Abstract
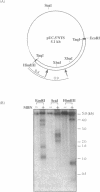
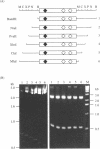
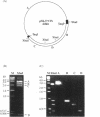
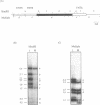
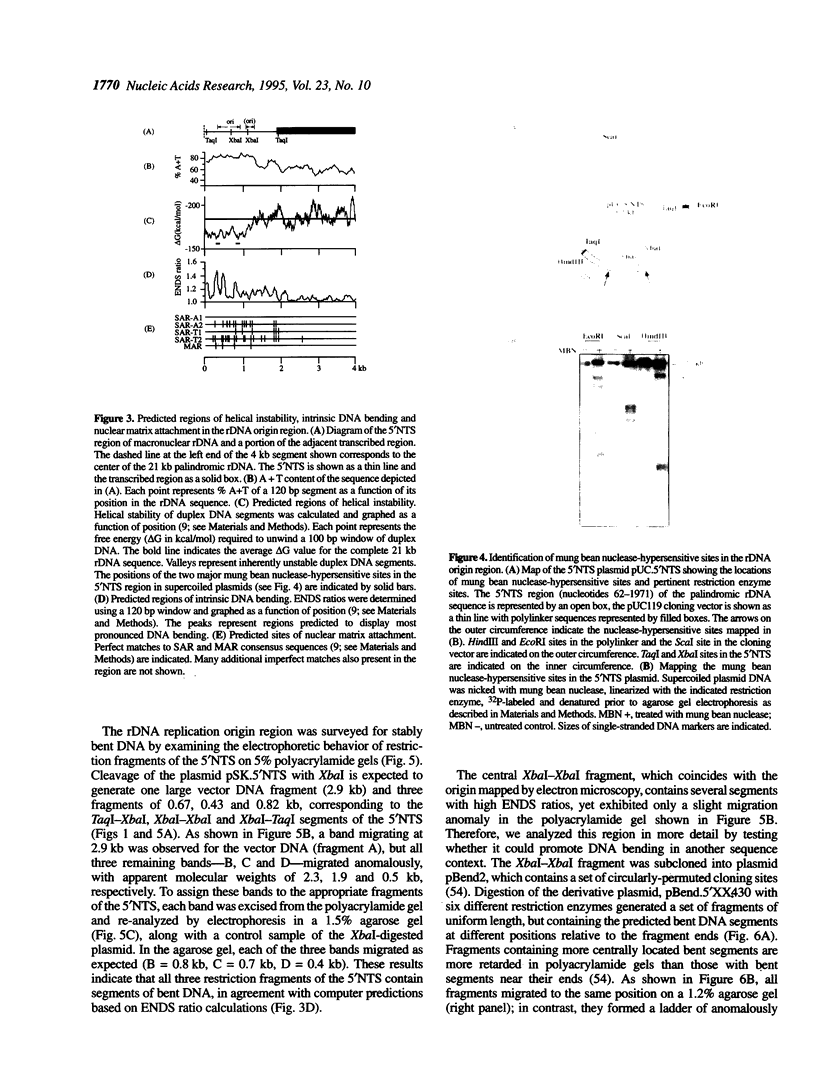
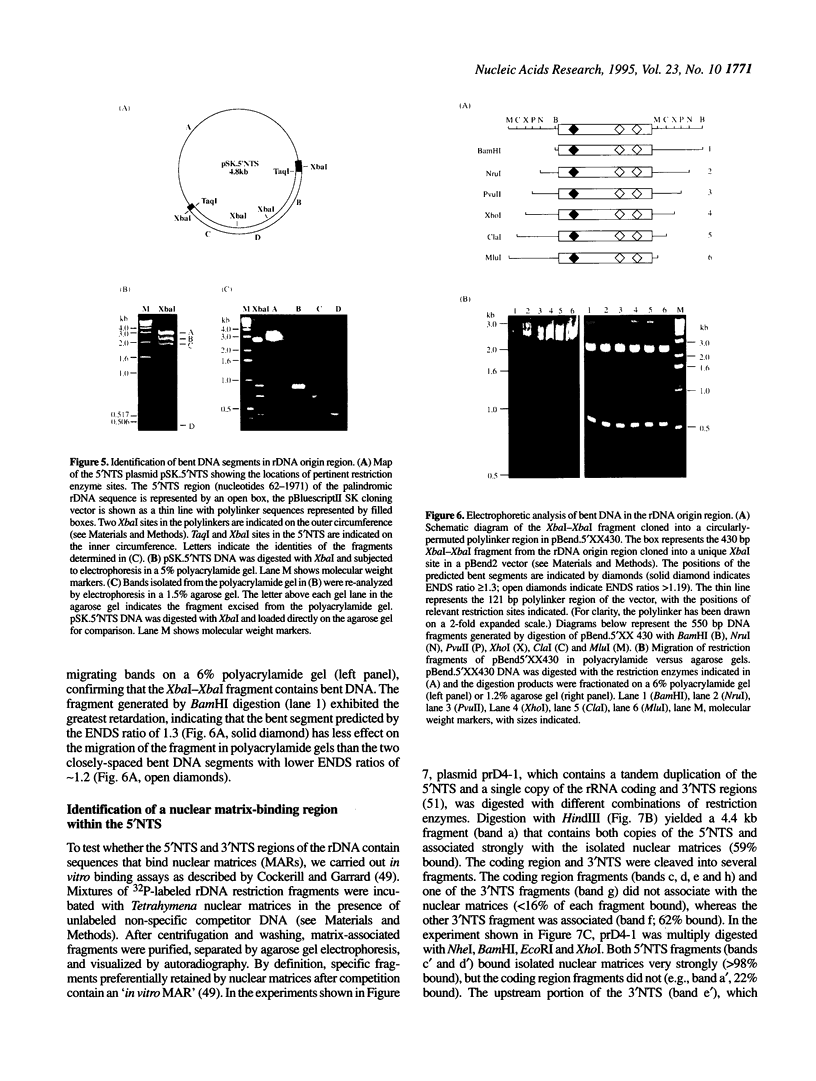
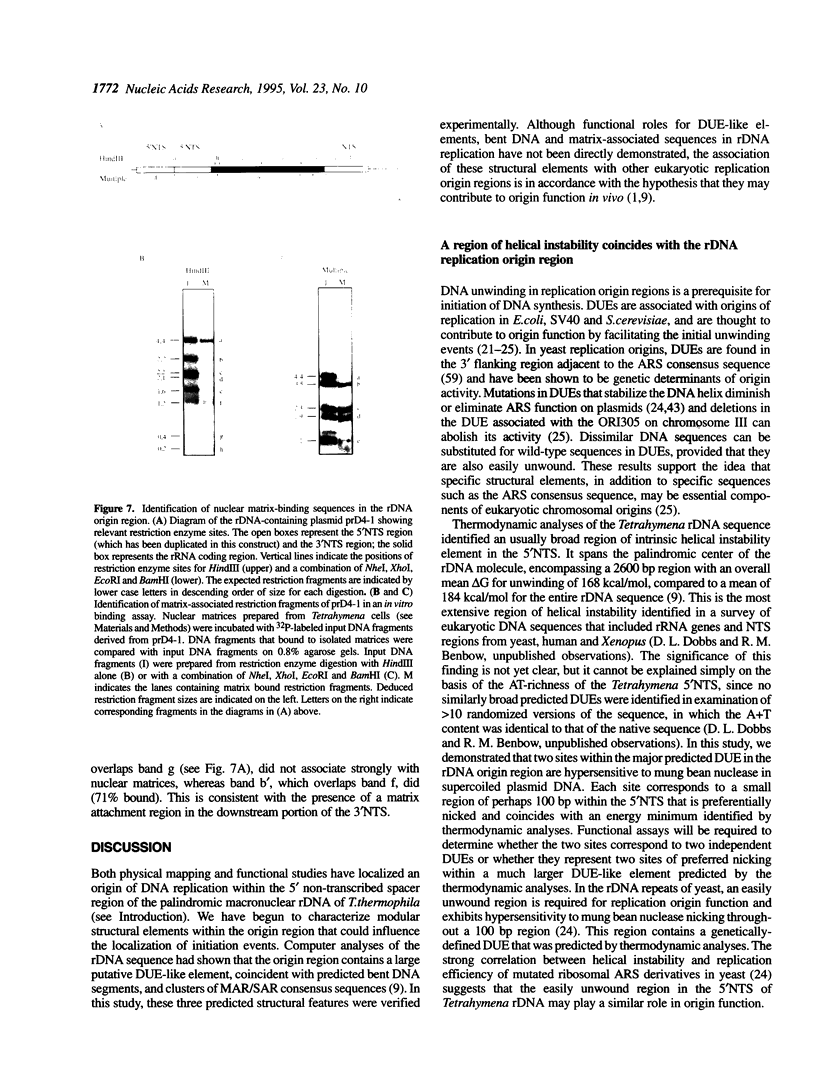
Computer analyses of the DNA replication origin region in the amplified rRNA genes of Tetrahymena thermophila identified a potential initiation zone in the 5'NTS [Dobbs, Shaiu and Benbow (1994), Nucleic Acids Res. 22, 2479-2489]. This region consists of a putative DNA unwinding element (DUE) aligned with predicted bent DNA segments, nuclear matrix or scaffold associated region (MAR/SAR) consensus sequences, and other common modular sequence elements previously shown to be clustered in eukaryotic chromosomal origin regions. In this study, two mung bean nuclease-hypersensitive sites in super-coiled plasmid DNA were localized within the major DUE-like element predicted by thermodynamic analyses. Three restriction fragments of the 5'NTS region predicted to contain bent DNA segments exhibited anomalous migration characteristic of bent DNA during electrophoresis on polyacrylamide gels. Restriction fragments containing the 5'NTS region bound Tetrahymena nuclear matrices in an in vitro binding assay, consistent with an association of the replication origin region with the nuclear matrix in vivo. The direct demonstration in a protozoan origin region of elements previously identified in Drosophila, chick and mammalian origin regions suggests that clusters of modular structural elements may be a conserved feature of eukaryotic chromosomal origins of replication.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Amati B. B., Gasser S. M. Chromosomal ARS and CEN elements bind specifically to the yeast nuclear scaffold. Cell. 1988 Sep 23;54(7):967–978. doi: 10.1016/0092-8674(88)90111-0. [DOI] [PubMed] [Google Scholar]
- Amati B., Pick L., Laroche T., Gasser S. M. Nuclear scaffold attachment stimulates, but is not essential for ARS activity in Saccharomyces cerevisiae: analysis of the Drosophila ftz SAR. EMBO J. 1990 Dec;9(12):4007–4016. doi: 10.1002/j.1460-2075.1990.tb07622.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amin A. A., Pearlman R. E. Autonomously replicating sequences from the non transcribed spacers of Tetrahymena thermophila ribosomal DNA. Nucleic Acids Res. 1985 Apr 11;13(7):2647–2659. doi: 10.1093/nar/13.7.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benbow R. M., Zhao J., Larson D. D. On the nature of origins of DNA replication in eukaryotes. Bioessays. 1992 Oct;14(10):661–670. doi: 10.1002/bies.950141004. [DOI] [PubMed] [Google Scholar]
