Abstract
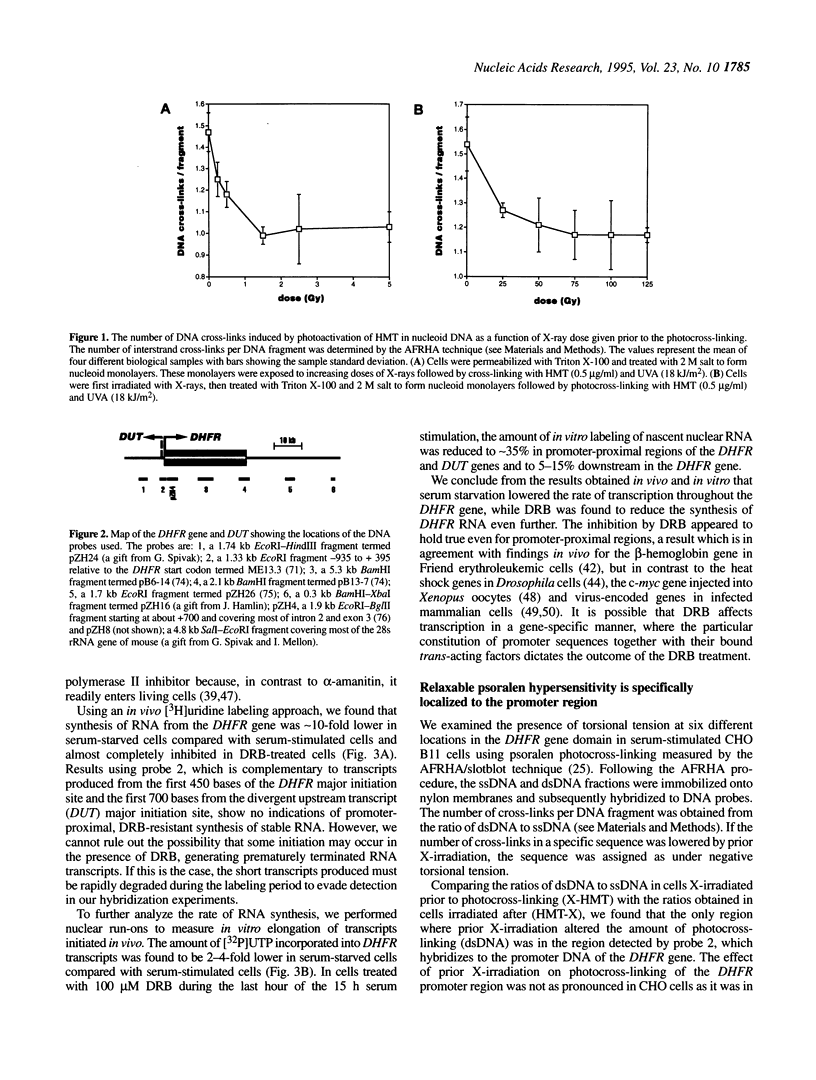
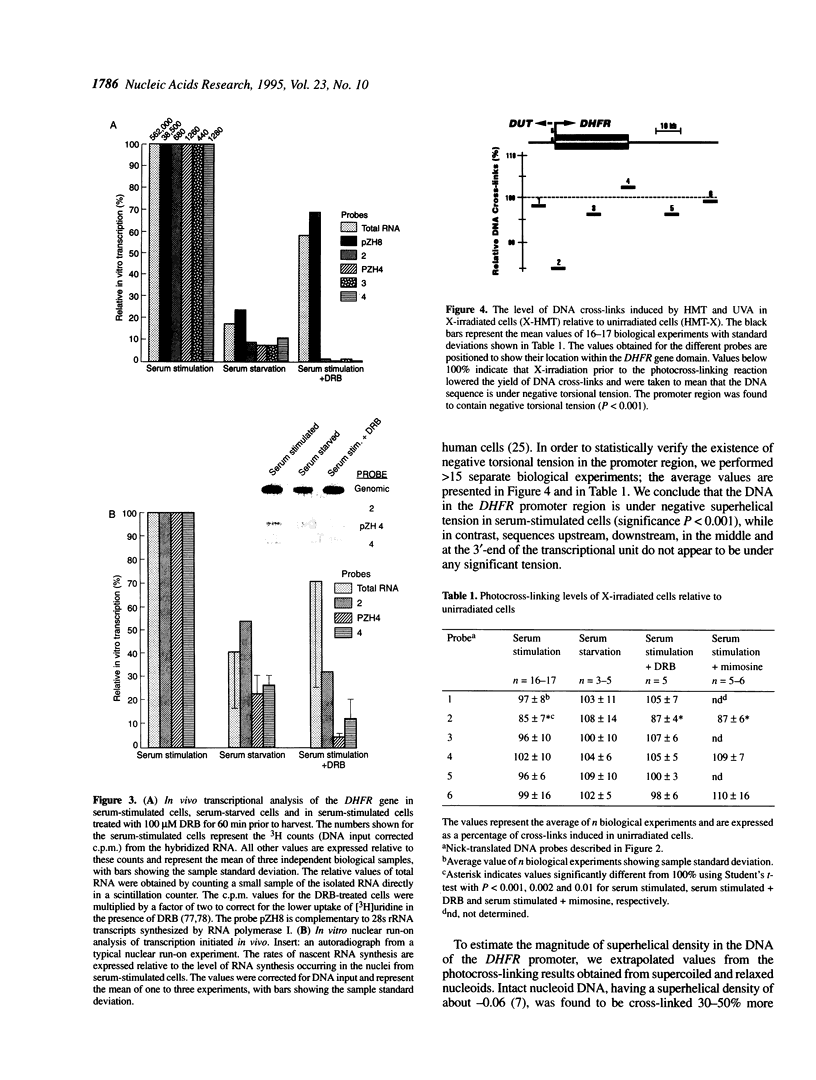
DNA topology has been suggested to play an important role in the process of transcription. Negative torsional tension has been shown to stimulate both pre-initiation complex formation and promoter clearance on plasmid DNA in vitro. We recently showed that genomic DNA in human cells contains localized torsional tension. In the present study we have further characterized and mapped torsional tension in the dihydrofolate reductase (DHFR) gene in Chinese hamster ovary (CHO) cells and investigated the effects of differential rates of transcription on the magnitude and location of this tension. Using psoralen photo-cross-linking in conjunction with X-irradiation, we found that relaxable psoralen hypersensitivity was specifically localized to the promoter region of the serum-regulated DHFR gene in serum-stimulated, but not in serum-starved, cells. Moreover, this hypersensitivity did not appear to be caused by transcription elongation, since it persisted in cells in which transcription of the DHFR gene had been reduced by the transcription inhibitor 5,6-dichloro-1-beta-D-ribofurano-sylbenzimidazole (DRB). We suggest that the generation of negative torsional tension in DNA may play an important role in gene regulation by poising genes for transcription.
