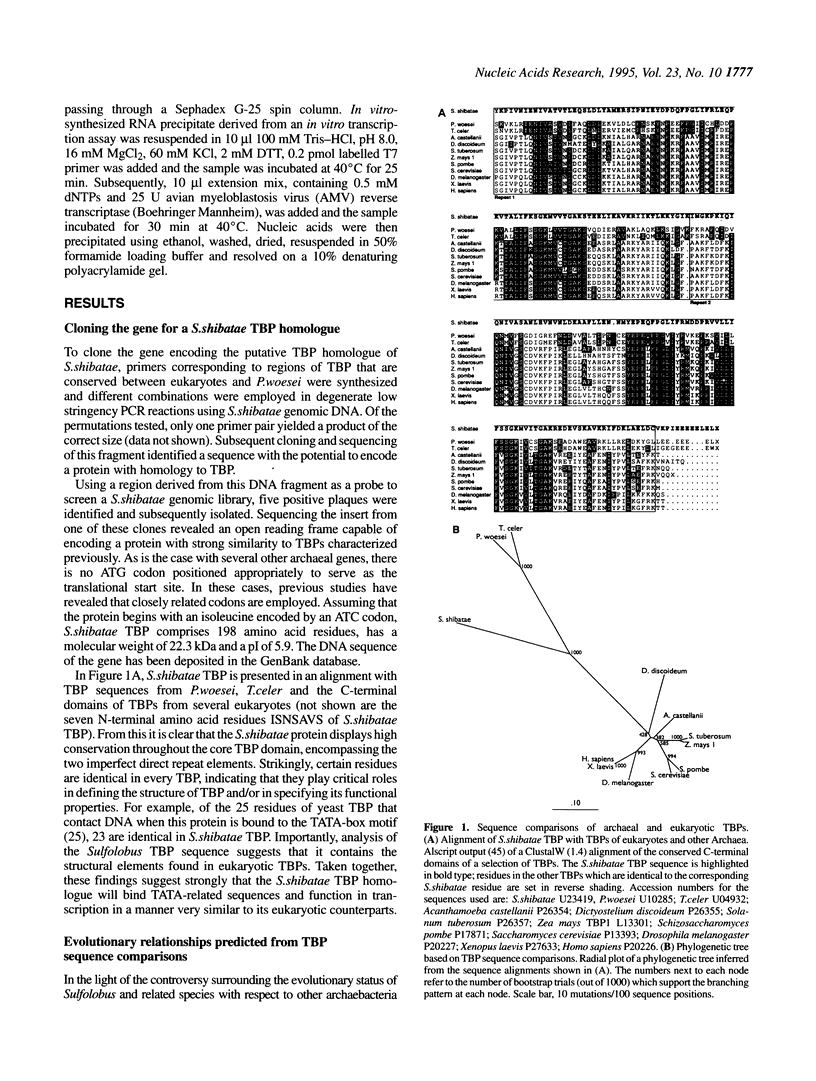
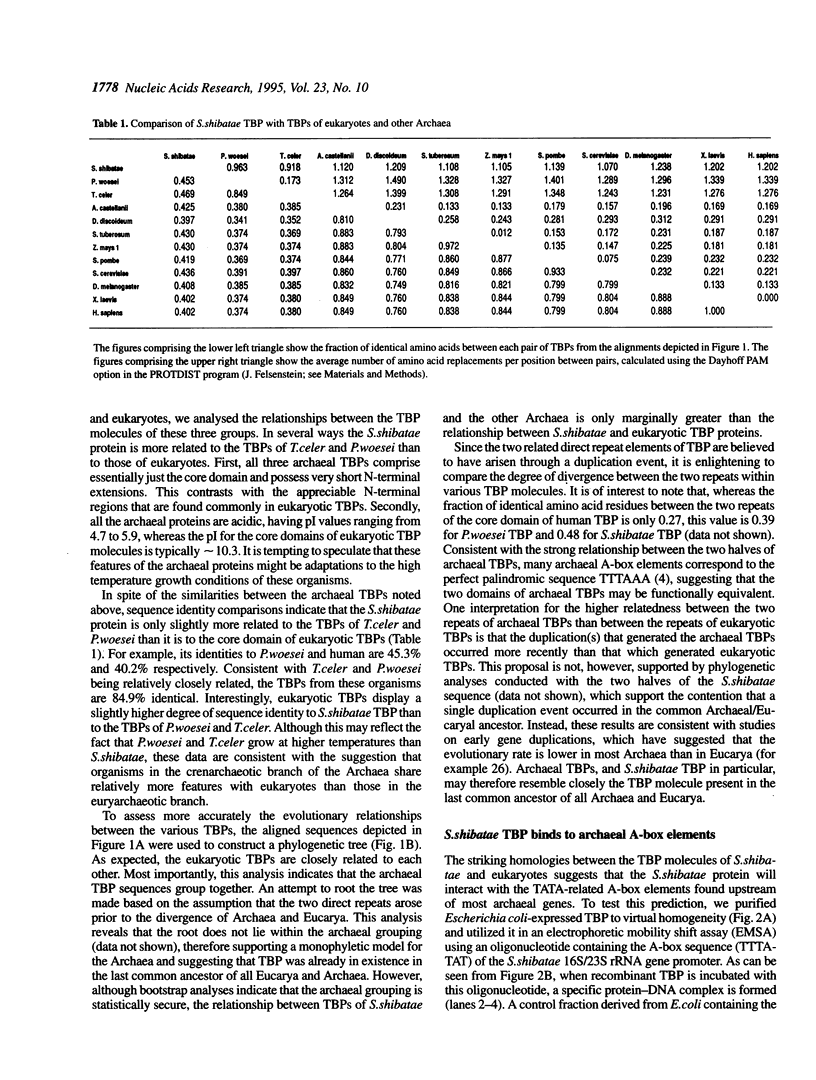
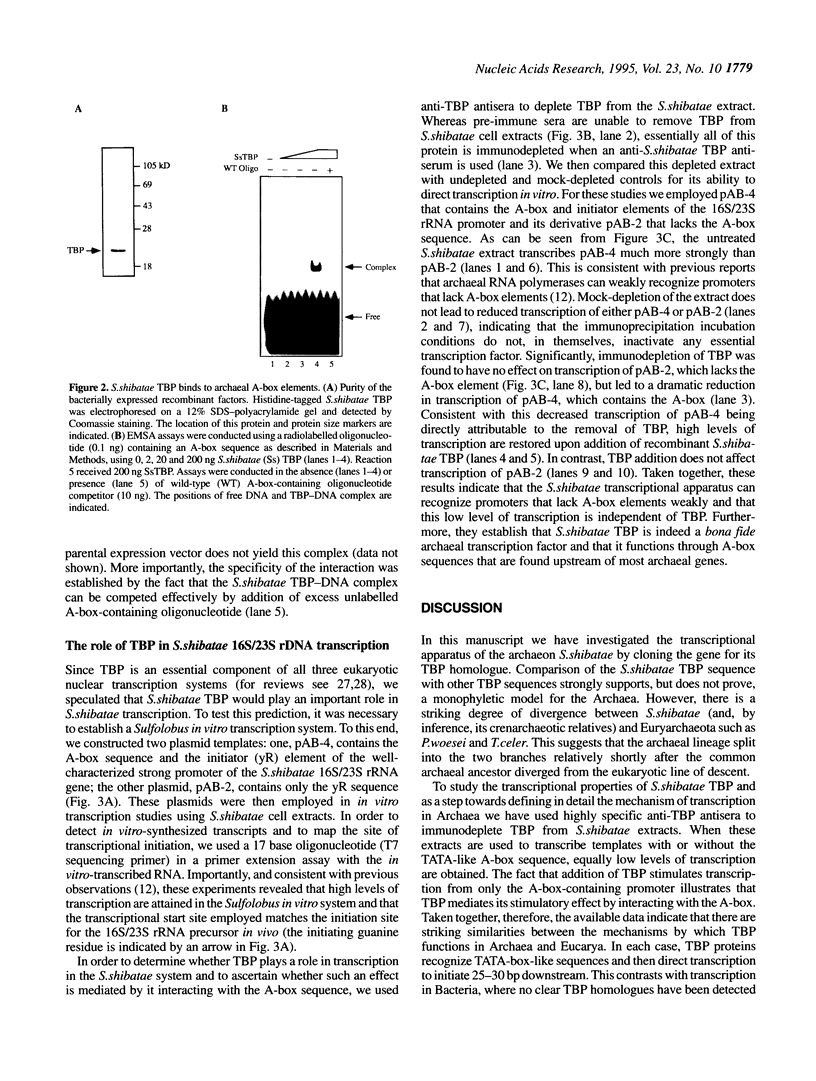
Abstract
Archaea (formerly archaebacteria) comprise a domain of life that is phylogenetically distinct from both Eucarya and Bacteria. Here we report the cloning of a gene from the Archaeon Sulfolobus shibatae that encodes a protein with strong homology to the TATA binding protein (TBP) of eukaryotes. Sulfolobus shibatae TBP is, however, almost as diverged from other archaeal TBPs that have been cloned as it is from eukaryotic TBPs. DNA binding studies indicate that S.shibatae TBP recognizes TATA-like A-box sequences that are present upstream of most archaeal genes. By quantitatively immunodepleting S.shibatae TBP from an in vitro transcription system, we demonstrate that Sulfolobus RNA polymerase is capable of transcribing the 16S/23S rRNA promoter weakly in the absence of TBP. Most significantly, we show that addition of recombinant S.shibatae TBP to this immunodepleted system leads to transcriptional stimulation and that this stimulation is dependent on the A-box sequence of the promoter. Taken together, these findings reveal fundamental similarities between the transcription machineries of Archaea and eukaryotes.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. Basic local alignment search tool. J Mol Biol. 1990 Oct 5;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Barton G. J. ALSCRIPT: a tool to format multiple sequence alignments. Protein Eng. 1993 Jan;6(1):37–40. doi: 10.1093/protein/6.1.37. [DOI] [PubMed] [Google Scholar]
- Berghöfer B., Kröckel L., Körtner C., Truss M., Schallenberg J., Klein A. Relatedness of archaebacterial RNA polymerase core subunits to their eubacterial and eukaryotic equivalents. Nucleic Acids Res. 1988 Aug 25;16(16):8113–8128. doi: 10.1093/nar/16.16.8113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J. W., Thomm M., Beckler G. S., Frey G., Stetter K. O., Reeve J. N. An archaebacterial RNA polymerase binding site and transcription initiation of the hisA gene in Methanococcus vannielii. Nucleic Acids Res. 1988 Jan 11;16(1):135–150. doi: 10.1093/nar/16.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busby S., Ebright R. H. Promoter structure, promoter recognition, and transcription activation in prokaryotes. Cell. 1994 Dec 2;79(5):743–746. doi: 10.1016/0092-8674(94)90063-9. [DOI] [PubMed] [Google Scholar]