- Bergemann A. D., Ma Z. W., Johnson E. M. Sequence of cDNA comprising the human pur gene and sequence-specific single-stranded-DNA-binding properties of the encoded protein. Mol Cell Biol. 1992 Dec;12(12):5673–5682. doi: 10.1128/mcb.12.12.5673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bode J., Kohwi Y., Dickinson L., Joh T., Klehr D., Mielke C., Kohwi-Shigematsu T. Biological significance of unwinding capability of nuclear matrix-associating DNAs. Science. 1992 Jan 10;255(5041):195–197. doi: 10.1126/science.1553545. [DOI] [PubMed] [Google Scholar]
- Bonven B. J., Gocke E., Westergaard O. A high affinity topoisomerase I binding sequence is clustered at DNAase I hypersensitive sites in Tetrahymena R-chromatin. Cell. 1985 Jun;41(2):541–551. doi: 10.1016/s0092-8674(85)80027-1. [DOI] [PubMed] [Google Scholar]
- Boulikas T. Nature of DNA sequences at the attachment regions of genes to the nuclear matrix. J Cell Biochem. 1993 May;52(1):14–22. doi: 10.1002/jcb.240520104. [DOI] [PubMed] [Google Scholar]
- Breslauer K. J., Frank R., Blöcker H., Marky L. A. Predicting DNA duplex stability from the base sequence. Proc Natl Acad Sci U S A. 1986 Jun;83(11):3746–3750. doi: 10.1073/pnas.83.11.3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brun C., Surdej P., Miassod R. Relationship between scaffold-attached regions, sequences replicating autonomously in yeast, and a chromosomal replication origin in the Drosophila rDNA. Exp Cell Res. 1993 Sep;208(1):104–114. doi: 10.1006/excr.1993.1227. [DOI] [PubMed] [Google Scholar]
- Caddle M. S., Lussier R. H., Heintz N. H. Intramolecular DNA triplexes, bent DNA and DNA unwinding elements in the initiation region of an amplified dihydrofolate reductase replicon. J Mol Biol. 1990 Jan 5;211(1):19–33. doi: 10.1016/0022-2836(90)90008-A. [DOI] [PubMed] [Google Scholar]
- Cech T. R., Brehm S. L. Replication of the extrachromosomal ribosomal RNA genes of Tetrahymena thermophilia. Nucleic Acids Res. 1981 Jul 24;9(14):3531–3543. doi: 10.1093/nar/9.14.3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challoner P. B., Amin A. A., Pearlman R. E., Blackburn E. H. Conserved arrangements of repeated DNA sequences in nontranscribed spacers of ciliate ribosomal RNA genes: evidence for molecular coevolution. Nucleic Acids Res. 1985 Apr 11;13(7):2661–2680. doi: 10.1093/nar/13.7.2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christiansen K., Bonven B. J., Westergaard O. Mapping of sequence-specific chromatin proteins by a novel method: topoisomerase I on Tetrahymena ribosomal chromatin. J Mol Biol. 1987 Feb 5;193(3):517–525. doi: 10.1016/0022-2836(87)90264-6. [DOI] [PubMed] [Google Scholar]
- Cockerill P. N., Garrard W. T. Chromosomal loop anchorage of the kappa immunoglobulin gene occurs next to the enhancer in a region containing topoisomerase II sites. Cell. 1986 Jan 31;44(2):273–282. doi: 10.1016/0092-8674(86)90761-0. [DOI] [PubMed] [Google Scholar]