Full text
PDF





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arroyo M., Bagchi S., Raychaudhuri P. Association of the human papillomavirus type 16 E7 protein with the S-phase-specific E2F-cyclin A complex. Mol Cell Biol. 1993 Oct;13(10):6537–6546. doi: 10.1128/mcb.13.10.6537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barsoum J., Varshavsky A. Preferential localization of variant nucleosomes near the 5'-end of the mouse dihydrofolate reductase gene. J Biol Chem. 1985 Jun 25;260(12):7688–7697. [PubMed] [Google Scholar]
- Benham C. J. Sites of predicted stress-induced DNA duplex destabilization occur preferentially at regulatory loci. Proc Natl Acad Sci U S A. 1993 Apr 1;90(7):2999–3003. doi: 10.1073/pnas.90.7.2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blake M. C., Azizkhan J. C. Transcription factor E2F is required for efficient expression of the hamster dihydrofolate reductase gene in vitro and in vivo. Mol Cell Biol. 1989 Nov;9(11):4994–5002. doi: 10.1128/mcb.9.11.4994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowater R. P., Chen D., Lilley D. M. Modulation of tyrT promoter activity by template supercoiling in vivo. EMBO J. 1994 Dec 1;13(23):5647–5655. doi: 10.1002/j.1460-2075.1994.tb06903.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brill S. J., Sternglanz R. Transcription-dependent DNA supercoiling in yeast DNA topoisomerase mutants. Cell. 1988 Jul 29;54(3):403–411. doi: 10.1016/0092-8674(88)90203-6. [DOI] [PubMed] [Google Scholar]
- Carothers A. M., Urlaub G., Ellis N., Chasin L. A. Structure of the dihydrofolate reductase gene in Chinese hamster ovary cells. Nucleic Acids Res. 1983 Apr 11;11(7):1997–2012. doi: 10.1093/nar/11.7.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen-Kiang S., Lavery D. J. Pulse labeling of heterogeneous nuclear RNA in isolated nuclei. Methods Enzymol. 1989;180:82–96. doi: 10.1016/0076-6879(89)80094-1. [DOI] [PubMed] [Google Scholar]
- Chen D., Bowater R., Dorman C. J., Lilley D. M. Activity of a plasmid-borne leu-500 promoter depends on the transcription and translation of an adjacent gene. Proc Natl Acad Sci U S A. 1992 Sep 15;89(18):8784–8788. doi: 10.1073/pnas.89.18.8784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimino G. D., Gamper H. B., Isaacs S. T., Hearst J. E. Psoralens as photoactive probes of nucleic acid structure and function: organic chemistry, photochemistry, and biochemistry. Annu Rev Biochem. 1985;54:1151–1193. doi: 10.1146/annurev.bi.54.070185.005443. [DOI] [PubMed] [Google Scholar]
- Dröge P. Transcription-driven site-specific DNA recombination in vitro. Proc Natl Acad Sci U S A. 1993 Apr 1;90(7):2759–2763. doi: 10.1073/pnas.90.7.2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois M. F., Nguyen V. T., Bellier S., Bensaude O. Inhibitors of transcription such as 5,6-dichloro-1-beta-D-ribofuranosylbenzimidazole and isoquinoline sulfonamide derivatives (H-8 and H-7) promote dephosphorylation of the carboxyl-terminal domain of RNA polymerase II largest subunit. J Biol Chem. 1994 May 6;269(18):13331–13336. [PubMed] [Google Scholar]
- Dunaway M., Ostrander E. A. Local domains of supercoiling activate a eukaryotic promoter in vivo. Nature. 1993 Feb 25;361(6414):746–748. doi: 10.1038/361746a0. [DOI] [PubMed] [Google Scholar]
- Egyhazi E. Initiation inhibition and reinitiation of the synthesis of heterogenous nuclear RNA in living cells. Nature. 1976 Jul 22;262(5566):319–321. doi: 10.1038/262319a0. [DOI] [PubMed] [Google Scholar]
- Esposito F., Sinden R. R. DNA supercoiling and eukaryotic gene expression. Oxf Surv Eukaryot Genes. 1988;5:1–50. [PubMed] [Google Scholar]
