Genetic systems in myxobacteria. Methods Enzymol. 1991;204:357–372. doi: 10.1016/0076-6879(91)04018-j. [DOI] [PubMed] [Google Scholar]
- Komano T., Franceschini T., Inouye S. Identification of a vegetative promoter in Myxococcus xanthus. A protein that has homology to histones. J Mol Biol. 1987 Aug 5;196(3):517–524. doi: 10.1016/0022-2836(87)90029-5. [DOI] [PubMed] [Google Scholar]
- Kushiro M., Shimizu M., Tomita K. Molecular cloning and sequence determination of the tuf gene coding for the elongation factor Tu of Thermus thermophilus HB8. Eur J Biochem. 1987 Dec 30;170(1-2):93–98. doi: 10.1111/j.1432-1033.1987.tb13671.x. [DOI] [PubMed] [Google Scholar]
- Leighton T. Sporulation-specific translational discrimination in Bacillus subtilis. J Mol Biol. 1974 Jul 15;86(4):855–863. doi: 10.1016/0022-2836(74)90358-1. [DOI] [PubMed] [Google Scholar]
- Lucas-Lenard J. Protein biosynthesis. Annu Rev Biochem. 1971;40:409–448. doi: 10.1146/annurev.bi.40.070171.002205. [DOI] [PubMed] [Google Scholar]
- McGowan S. J., Gorham H. C., Hodgson D. A. Light-induced carotenogenesis in Myxococcus xanthus: DNA sequence analysis of the carR region. Mol Microbiol. 1993 Nov;10(4):713–735. doi: 10.1111/j.1365-2958.1993.tb00943.x. [DOI] [PubMed] [Google Scholar]
- Moazed D., Noller H. F. Intermediate states in the movement of transfer RNA in the ribosome. Nature. 1989 Nov 9;342(6246):142–148. doi: 10.1038/342142a0. [DOI] [PubMed] [Google Scholar]
- Pedersen S., Bloch P. L., Reeh S., Neidhardt F. C. Patterns of protein synthesis in E. coli: a catalog of the amount of 140 individual proteins at different growth rates. Cell. 1978 May;14(1):179–190. doi: 10.1016/0092-8674(78)90312-4. [DOI] [PubMed] [Google Scholar]
- Ross W., Gosink K. K., Salomon J., Igarashi K., Zou C., Ishihama A., Severinov K., Gourse R. L. A third recognition element in bacterial promoters: DNA binding by the alpha subunit of RNA polymerase. Science. 1993 Nov 26;262(5138):1407–1413. doi: 10.1126/science.8248780. [DOI] [PubMed] [Google Scholar]
- Sancar A., Hack A. M., Rupp W. D. Simple method for identification of plasmid-coded proteins. J Bacteriol. 1979 Jan;137(1):692–693. doi: 10.1128/jb.137.1.692-693.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh M., Tanaka T., Kushiro A., Hakoshima T., Tomita K. Molecular cloning, nucleotide sequence and expression of the tufB gene encoding elongation factor Tu from Thermus thermophilus HB8. FEBS Lett. 1991 Aug 19;288(1-2):98–100. doi: 10.1016/0014-5793(91)81011-v. [DOI] [PubMed] [Google Scholar]
- Shimkets L. J. Social and developmental biology of the myxobacteria. Microbiol Rev. 1990 Dec;54(4):473–501. doi: 10.1128/mr.54.4.473-501.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skladny H., Heidelbach M., Schairer H. U. Cloning and characterization of the gene encoding the major sigma factor of Stigmatella aurantiaca. Gene. 1994 May 27;143(1):123–127. doi: 10.1016/0378-1119(94)90616-5. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Starich T., Zissler J. Movement of multiple DNA units between Myxococcus xanthus cells. J Bacteriol. 1989 May;171(5):2323–2336. doi: 10.1128/jb.171.5.2323-2336.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tojo N., Inouye S., Komano T. Cloning and nucleotide sequence of the Myxococcus xanthus lon gene: indispensability of lon for vegetative growth. J Bacteriol. 1993 Apr;175(8):2271–2277. doi: 10.1128/jb.175.8.2271-2277.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiborg O., Andersen C., Knudsen C. R., Kristensen T. J., Clark B. F. Towards an understanding of structure-function relationships of elongation factor Tu. Biotechnol Appl Biochem. 1994 Feb;19(Pt 1):3–15. [PubMed] [Google Scholar]
- Yokota T., Sugisaki H., Takanami M., Kaziro Y. The nucleotide sequence of the cloned tufA gene of Escherichia coli. Gene. 1980 Dec;12(1-2):25–31. doi: 10.1016/0378-1119(80)90012-8. [DOI] [PubMed] [Google Scholar]