- DePamphili M. L. How transcription factors regulate origins of DNA replication in eukaryotic cells. Trends Cell Biol. 1993 May;3(5):161–167. doi: 10.1016/0962-8924(93)90137-p. [DOI] [PubMed] [Google Scholar]
- DePamphilis M. L. Eukaryotic DNA replication: anatomy of an origin. Annu Rev Biochem. 1993;62:29–63. doi: 10.1146/annurev.bi.62.070193.000333. [DOI] [PubMed] [Google Scholar]
- Deb S., DeLucia A. L., Koff A., Tsui S., Tegtmeyer P. The adenine-thymine domain of the simian virus 40 core origin directs DNA bending and coordinately regulates DNA replication. Mol Cell Biol. 1986 Dec;6(12):4578–4584. doi: 10.1128/mcb.6.12.4578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dijkwel P. A., Hamlin J. L. Matrix attachment regions are positioned near replication initiation sites, genes, and an interamplicon junction in the amplified dihydrofolate reductase domain of Chinese hamster ovary cells. Mol Cell Biol. 1988 Dec;8(12):5398–5409. doi: 10.1128/mcb.8.12.5398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dijkwel P. A., Vaughn J. P., Hamlin J. L. Mapping of replication initiation sites in mammalian genomes by two-dimensional gel analysis: stabilization and enrichment of replication intermediates by isolation on the nuclear matrix. Mol Cell Biol. 1991 Aug;11(8):3850–3859. doi: 10.1128/mcb.11.8.3850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobbs D. L., Shaiu W. L., Benbow R. M. Modular sequence elements associated with origin regions in eukaryotic chromosomal DNA. Nucleic Acids Res. 1994 Jul 11;22(13):2479–2489. doi: 10.1093/nar/22.13.2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckdahl T. T., Anderson J. N. Computer modelling of DNA structures involved in chromosome maintenance. Nucleic Acids Res. 1987 Oct 26;15(20):8531–8545. doi: 10.1093/nar/15.20.8531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckdahl T. T., Anderson J. N. Conserved DNA structures in origins of replication. Nucleic Acids Res. 1990 Mar 25;18(6):1609–1612. doi: 10.1093/nar/18.6.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engberg J., Nielsen H. Complete sequence of the extrachromosomal rDNA molecule from the ciliate Tetrahymena thermophila strain B1868VII. Nucleic Acids Res. 1990 Dec 11;18(23):6915–6919. doi: 10.1093/nar/18.23.6915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engberg J. The ribosomal RNA genes of Tetrahymena: structure and function. Eur J Cell Biol. 1985 Jan;36(1):133–151. [PubMed] [Google Scholar]