- Freeman L. A., Garrard W. T. DNA supercoiling in chromatin structure and gene expression. Crit Rev Eukaryot Gene Expr. 1992;2(2):165–209. [PubMed] [Google Scholar]
- Gartenberg M. R., Wang J. C. Positive supercoiling of DNA greatly diminishes mRNA synthesis in yeast. Proc Natl Acad Sci U S A. 1992 Dec 1;89(23):11461–11465. doi: 10.1073/pnas.89.23.11461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh R., Amstad P., Cerutti P. UVB-induced DNA breaks interfere with transcriptional induction of c-fos. Mol Cell Biol. 1993 Nov;13(11):6992–6999. doi: 10.1128/mcb.13.11.6992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaever G. N., Wang J. C. Supercoiling of intracellular DNA can occur in eukaryotic cells. Cell. 1988 Dec 2;55(5):849–856. doi: 10.1016/0092-8674(88)90140-7. [DOI] [PubMed] [Google Scholar]
- Giardina C., Lis J. T. Polymerase processivity and termination on Drosophila heat shock genes. J Biol Chem. 1993 Nov 15;268(32):23806–23811. [PubMed] [Google Scholar]
- Gilmour D. S., Elgin S. C. Localization of specific topoisomerase I interactions within the transcribed region of active heat shock genes by using the inhibitor camptothecin. Mol Cell Biol. 1987 Jan;7(1):141–148. doi: 10.1128/mcb.7.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich J. A., Tjian R. Transcription factors IIE and IIH and ATP hydrolysis direct promoter clearance by RNA polymerase II. Cell. 1994 Apr 8;77(1):145–156. doi: 10.1016/0092-8674(94)90242-9. [DOI] [PubMed] [Google Scholar]
- Harland R. M., Weintraub H., McKnight S. L. Transcription of DNA injected into Xenopus oocytes is influenced by template topology. Nature. 1983 Mar 3;302(5903):38–43. doi: 10.1038/302038a0. [DOI] [PubMed] [Google Scholar]
- Helin K., Wu C. L., Fattaey A. R., Lees J. A., Dynlacht B. D., Ngwu C., Harlow E. Heterodimerization of the transcription factors E2F-1 and DP-1 leads to cooperative trans-activation. Genes Dev. 1993 Oct;7(10):1850–1861. doi: 10.1101/gad.7.10.1850. [DOI] [PubMed] [Google Scholar]
- Hirose S., Suzuki Y. In vitro transcription of eukaryotic genes is affected differently by the degree of DNA supercoiling. Proc Natl Acad Sci U S A. 1988 Feb;85(3):718–722. doi: 10.1073/pnas.85.3.718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho L., Bohr V. A., Hanawalt P. C. Demethylation enhances removal of pyrimidine dimers from the overall genome and from specific DNA sequences in Chinese hamster ovary cells. Mol Cell Biol. 1989 Apr;9(4):1594–1603. doi: 10.1128/mcb.9.4.1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyde J. E., Hearst J. E. Binding of psoralen derivatives to DNA and chromatin: influence of the ionic environment on dark binding and photoreactivity. Biochemistry. 1978 Apr 4;17(7):1251–1257. doi: 10.1021/bi00600a019. [DOI] [PubMed] [Google Scholar]
- Jackson D. A., Dickinson P., Cook P. R. The size of chromatin loops in HeLa cells. EMBO J. 1990 Feb;9(2):567–571. doi: 10.1002/j.1460-2075.1990.tb08144.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jupe E. R., Sinden R. R., Cartwright I. L. Stably maintained microdomain of localized unrestrained supercoiling at a Drosophila heat shock gene locus. EMBO J. 1993 Mar;12(3):1067–1075. doi: 10.1002/j.1460-2075.1993.tb05748.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman R. J., Schimke R. T. Amplification and loss of dihydrofolate reductase genes in a Chinese hamster ovary cell line. Mol Cell Biol. 1981 Dec;1(12):1069–1076. doi: 10.1128/mcb.1.12.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwano M., Ikehara Y. Inhibition by alpha-amanitin of messenger RNA formation in cultured fibroblasts: potentiation by amphotericin B. Exp Cell Res. 1973 Dec;82(2):454–457. doi: 10.1016/0014-4827(73)90365-0. [DOI] [PubMed] [Google Scholar]