- Chen J. L., Attardi L. D., Verrijzer C. P., Yokomori K., Tjian R. Assembly of recombinant TFIID reveals differential coactivator requirements for distinct transcriptional activators. Cell. 1994 Oct 7;79(1):93–105. doi: 10.1016/0092-8674(94)90403-0. [DOI] [PubMed] [Google Scholar]
- Creti R., Londei P., Cammarano P. Complete nucleotide sequence of an archaeal (Pyrococcus woesei) gene encoding a homolog of eukaryotic transcription factor IIB (TFIIB). Nucleic Acids Res. 1993 Jun 25;21(12):2942–2942. doi: 10.1093/nar/21.12.2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eick D., Wedel A., Heumann H. From initiation to elongation: comparison of transcription by prokaryotic and eukaryotic RNA polymerases. Trends Genet. 1994 Aug;10(8):292–296. doi: 10.1016/0168-9525(90)90013-v. [DOI] [PubMed] [Google Scholar]
- Fox G. E., Stackebrandt E., Hespell R. B., Gibson J., Maniloff J., Dyer T. A., Wolfe R. S., Balch W. E., Tanner R. S., Magrum L. J. The phylogeny of prokaryotes. Science. 1980 Jul 25;209(4455):457–463. doi: 10.1126/science.6771870. [DOI] [PubMed] [Google Scholar]
- Frey G., Thomm M., Brüdigam B., Gohl H. P., Hausner W. An archaebacterial cell-free transcription system. The expression of tRNA genes from Methanococcus vannielii is mediated by a transcription factor. Nucleic Acids Res. 1990 Mar 25;18(6):1361–1367. doi: 10.1093/nar/18.6.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich J. A., Tjian R. TBP-TAF complexes: selectivity factors for eukaryotic transcription. Curr Opin Cell Biol. 1994 Jun;6(3):403–409. doi: 10.1016/0955-0674(94)90033-7. [DOI] [PubMed] [Google Scholar]
- Ha I., Roberts S., Maldonado E., Sun X., Kim L. U., Green M., Reinberg D. Multiple functional domains of human transcription factor IIB: distinct interactions with two general transcription factors and RNA polymerase II. Genes Dev. 1993 Jun;7(6):1021–1032. doi: 10.1101/gad.7.6.1021. [DOI] [PubMed] [Google Scholar]
- Hausner W., Frey G., Thomm M. Control regions of an archaeal gene. A TATA box and an initiator element promote cell-free transcription of the tRNA(Val) gene of Methanococcus vannielii. J Mol Biol. 1991 Dec 5;222(3):495–508. doi: 10.1016/0022-2836(91)90492-o. [DOI] [PubMed] [Google Scholar]