- Gaertig J., Gu L., Hai B., Gorovsky M. A. High frequency vector-mediated transformation and gene replacement in Tetrahymena. Nucleic Acids Res. 1994 Dec 11;22(24):5391–5398. doi: 10.1093/nar/22.24.5391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gale J. M., Tobey R. A., D'Anna J. A. Localization and DNA sequence of a replication origin in the rhodopsin gene locus of Chinese hamster cells. J Mol Biol. 1992 Mar 20;224(2):343–358. doi: 10.1016/0022-2836(92)90999-z. [DOI] [PubMed] [Google Scholar]
- Gasser S. M., Amati B. B., Cardenas M. E., Hofmann J. F. Studies on scaffold attachment sites and their relation to genome function. Int Rev Cytol. 1989;119:57–96. doi: 10.1016/s0074-7696(08)60649-x. [DOI] [PubMed] [Google Scholar]
- Gasser S. M., Laemmli U. K. Cohabitation of scaffold binding regions with upstream/enhancer elements of three developmentally regulated genes of D. melanogaster. Cell. 1986 Aug 15;46(4):521–530. doi: 10.1016/0092-8674(86)90877-9. [DOI] [PubMed] [Google Scholar]
- Hamlin J. L. Initiation of replication in mammalian chromosomes. Crit Rev Eukaryot Gene Expr. 1992;2(4):359–381. [PubMed] [Google Scholar]
- Heintz N. H. Transcription factors and the control of DNA replication. Curr Opin Cell Biol. 1992 Jun;4(3):459–467. doi: 10.1016/0955-0674(92)90012-2. [DOI] [PubMed] [Google Scholar]
- Held P. G., Heintz N. H. Eukaryotic replication origins. Biochim Biophys Acta. 1992 Apr 6;1130(3):235–246. doi: 10.1016/0167-4781(92)90435-3. [DOI] [PubMed] [Google Scholar]
- Hernández P., Martín-Parras L., Martínez-Robles M. L., Schvartzman J. B. Conserved features in the mode of replication of eukaryotic ribosomal RNA genes. EMBO J. 1993 Apr;12(4):1475–1485. doi: 10.1002/j.1460-2075.1993.tb05791.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashinakagawa T., Narushima-Iio M., Saiga H., Kondo S., Mita T. Properties of isolated extrachromosomal nucleoli from Tetrahymena pyriformis. Chromosoma. 1992 Apr;101(7):413–419. doi: 10.1007/BF00582835. [DOI] [PubMed] [Google Scholar]
- Homberger H. P. Bent DNA is a structural feature of scaffold-attached regions in Drosophila melanogaster interphase nuclei. Chromosoma. 1989 Aug;98(2):99–104. doi: 10.1007/BF00291044. [DOI] [PubMed] [Google Scholar]
- Hou Z., Umthun A. R., Dobbs D. L. A single-stranded DNA binding protein that specifically recognizes cis-acting sequences in the replication origin and transcriptional promoter region of Tetrahymena rDNA. Biochemistry. 1995 Apr 11;34(14):4583–4592. doi: 10.1021/bi00014a011. [DOI] [PubMed] [Google Scholar]
- Hozák P., Hassan A. B., Jackson D. A., Cook P. R. Visualization of replication factories attached to nucleoskeleton. Cell. 1993 Apr 23;73(2):361–373. doi: 10.1016/0092-8674(93)90235-i. [DOI] [PubMed] [Google Scholar]