- Leonard M. W., Patient R. K. Evidence for torsional stress in transcriptionally activated chromatin. Mol Cell Biol. 1991 Dec;11(12):6128–6138. doi: 10.1128/mcb.11.12.6128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L. F., Wang J. C. Supercoiling of the DNA template during transcription. Proc Natl Acad Sci U S A. 1987 Oct;84(20):7024–7027. doi: 10.1073/pnas.84.20.7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ljungman M., Hanawalt P. C. Efficient protection against oxidative DNA damage in chromatin. Mol Carcinog. 1992;5(4):264–269. doi: 10.1002/mc.2940050406. [DOI] [PubMed] [Google Scholar]
- Ljungman M., Hanawalt P. C. Localized torsional tension in the DNA of human cells. Proc Natl Acad Sci U S A. 1992 Jul 1;89(13):6055–6059. doi: 10.1073/pnas.89.13.6055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ljungman M., Nyberg S., Nygren J., Eriksson M., Ahnström G. DNA-bound proteins contribute much more than soluble intracellular compounds to the intrinsic protection against radiation-induced DNA strand breaks in human cells. Radiat Res. 1991 Aug;127(2):171–176. [PubMed] [Google Scholar]
- Ljungman M. Pretreatment with UV light renders the chromatin in human fibroblasts more susceptible to the DNA-damaging agents bleomycin, gamma radiation and 8-methoxypsoralen. Carcinogenesis. 1989 Mar;10(3):447–451. doi: 10.1093/carcin/10.3.447. [DOI] [PubMed] [Google Scholar]
- Ljungman M. The influence of chromatin structure on the frequency of radiation-induced DNA strand breaks: a study using nuclear and nucleoid monolayers. Radiat Res. 1991 Apr;126(1):58–64. [PubMed] [Google Scholar]
- Luchnik A. N., Hisamutdinov T. A., Georgiev G. P. Inhibition of transcription in eukaryotic cells by X-irradiation: relation to the loss of topological constraint in closed DNA loops. Nucleic Acids Res. 1988 Jun 10;16(11):5175–5190. doi: 10.1093/nar/16.11.5175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutter L. C., Judis L., Paretti R. F. Effects of histone acetylation on chromatin topology in vivo. Mol Cell Biol. 1992 Nov;12(11):5004–5014. doi: 10.1128/mcb.12.11.5004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellon I., Spivak G., Hanawalt P. C. Selective removal of transcription-blocking DNA damage from the transcribed strand of the mammalian DHFR gene. Cell. 1987 Oct 23;51(2):241–249. doi: 10.1016/0092-8674(87)90151-6. [DOI] [PubMed] [Google Scholar]
- Mitchell P. J., Carothers A. M., Han J. H., Harding J. D., Kas E., Venolia L., Chasin L. A. Multiple transcription start sites, DNase I-hypersensitive sites, and an opposite-strand exon in the 5' region of the CHO dhfr gene. Mol Cell Biol. 1986 Feb;6(2):425–440. doi: 10.1128/mcb.6.2.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizutani M., Ohta T., Watanabe H., Handa H., Hirose S. Negative supercoiling of DNA facilitates an interaction between transcription factor IID and the fibroin gene promoter. Proc Natl Acad Sci U S A. 1991 Feb 1;88(3):718–722. doi: 10.1073/pnas.88.3.718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizutani M., Ura K., Hirose S. DNA superhelicity affects the formation of transcription preinitiation complex on eukaryotic genes differently. Nucleic Acids Res. 1991 Jun 11;19(11):2907–2911. doi: 10.1093/nar/19.11.2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee R., Molloy G. R. 5,6-Dichloro-1-beta-D-ribofuranosylbenzimidazole inhibits transcription of the beta-hemoglobin gene in vivo at initiation. J Biol Chem. 1987 Oct 5;262(28):13697–13705. [PubMed] [Google Scholar]
