Abstract
The ARV1-encoded protein mediates sterol transport from the endoplasmic reticulum (ER) to the plasma membrane. Yeast ARV1 mutants accumulate multiple lipids in the ER and are sensitive to pharmacological modulators of both sterol and sphingolipid metabolism. Using fluorescent and electron microscopy, we demonstrate sterol accumulation, subcellular membrane expansion, elevated lipid droplet formation, and vacuolar fragmentation in ARV1 mutants. Motif-based regression analysis of ARV1 deletion transcription profiles indicates activation of Hac1p, an integral component of the unfolded protein response (UPR). Accordingly, we show constitutive splicing of HAC1 transcripts, induction of a UPR reporter, and elevated expression of UPR targets in ARV1 mutants. IRE1, encoding the unfolded protein sensor in the ER lumen, exhibits a lethal genetic interaction with ARV1, indicating a viability requirement for the UPR in cells lacking ARV1. Surprisingly, ARV1 mutants expressing a variant of Ire1p defective in sensing unfolded proteins are viable. Moreover, these strains also exhibit constitutive HAC1 splicing that interacts with DTT-mediated perturbation of protein folding. These data suggest that a component of UPR induction in arv1Δ strains is distinct from protein misfolding. Decreased ARV1 expression in murine macrophages also results in UPR induction, particularly up-regulation of activating transcription factor-4, CHOP (C/EBP homologous protein), and apoptosis. Cholesterol loading or inhibition of cholesterol esterification further elevated CHOP expression in ARV1 knockdown cells. Thus, loss or down-regulation of ARV1 disturbs membrane and lipid homeostasis, resulting in a disruption of ER integrity, one consequence of which is induction of the UPR.
Keywords: Apoptosis, Cholesterol Metabolism, Endoplasmic Reticulum (ER), ER Stress, Protein Folding
Introduction
The organization of lipids within and between organelle membranes is essential for many cellular processes, including protein folding and trafficking, osmotic integrity, and signal transduction. Sterols, such as cholesterol, are often physically coupled with sphingolipids in the lipid bilayer of membranes, where they act as foci for many important processes (1). By contrast, excess cholesterol is cytotoxic (2) and tightly regulated by the combined impact of biosynthesis, influx, efflux, and esterification (3). Many of these events initiate at the endoplasmic reticulum (ER).11 Consequently, the sterol content of the ER is maintained at a critical threshold of 5% (molar basis) of total ER lipids (4). This is a minor fraction of cellular cholesterol relative to the other membranes, but its impact on the cell is significant.
In mammals, cellular cholesterol and fatty acids are primarily sensed and controlled via the ER-associated sterol regulatory element-binding protein and its co-factors. Similarly, upon accumulation of unfolded proteins in the ER lumen, the membrane-associated stress transducer, IRE1 (inositol-requiring enzyme-1), initiates a cytoprotective cascade called the unfolded protein response (UPR) (5). Protein misfolding in the ER lumen initiates dimerization, trans-autophosphorylation, and activation of the cytosolic endoribonuclease domain of Ire1p, leading to removal of a 26-base pair intron from either the HAC1 (yeast) or XBP1 (higher eukaryotes) transcript. The spliced transcript is translated into a transcription factor that activates expression of UPR target genes. The IRE1-mediated branch of the UPR is conserved in all eukaryotes. However, in mammalian cells, there are two additional ER membrane-bound stress transducers: ATF6 (activating transcription factor-6) and PERK (protein kinase RNA (PKR)-like ER kinase) (6). Prolonged ER stress triggers apoptosis, due to PERK-mediated activation of the cell death effector CHOP (C/EBP homologous protein) (7).
Although the UPR is considered a protective reaction to misfolded proteins, perturbations in lipid homeostasis also activate the UPR in both yeast and mammalian systems. Cholesterol accumulation in murine macrophages induces IRE1- and PERK-mediated UPR induction, liberation of ER calcium stores, and CHOP-mediated apoptosis, contributing to macrophage death and atherosclerosis (2). The UPR is also induced by alterations in sphingolipid biosynthesis, such as knockdown of ceramide synthases in mammalian cells (8) or loss of Orm1p and Orm2p in yeast (9). Clearly, there is an intimate connection between the triad of lipid metabolism, protein folding, and ER function, although the mechanism and hierarchies of these relationships are unresolved.