- Huang R. Y., Kowalski D. A DNA unwinding element and an ARS consensus comprise a replication origin within a yeast chromosome. EMBO J. 1993 Dec;12(12):4521–4531. doi: 10.1002/j.1460-2075.1993.tb06141.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapler G. M. Developmentally regulated processing and replication of the Tetrahymena rDNA minichromosome. Curr Opin Genet Dev. 1993 Oct;3(5):730–735. doi: 10.1016/s0959-437x(05)80091-7. [DOI] [PubMed] [Google Scholar]
- Kim J., Zwieb C., Wu C., Adhya S. Bending of DNA by gene-regulatory proteins: construction and use of a DNA bending vector. Gene. 1989 Dec 21;85(1):15–23. doi: 10.1016/0378-1119(89)90459-9. [DOI] [PubMed] [Google Scholar]
- Koo H. S., Wu H. M., Crothers D. M. DNA bending at adenine . thymine tracts. Nature. 1986 Apr 10;320(6062):501–506. doi: 10.1038/320501a0. [DOI] [PubMed] [Google Scholar]
- Kowalski D., Eddy M. J. The DNA unwinding element: a novel, cis-acting component that facilitates opening of the Escherichia coli replication origin. EMBO J. 1989 Dec 20;8(13):4335–4344. doi: 10.1002/j.1460-2075.1989.tb08620.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson D. D., Blackburn E. H., Yaeger P. C., Orias E. Control of rDNA replication in Tetrahymena involves a cis-acting upstream repeat of a promoter element. Cell. 1986 Oct 24;47(2):229–240. doi: 10.1016/0092-8674(86)90445-9. [DOI] [PubMed] [Google Scholar]
- Larson D. D., Umthun A. R., Shaiu W. L. Copy number control in the Tetrahymena macronuclear genome. J Protozool. 1991 May-Jun;38(3):258–263. doi: 10.1111/j.1550-7408.1991.tb04439.x. [DOI] [PubMed] [Google Scholar]
- Lin S., Kowalski D. DNA helical instability facilitates initiation at the SV40 replication origin. J Mol Biol. 1994 Jan 14;235(2):496–507. doi: 10.1006/jmbi.1994.1009. [DOI] [PubMed] [Google Scholar]
- Linial M., Shlomai J. Bent DNA structures associated with several origins of replication are recognized by a unique enzyme from trypanosomatids. Nucleic Acids Res. 1988 Jul 25;16(14A):6477–6492. doi: 10.1093/nar/16.14.6477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linial M., Shlomai J. Sequence-directed bent DNA helix is the specific binding site for Crithidia fasciculata nicking enzyme. Proc Natl Acad Sci U S A. 1987 Dec;84(23):8205–8209. doi: 10.1073/pnas.84.23.8205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linskens M. H., Huberman J. A. The two faces of higher eukaryotic DNA replication origins. Cell. 1990 Sep 7;62(5):845–847. doi: 10.1016/0092-8674(90)90258-g. [DOI] [PubMed] [Google Scholar]
- Ludérus M. E., den Blaauwen J. L., de Smit O. J., Compton D. A., van Driel R. Binding of matrix attachment regions to lamin polymers involves single-stranded regions and the minor groove. Mol Cell Biol. 1994 Sep;14(9):6297–6305. doi: 10.1128/mcb.14.9.6297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marahrens Y., Stillman B. A yeast chromosomal origin of DNA replication defined by multiple functional elements. Science. 1992 Feb 14;255(5046):817–823. doi: 10.1126/science.1536007. [DOI] [PubMed] [Google Scholar]