- Norton V. G., Imai B. S., Yau P., Bradbury E. M. Histone acetylation reduces nucleosome core particle linking number change. Cell. 1989 May 5;57(3):449–457. doi: 10.1016/0092-8674(89)90920-3. [DOI] [PubMed] [Google Scholar]
- Ohta T., Hirose S. Purification of a DNA supercoiling factor from the posterior silk gland of Bombyx mori. Proc Natl Acad Sci U S A. 1990 Jul;87(14):5307–5311. doi: 10.1073/pnas.87.14.5307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne B. I., Guarente L. Transcription by RNA polymerase II induces changes of DNA topology in yeast. Genes Dev. 1988 Jun;2(6):766–772. doi: 10.1101/gad.2.6.766. [DOI] [PubMed] [Google Scholar]
- Parvin J. D., Sharp P. A. DNA topology and a minimal set of basal factors for transcription by RNA polymerase II. Cell. 1993 May 7;73(3):533–540. doi: 10.1016/0092-8674(93)90140-l. [DOI] [PubMed] [Google Scholar]
- Roberts S., Bentley D. L. Distinct modes of transcription read through or terminate at the c-myc attenuator. EMBO J. 1992 Mar;11(3):1085–1093. doi: 10.1002/j.1460-2075.1992.tb05147.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodi C. P., Sauerbier W. Structure of transcriptionally active chromatin: radiological evidence for requirement of torsionally constrained DNA. J Cell Physiol. 1989 Nov;141(2):346–352. doi: 10.1002/jcp.1041410216. [DOI] [PubMed] [Google Scholar]
- Schultz M. C., Brill S. J., Ju Q., Sternglanz R., Reeder R. H. Topoisomerases and yeast rRNA transcription: negative supercoiling stimulates initiation and topoisomerase activity is required for elongation. Genes Dev. 1992 Jul;6(7):1332–1341. doi: 10.1101/gad.6.7.1332. [DOI] [PubMed] [Google Scholar]
- Sehgal P. B., Darnell J. E., Jr, Tamm I. The inhibition by DRB (5,6-dichloro-1-beta-D-ribofuranosylbenzimidazole) of hnRNA and mRNA production in HeLa cells. Cell. 1976 Nov;9(3):473–480. doi: 10.1016/0092-8674(76)90092-1. [DOI] [PubMed] [Google Scholar]
- Sehgal P. B., Derman E., Molloy G. R., Tamm I., Darnell J. E. 5,6-Dichloro-1-Beta-D-ribofuranosylbenzimidazole inhibits initiation of nuclear heterogeneous RNA chains in HeLa cells. Science. 1976 Oct 22;194(4263):431–433. doi: 10.1126/science.982026. [DOI] [PubMed] [Google Scholar]
- Sehgal P. B., Fraser N. W., Darnell J. E., Jr Early Ad-2 transcription units: only promoter-proximal RNA continues to be made in the presence of DRB. Virology. 1979 Apr 15;94(1):185–191. doi: 10.1016/0042-6822(79)90448-3. [DOI] [PubMed] [Google Scholar]
- Sekiguchi J. M., Swank R. A., Kmiec E. B. Changes in DNA topology can modulate in vitro transcription of certain RNA polymerase III genes. Mol Cell Biochem. 1989 Feb 21;85(2):123–133. doi: 10.1007/BF00577108. [DOI] [PubMed] [Google Scholar]
- Shimada T., Nienhuis A. W. Only the promoter region of the constitutively expressed normal and amplified human dihydrofolate reductase gene is DNase I hypersensitive and undermethylated. J Biol Chem. 1985 Feb 25;260(4):2468–2474. [PubMed] [Google Scholar]
- Sinden R. R., Carlson J. O., Pettijohn D. E. Torsional tension in the DNA double helix measured with trimethylpsoralen in living E. coli cells: analogous measurements in insect and human cells. Cell. 1980 Oct;21(3):773–783. doi: 10.1016/0092-8674(80)90440-7. [DOI] [PubMed] [Google Scholar]