The ARV1 (ACAT-related enzyme-2 required for viability 1) gene encodes an ER-localized protein that is a key component of lipid homeostasis in yeast and mammals (10, 11). Deletion of yeast ARV1 results in accumulation of sterols and ceramide in the ER (10, 12), consistent with a generalized role for this protein in anterograde lipid transport. Yeast ARV1 is also required for the maturation of glycosylphosphatidylinositol (GPI) anchors in the ER (13). The ARV1 gene is conserved throughout eukaryotic evolution. The majority of conservation in this membrane-associated protein lies in a 61-residue sequence known as the ARV1 homology domain. Importantly, expression of human ARV1 fully restores lipid transport and viability to yeast cells deleted for the endogenous gene (10, 12). In keeping with conservation, the antisense oligonucleotide (ASO)-mediated down-regulation of ARV1 in mice leads to elevated ER and serum cholesterol and misregulated bile acid metabolism (11).
In this study, we show that one consequence of loss of ARV1 in yeast and mammalian cells is induction of the UPR. We also demonstrate that induction of the UPR in ARV1 mutants is unusual in its severity and is synergistic with protein misfolding. This suggests a coordinated role for Arv1p in ER stress and lipid metabolism that is conserved throughout eukaryotic evolution.
EXPERIMENTAL PROCEDURES
General
Complete yeast media (YEPD) were prepared as described (14). Selection media were prepared with the appropriate drug(s), G418 (200 mg/liter; Invitrogen) and clonNAT (100 mg/liter; Werner BioAgents), as described previously (15). Yeast molecular and genetic techniques were performed according to conventional protocols (14). The Institutional Review Board at Columbia University Medical Center approved all animal protocols used in this paper.
Yeast Strains, Plasmids, Transformation, and Growth
Yeast strains (supplemental Table S1) were derived from W303-1A (16) or S288C (17) backgrounds. Deletion mutants were purchased from Open Biosystems or generated using one-step PCR-mediated gene disruption (18). Ire1p cLD plasmids (19) and the 4× unfolded protein response element (UPRE)-GFP reporter (20) were from Peter Walter and Randy Hampton, respectively. PCR products and plasmids were introduced into the host strain via lithium acetate transformation (21), followed by selection and confirmation by PCR. Spot assay random spore analysis was carried out as described (22). Aliquots from 2-ml cultures were plated as serial dilutions on defined media to assess cell viability. Growth curves were obtained using a Microbiology Workstation Bioscreen C (Thermo Electron Corp.).
Isolation and Transfection of Mouse Peritoneal Macrophages
Primary macrophages from adult female C57BL6J mice were harvested by peritoneal lavage after intraperitoneal injection of methyl-BSA as described previously (23). The acyl-coenzyme A-cholesterol acyltransferase (ACAT) inhibitor 58035 (3-[decyldimethylsilyl]-N-[2-(4-methylphenyl)-1-phenylethyl]propanamide) and acetylated LDL (AcLDL) were prepared as previously described (23). Macrophages were grown in media containing DMEM, 10% FBS, 1% penicillin/streptomycin, and 20% L-cell conditioned media to 50–60% confluence. On the day of the experiment, cells were treated with 250 nm control or ARV1 ASO, complexed with Lipofectamine 2000 (Invitrogen).
Microarray and Bioinformatics
Biotin-labeled and fragmented cRNA was hybridized to the Ye6100 or S98 Yeast GeneChip arrays (Affymetrix) (24). All data mining was performed as described previously (24). The data discussed in this publication have been deposited in the NCBI Gene Expression Omnibus (25) and are accessible through GEO series accession number GSE26801. Bioinformatics and gene ontology (GO) analyses were performed as described in the supplemental material.