- Hausner W., Thomm M. Purification and characterization of a general transcription factor, aTFB, from the archaeon Methanococcus thermolithotrophicus. J Biol Chem. 1993 Nov 15;268(32):24047–24052. [PubMed] [Google Scholar]
- Hernandez N. TBP, a universal eukaryotic transcription factor? Genes Dev. 1993 Jul;7(7B):1291–1308. doi: 10.1101/gad.7.7b.1291. [DOI] [PubMed] [Google Scholar]
- Hüdepohl U., Reiter W. D., Zillig W. In vitro transcription of two rRNA genes of the archaebacterium Sulfolobus sp. B12 indicates a factor requirement for specific initiation. Proc Natl Acad Sci U S A. 1990 Aug;87(15):5851–5855. doi: 10.1073/pnas.87.15.5851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwabe N., Kuma K., Hasegawa M., Osawa S., Miyata T. Evolutionary relationship of archaebacteria, eubacteria, and eukaryotes inferred from phylogenetic trees of duplicated genes. Proc Natl Acad Sci U S A. 1989 Dec;86(23):9355–9359. doi: 10.1073/pnas.86.23.9355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassavetis G. A., Joazeiro C. A., Pisano M., Geiduschek E. P., Colbert T., Hahn S., Blanco J. A. The role of the TATA-binding protein in the assembly and function of the multisubunit yeast RNA polymerase III transcription factor, TFIIIB. Cell. 1992 Dec 11;71(6):1055–1064. doi: 10.1016/0092-8674(92)90399-w. [DOI] [PubMed] [Google Scholar]
- Kaufmann J., Smale S. T. Direct recognition of initiator elements by a component of the transcription factor IID complex. Genes Dev. 1994 Apr 1;8(7):821–829. doi: 10.1101/gad.8.7.821. [DOI] [PubMed] [Google Scholar]
- Khoo B., Brophy B., Jackson S. P. Conserved functional domains of the RNA polymerase III general transcription factor BRF. Genes Dev. 1994 Dec 1;8(23):2879–2890. doi: 10.1101/gad.8.23.2879. [DOI] [PubMed] [Google Scholar]
- Kim Y., Geiger J. H., Hahn S., Sigler P. B. Crystal structure of a yeast TBP/TATA-box complex. Nature. 1993 Oct 7;365(6446):512–520. doi: 10.1038/365512a0. [DOI] [PubMed] [Google Scholar]