- Marini J. C., Levene S. D., Crothers D. M., Englund P. T. Bent helical structure in kinetoplast DNA. Proc Natl Acad Sci U S A. 1982 Dec;79(24):7664–7668. doi: 10.1073/pnas.79.24.7664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller C. A., Kowalski D. cis-acting components in the replication origin from ribosomal DNA of Saccharomyces cerevisiae. Mol Cell Biol. 1993 Sep;13(9):5360–5369. doi: 10.1128/mcb.13.9.5360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyahara K., Hashimoto N., Higashinakagawa T., Pearlman R. E. Common sequence elements are important for transcription and replication of the extrachromosomal rRNA-encoding genes of Tetrahymena. Gene. 1993 May 30;127(2):209–213. doi: 10.1016/0378-1119(93)90721-e. [DOI] [PubMed] [Google Scholar]
- Nakayasu H., Berezney R. Mapping replicational sites in the eucaryotic cell nucleus. J Cell Biol. 1989 Jan;108(1):1–11. doi: 10.1083/jcb.108.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natale D. A., Schubert A. E., Kowalski D. DNA helical stability accounts for mutational defects in a yeast replication origin. Proc Natl Acad Sci U S A. 1992 Apr 1;89(7):2654–2658. doi: 10.1073/pnas.89.7.2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natale D. A., Umek R. M., Kowalski D. Ease of DNA unwinding is a conserved property of yeast replication origins. Nucleic Acids Res. 1993 Feb 11;21(3):555–560. doi: 10.1093/nar/21.3.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newlon C. S., Theis J. F. The structure and function of yeast ARS elements. Curr Opin Genet Dev. 1993 Oct;3(5):752–758. doi: 10.1016/s0959-437x(05)80094-2. [DOI] [PubMed] [Google Scholar]
- Niles E. G., Sutiphong J., Haque S. Structure of the Tetrahymena pyriformis rRNA gene. Nucleotide sequence of the transcription initiation region. J Biol Chem. 1981 Dec 25;256(24):12849–12856. [PubMed] [Google Scholar]
- Palen T. E., Cech T. R. Chromatin structure at the replication origins and transcription-initiation regions of the ribosomal RNA genes of Tetrahymena. Cell. 1984 Apr;36(4):933–942. doi: 10.1016/0092-8674(84)90043-6. [DOI] [PubMed] [Google Scholar]
- Pan W. J., Blackburn E. H. Tandem repeats of the 5' non-transcribed spacer of Tetrahymena rDNA function as high copy number autonomous replicons in the macronucleus but do not prevent rRNA gene dosage regulation. Nucleic Acids Res. 1995 May 11;23(9):1561–1569. doi: 10.1093/nar/23.9.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao B. S., Zannis-Hadjopoulos M., Price G. B., Reitman M., Martin R. G. Sequence similarities among monkey ori-enriched (ors) fragments. Gene. 1990 Mar 15;87(2):233–242. doi: 10.1016/0378-1119(90)90307-d. [DOI] [PubMed] [Google Scholar]