Gene Expression Analyses
For Northern blot analyses, 10 μg of RNA was run on a 1.2% formaldehyde gel and transferred to a nylon membrane. Membranes were hybridized with PCR-generated gene-specific probes and analyzed by audioradiography and phosphorimaging (GE Healthcare) (n = 3). For real-time PCR, RNA was isolated from cells using the RNAeasy kit (Qiagen). 100 ng of RNA was reverse transcribed using the SuperScript first strand system for RT-PCR (Invitrogen). Real-time PCRs were performed with the MyIQ single-color real time PCR detection system (Bio-Rad), SYBR Green 2× Supermix (Bio-Rad), and the primers described (supplemental Table S2). Expression levels were calculated relative to a housekeeping gene as described previously (26).
Immunoblots
Cells were washed two times in cold PBS prior to lysis. Whole cell lysates were isolated in Laemmli sample buffer (Bio-Rad) containing 0.5% β-mercaptoethanol. 50 μg of lysate was run on a 4–20% Tris-HCl gel (Bio-Rad). Protein was electrotransferred onto a 0.45-μm nitrocellulose membrane (Bio-Rad) and incubated with the primary antibody overnight. Protein bands were detected with HRP-conjugated secondary antibodies (Bio-Rad) and SuperSignal West Femto enhanced chemiluminescent solution (Pierce).
Microscopy
For electron microscopy, cells were grown to mid-log phase, prefixed in glutaraldehyde, postfixed in potassium permanganate, dehydrated by graded ethanol series, and embedded in Spurr's resin (27). Cells were thin sectioned, stained with Reynold's lead citrate (28), and visualized on a transmission electron microscope. Fluorescent imaging in yeast was carried out on a Zeiss Axiovert 200M using a 63× oil immersion objective. All yeast images were taken using a Hamamatsu Orca-ER camera. Nile Red (Sigma), FM4-64 (Invitrogen), and 22-(N-(7-nitrobenz-2-oxa-1,3-diazol-4-yl)amino)-23,24-bisnor-5-cholen-3β-ol (NBD-cholesterol) (Invitrogen) were used at the indicated concentrations. To examine sterol distribution, live cells were grown to saturation in YEPD, resuspended in YEPD containing 100 μg/ml Filipin (Polysciences, Inc.) and imaged immediately. Macrophages were assayed for apoptosis by staining with Alexa 488-conjugated Annexin V and propidium iodide as previously described (23).
RESULTS
Loss of ARV1 in Yeast Is Pleiotropic
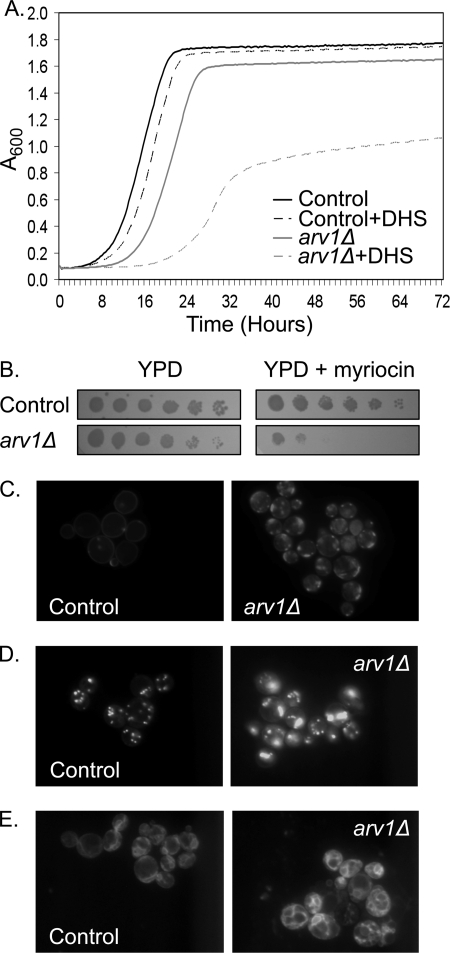
The independent identification of mutations in ARV1 from screens that perturbed sterol (10) or sphingolipid (12) metabolism suggests alterations in lipid distribution and membrane homeostasis in ARV1 deficient cells. Indeed, an established biochemical feature of ARV1 mutants is accumulation of ER ceramide at the expense of complex sphingolipid formation (10). Consistent with a growth inhibitory blockade in ER export of ceramide, we found that ARV1 mutants are sensitive to exogenous dihydrosphingosine, a substrate for ceramide synthases (Fig. 1A). Repression of the entire sphingolipid biosynthetic pathway by treatment with the serine palmitoyltransferase inhibitor myriocin is also more toxic in ARV1 mutants than controls (Fig. 1B), suggesting that formation of sphingolipids is especially critical in these strains.