- Klenk H. P., Palm P., Lottspeich F., Zillig W. Component H of the DNA-dependent RNA polymerases of Archaea is homologous to a subunit shared by the three eucaryal nuclear RNA polymerases. Proc Natl Acad Sci U S A. 1992 Jan 1;89(1):407–410. doi: 10.1073/pnas.89.1.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lake J. A. Origin of the eukaryotic nucleus determined by rate-invariant analysis of rRNA sequences. Nature. 1988 Jan 14;331(6152):184–186. doi: 10.1038/331184a0. [DOI] [PubMed] [Google Scholar]
- Marsh T. L., Reich C. I., Whitelock R. B., Olsen G. J. Transcription factor IID in the Archaea: sequences in the Thermococcus celer genome would encode a product closely related to the TATA-binding protein of eukaryotes. Proc Natl Acad Sci U S A. 1994 May 10;91(10):4180–4184. doi: 10.1073/pnas.91.10.4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouzounis C., Sander C. TFIIB, an evolutionary link between the transcription machineries of archaebacteria and eukaryotes. Cell. 1992 Oct 16;71(2):189–190. doi: 10.1016/0092-8674(92)90347-f. [DOI] [PubMed] [Google Scholar]
- Pearson W. R., Lipman D. J. Improved tools for biological sequence comparison. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon D., Weil P. A. Immunopurification of yeast TATA-binding protein and associated factors. Presence of transcription factor IIIB transcriptional activity. J Biol Chem. 1993 Jul 25;268(21):15325–15328. [PubMed] [Google Scholar]
- Purnell B. A., Emanuel P. A., Gilmour D. S. TFIID sequence recognition of the initiator and sequences farther downstream in Drosophila class II genes. Genes Dev. 1994 Apr 1;8(7):830–842. doi: 10.1101/gad.8.7.830. [DOI] [PubMed] [Google Scholar]
- Pühler G., Lottspeich F., Zillig W. Organization and nucleotide sequence of the genes encoding the large subunits A, B and C of the DNA-dependent RNA polymerase of the archaebacterium Sulfolobus acidocaldarius. Nucleic Acids Res. 1989 Jun 26;17(12):4517–4534. doi: 10.1093/nar/17.12.4517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter W. D., Hüdepohl U., Zillig W. Mutational analysis of an archaebacterial promoter: essential role of a TATA box for transcription efficiency and start-site selection in vitro. Proc Natl Acad Sci U S A. 1990 Dec;87(24):9509–9513. doi: 10.1073/pnas.87.24.9509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera M. C., Lake J. A. Evidence that eukaryotes and eocyte prokaryotes are immediate relatives. Science. 1992 Jul 3;257(5066):74–76. doi: 10.1126/science.1621096. [DOI] [PubMed] [Google Scholar]