- Rivier D. H., Pillus L. Silencing speaks up. Cell. 1994 Mar 25;76(6):963–966. doi: 10.1016/0092-8674(94)90373-5. [DOI] [PubMed] [Google Scholar]
- Roberge M., Gasser S. M. DNA loops: structural and functional properties of scaffold-attached regions. Mol Microbiol. 1992 Feb;6(4):419–423. doi: 10.1111/j.1365-2958.1992.tb01485.x. [DOI] [PubMed] [Google Scholar]
- Rowley A., Dowell S. J., Diffley J. F. Recent developments in the initiation of chromosomal DNA replication: a complex picture emerges. Biochim Biophys Acta. 1994 Apr 6;1217(3):239–256. doi: 10.1016/0167-4781(94)90283-6. [DOI] [PubMed] [Google Scholar]
- Ryder K., Silver S., DeLucia A. L., Fanning E., Tegtmeyer P. An altered DNA conformation in origin region I is a determinant for the binding of SV40 large T antigen. Cell. 1986 Mar 14;44(5):719–725. doi: 10.1016/0092-8674(86)90838-x. [DOI] [PubMed] [Google Scholar]
- Shinomiya T., Ina S. Mapping an initiation region of DNA replication at a single-copy chromosomal locus in Drosophila melanogaster cells by two-dimensional gel methods and PCR-mediated nascent-strand analysis: multiple replication origins in a broad zone. Mol Cell Biol. 1994 Nov;14(11):7394–7403. doi: 10.1128/mcb.14.11.7394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder M., Buchman A. R., Davis R. W. Bent DNA at a yeast autonomously replicating sequence. Nature. 1986 Nov 6;324(6092):87–89. doi: 10.1038/324087a0. [DOI] [PubMed] [Google Scholar]
- Stenzel T. T., Patel P., Bastia D. The integration host factor of Escherichia coli binds to bent DNA at the origin of replication of the plasmid pSC101. Cell. 1987 Jun 5;49(5):709–717. doi: 10.1016/0092-8674(87)90547-2. [DOI] [PubMed] [Google Scholar]
- Suzuki M., Izuta S., Savoysky E., Sakurai T., Simbulan C., Tatebe M., Kojima K., Yoshida S. Deoxypyrimidine cluster mediates the priming by calf thymus DNA primase subunit. Biochem Mol Biol Int. 1993 Mar;29(4):645–652. [PubMed] [Google Scholar]
- Suzuki M., Savoysky E., Izuta S., Tatebe M., Okajima T., Yoshida S. RNA priming coupled with DNA synthesis on natural template by calf thymus DNA polymerase alpha-primase. Biochemistry. 1993 Nov 30;32(47):12782–12792. doi: 10.1021/bi00210a030. [DOI] [PubMed] [Google Scholar]
- Tasheva E. S., Roufa D. J. A mammalian origin of bidirectional DNA replication within the Chinese hamster RPS14 locus. Mol Cell Biol. 1994 Sep;14(9):5628–5635. doi: 10.1128/mcb.14.9.5628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trempe J. P., Lindstrom Y. I., Leffak M. Opposite replication polarities of transcribed and nontranscribed histone H5 genes. Mol Cell Biol. 1988 Apr;8(4):1657–1663. doi: 10.1128/mcb.8.4.1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umek R. M., Kowalski D. The ease of DNA unwinding as a determinant of initiation at yeast replication origins. Cell. 1988 Feb 26;52(4):559–567. doi: 10.1016/0092-8674(88)90469-2. [DOI] [PubMed] [Google Scholar]