FIGURE 1.
Loss of Arv1p disrupts lipid homeostasis. A, growth of control and arv1Δ yeast strains in YPD with and without 1 μm dihydrosphingosine (DHS); n = 3 for each condition. B, serial dilutions of yeast strains grown with and without 500 ng/ml myriocin. Filipin (100 μg/ml) (C) or Nile Red (1 μg/ml) (D) staining of control and arv1Δ strains (both in a upc2-1 background) grown for 24 h in YPD + 5 μg/ml cholesterol. E, aerobic uptake of NBD-cholesterol (50 μg/ml) in control and arv1Δ strains (both in a upc2-1 background).
Under standard growth conditions, loss of ARV1 results in subcellular accumulation (29, 30) and esterification (10, 12) of endogenously synthesized sterol. To specifically assess the impact of ARV1 upon anterograde sterol transport from the ER, we inspected ARV1 mutants with lipid-specific fluorescent dyes under sterol-loading conditions. Deletion of ARV1 results in anaerobic inviability, precluding the analysis of exogenous sterol uptake. To overcome this, we assessed the role of ARV1 in the presence of a mutation in the UPC2 transcription factor that activates aerobic sterol uptake (24). ARV1 mutants exhibited subcellular sterol accumulation as detected by filipin staining (Fig. 1C). Interestingly, this staining pattern occurred in 43% of the ARV1 mutant population and was completely absent from control cells. Consistent with an accumulation of acyltransferase substrates at the ER, steryl ester lipid droplet formation was also increased in sterol-loaded arv1Δ strains stained with Nile Red (Fig. 1D). These strains were further profiled using the fluorescent sterol probe NBD-cholesterol, which also accumulated in arv1Δ strains as a marked proliferation of subcellular membranes (Fig. 1E).
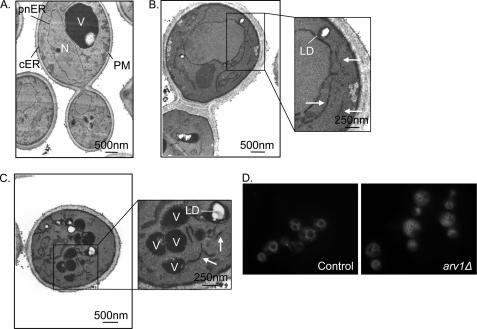
We then investigated the ultrastructure of ARV1 mutants by transmission electron microscopy. The ER in control Saccharomyces cerevisiae strains (Fig. 2A) is contiguous with the nuclear (perinuclear (pnER)) and plasma membranes (cortical (cER)). By contrast, arv1Δ strains exhibit abundant cytoplasmic double membrane stretches (Fig. 2, B and C). Consistent with the accumulation of neutral lipids described above, ARV1 mutants also exhibited more cytoplasmic lipid droplets than control cells. These droplets were engulfed in a membranous structure that was absent from control strains. In addition, we observed vacuolar fragmentation in arv1Δ strains (Fig. 2C) that was confirmed by fluorescent imaging with the vacuole-specific dye FM4-64 (30) (Fig. 2D).
FIGURE 2.
Loss of Arv1p disrupts organelle integrity. Thin section EM of control (A) and arv1Δ strains (B and C). N, nucleus; V, vacuole; pnER and cER, perinuclear and cortical endoplasmic reticulum, respectively; PM, plasma membrane; LD, lipid droplet. The arrows indicate membrane proliferation seen in ARV1 mutants. D, FM4-64 (20 μg/ml) staining of cells.