- Rowlands T., Baumann P., Jackson S. P. The TATA-binding protein: a general transcription factor in eukaryotes and archaebacteria. Science. 1994 May 27;264(5163):1326–1329. doi: 10.1126/science.8191287. [DOI] [PubMed] [Google Scholar]
- Smale S. T., Baltimore D. The "initiator" as a transcription control element. Cell. 1989 Apr 7;57(1):103–113. doi: 10.1016/0092-8674(89)90176-1. [DOI] [PubMed] [Google Scholar]
- Thomm M., Wich G. An archaebacterial promoter element for stable RNA genes with homology to the TATA box of higher eukaryotes. Nucleic Acids Res. 1988 Jan 11;16(1):151–163. doi: 10.1093/nar/16.1.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J. D., Higgins D. G., Gibson T. J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994 Nov 11;22(22):4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner M., Chaussivert N., Willis I. M., Sentenac A. Interaction between a complex of RNA polymerase III subunits and the 70-kDa component of transcription factor IIIB. J Biol Chem. 1993 Oct 5;268(28):20721–20724. [PubMed] [Google Scholar]
- White R. J., Gottlieb T. M., Downes C. S., Jackson S. P. Mitotic regulation of a TATA-binding-protein-containing complex. Mol Cell Biol. 1995 Apr;15(4):1983–1992. doi: 10.1128/mcb.15.4.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White R. J., Jackson S. P. The TATA-binding protein: a central role in transcription by RNA polymerases I, II and III. Trends Genet. 1992 Aug;8(8):284–288. doi: 10.1016/0168-9525(92)90255-3. [DOI] [PubMed] [Google Scholar]
- Woese C. R., Fox G. E. Phylogenetic structure of the prokaryotic domain: the primary kingdoms. Proc Natl Acad Sci U S A. 1977 Nov;74(11):5088–5090. doi: 10.1073/pnas.74.11.5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C. R., Kandler O., Wheelis M. L. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci U S A. 1990 Jun;87(12):4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita S., Hisatake K., Kokubo T., Doi K., Roeder R. G., Horikoshi M., Nakatani Y. Transcription factor TFIIB sites important for interaction with promoter-bound TFIID. Science. 1993 Jul 23;261(5120):463–466. doi: 10.1126/science.8332911. [DOI] [PubMed] [Google Scholar]