- Umthun A. R., Hou Z., Sibenaller Z. A., Shaiu W. L., Dobbs D. L. Identification of DNA-binding proteins that recognize a conserved type I repeat sequence in the replication origin region of Tetrahymena rDNA. Nucleic Acids Res. 1994 Oct 25;22(21):4432–4440. doi: 10.1093/nar/22.21.4432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanWye J. D., Bronson E. C., Anderson J. N. Species-specific patterns of DNA bending and sequence. Nucleic Acids Res. 1991 Oct 11;19(19):5253–5261. doi: 10.1093/nar/19.19.5253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughn J. P., Dijkwel P. A., Mullenders L. H., Hamlin J. L. Replication forks are associated with the nuclear matrix. Nucleic Acids Res. 1990 Apr 25;18(8):1965–1969. doi: 10.1093/nar/18.8.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virta-Pearlman V. J., Gunaratne P. H., Chinault A. C. Analysis of a replication initiation sequence from the adenosine deaminase region of the mouse genome. Mol Cell Biol. 1993 Oct;13(10):5931–5942. doi: 10.1128/mcb.13.10.5931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams J. S., Eckdahl T. T., Anderson J. N. Bent DNA functions as a replication enhancer in Saccharomyces cerevisiae. Mol Cell Biol. 1988 Jul;8(7):2763–2769. doi: 10.1128/mcb.8.7.2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaeger P. C., Orias E., Shaiu W. L., Larson D. D., Blackburn E. H. The replication advantage of a free linear rRNA gene is restored by somatic recombination in Tetrahymena thermophila. Mol Cell Biol. 1989 Feb;9(2):452–460. doi: 10.1128/mcb.9.2.452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G. L., Blackburn E. H. Amplification of tandemly repeated origin control sequences confers a replication advantage on rDNA replicons in Tetrahymena thermophila. Mol Cell Biol. 1990 May;10(5):2070–2080. doi: 10.1128/mcb.10.5.2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G. L., Blackburn E. H. Transformation of Tetrahymena thermophila with a mutated circular ribosomal DNA plasmid vector. Proc Natl Acad Sci U S A. 1989 Nov;86(21):8487–8491. doi: 10.1073/pnas.86.21.8487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahn K., Blattner F. R. Direct evidence for DNA bending at the lambda replication origin. Science. 1987 Apr 24;236(4800):416–422. doi: 10.1126/science.2951850. [DOI] [PubMed] [Google Scholar]
- von Kries J. P., Phi-Van L., Diekmann S., Strätling W. H. A non-curved chicken lysozyme 5' matrix attachment site is 3' followed by a strongly curved DNA sequence. Nucleic Acids Res. 1990 Jul 11;18(13):3881–3885. doi: 10.1093/nar/18.13.3881. [DOI] [PMC free article] [PubMed] [Google Scholar]