- Sinden R. R., Ussery D. W. Analysis of DNA structure in vivo using psoralen photobinding: measurement of supercoiling, topological domains, and DNA-protein interactions. Methods Enzymol. 1992;212:319–335. doi: 10.1016/0076-6879(92)12020-q. [DOI] [PubMed] [Google Scholar]
- Slansky J. E., Li Y., Kaelin W. G., Farnham P. J. A protein synthesis-dependent increase in E2F1 mRNA correlates with growth regulation of the dihydrofolate reductase promoter. Mol Cell Biol. 1993 Mar;13(3):1610–1618. doi: 10.1128/mcb.13.3.1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart A. F., Schütz G. Camptothecin-induced in vivo topoisomerase I cleavages in the transcriptionally active tyrosine aminotransferase gene. Cell. 1987 Sep 25;50(7):1109–1117. doi: 10.1016/0092-8674(87)90177-2. [DOI] [PubMed] [Google Scholar]
- Tamm I., Hand R., Caliguiri L. A. Action of dichlorobenzimidazole riboside on RNA synthesis in L-929 and HeLa cells. J Cell Biol. 1976 May;69(2):229–240. doi: 10.1083/jcb.69.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomsen B., Bendixen C., Westergaard O. Histone hyperacetylation is accompanied by changes in DNA topology in vivo. Eur J Biochem. 1991 Oct 1;201(1):107–111. doi: 10.1111/j.1432-1033.1991.tb16262.x. [DOI] [PubMed] [Google Scholar]
- Timmers H. T. Transcription initiation by RNA polymerase II does not require hydrolysis of the beta-gamma phosphoanhydride bond of ATP. EMBO J. 1994 Jan 15;13(2):391–399. doi: 10.1002/j.1460-2075.1994.tb06273.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjian R., Maniatis T. Transcriptional activation: a complex puzzle with few easy pieces. Cell. 1994 Apr 8;77(1):5–8. doi: 10.1016/0092-8674(94)90227-5. [DOI] [PubMed] [Google Scholar]
- Tsao Y. P., Wu H. Y., Liu L. F. Transcription-driven supercoiling of DNA: direct biochemical evidence from in vitro studies. Cell. 1989 Jan 13;56(1):111–118. doi: 10.1016/0092-8674(89)90989-6. [DOI] [PubMed] [Google Scholar]
- Ura K., Hirose S. Possible role of DNA topoisomerase II on transcription of the homeobox gene Hox-2.1 in F9 embryonal carcinoma cells. Nucleic Acids Res. 1991 Nov 25;19(22):6087–6092. doi: 10.1093/nar/19.22.6087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vennström B., Persson H., Pettersson U., Philipson L. A DRB (5,6 dichloro-beta-D-ribofuranosylbenzimidazole)-resistant adenovirus mRNA. Nucleic Acids Res. 1979 Nov 24;7(6):1405–1418. doi: 10.1093/nar/7.6.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villeponteau B., Lundell M., Martinson H. Torsional stress promotes the DNAase I sensitivity of active genes. Cell. 1984 Dec;39(3 Pt 2):469–478. doi: 10.1016/0092-8674(84)90454-9. [DOI] [PubMed] [Google Scholar]
- Villeponteau B., Martinson H. G. Gamma rays and bleomycin nick DNA and reverse the DNase I sensitivity of beta-globin gene chromatin in vivo. Mol Cell Biol. 1987 May;7(5):1917–1924. doi: 10.1128/mcb.7.5.1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub H., Cheng P. F., Conrad K. Expression of transfected DNA depends on DNA topology. Cell. 1986 Jul 4;46(1):115–122. doi: 10.1016/0092-8674(86)90865-2. [DOI] [PubMed] [Google Scholar]
- Zandomeni R., Mittleman B., Bunick D., Ackerman S., Weinmann R. Mechanism of action of dichloro-beta-D-ribofuranosylbenzimidazole: effect on in vitro transcription. Proc Natl Acad Sci U S A. 1982 May;79(10):3167–3170. doi: 10.1073/pnas.79.10.3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zandomeni R., Zandomeni M. C., Shugar D., Weinmann R. Casein kinase type II is involved in the inhibition by 5,6-dichloro-1-beta-D-ribofuranosylbenzimidazole of specific RNA polymerase II transcription. J Biol Chem. 1986 Mar 5;261(7):3414–3419. [PubMed] [Google Scholar]