Transcriptional Response to Loss of ARV1
Deletion of ARV1 has numerous consequences, including morphological changes in organelles and inviability in response to a variety of lipid-related stresses. These aberrancies are consistent with redistribution of sterols and sphingolipids within the cell. This pleiotropy led us to hypothesize that multiple homeostatic mechanisms are disturbed by loss of ARV1. We therefore profiled the response to ARV1 deletion using transcription microarrays. 273 genes were misregulated greater than 2-fold upon ARV1 deletion (supplemental Table S3). The gene with the greatest increase in expression was ULI1, with a 48-fold change in expression. ULI1 encodes a protein of unknown function that is up-regulated during the UPR (31). The scope of expression changes in arv1Δ strains is reflected in a GO analysis in which 57 GO categories were significantly enriched in the mutants (supplemental Table S3). The GO categories describe cellular processes consistent with disrupted ER homeostasis in arv1Δ strains. These include down-regulation of translation (negative t values; e.g. ribosome biogenesis (GO:0042254) and translation (GO:0006412)) and up-regulation of ER stress responses (positive t values; e.g. ER-associated protein catabolism (GO:0030433) and ubiquitin-dependent protein catabolism (GO:0006511)).
To identify the transcriptional programs altered in ARV1 mutants, we performed motif-based regression analysis of the microarray data (32). This method infers changes in transcription factor activity by relating differential mRNA expression to the affinity with which the factor binds to the promoter region of each gene. Three transcription factors are highly differentially activated in response to arv1Δ. The activity of each transcription factor is defined by its t value; a significant p value confirms that the activity of the transcription factor (t value) is significantly different in the ARV1 deletion condition. Transcriptional changes due to Thi2p (t value = 5.42, p value 2.92E−08 (33)) and Hap2p (t value = 4.27, p value = 9.80E−06 (34)) activation were significant and confirmed by real-time PCR of representative targets (THI20 and THI4, and YCR100C and ARH1, respectively; data not shown). However, Hac1p, the transcriptional regulator of the UPR in yeast, exhibited the greatest change in differential activity in ARV1 deletion strains (t value = 6.53, p value = 3.25E−11), relative to controls. These data suggest induction of the UPR in ARV1 mutants.
Loss of ARV1 Constitutively Induces the UPR
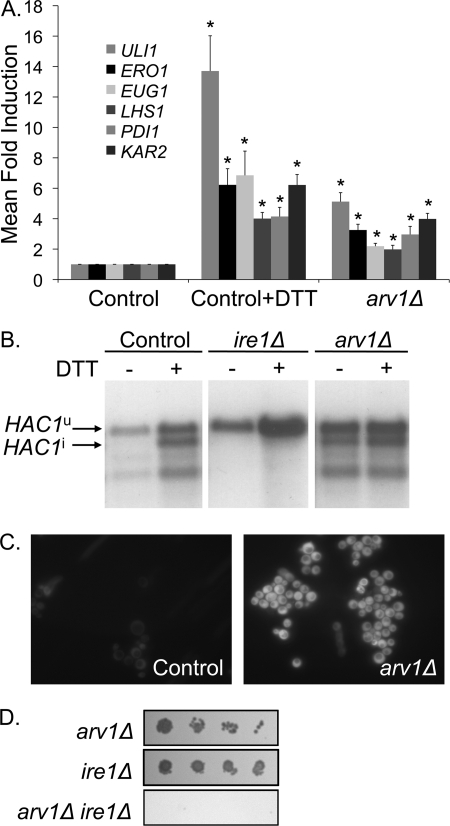
To confirm activation of the UPR in arv1Δ strains, we performed real-time expression analyses of canonical UPR target genes (the adenyl-nucleotide exchange factor LHS1, the thiol oxidase ERO1, the protein-disulfide isomerases EUG1 and PDI1, and the HSP70 family molecular chaperone encoded by the KAR2 gene (35)). arv1Δ strains exhibit induction of the UPR target genes (Fig. 3A) of similar magnitude to control cells treated with the reducing agent dithiothreitol (DTT), an established and marked inducer of the UPR.
FIGURE 3.
The UPR is activated in arv1Δ strains. A, real-time PCR measurements of UPR target gene expression due to 2 mm DTT treatment for 45 min or arv1Δ. *, statistically significant (p < 0.05) differences of the mean -fold induction (ΔΔCt) ± S.D. (error bars) in comparison with controls (n = 3). B, Northern blot analysis of HAC1 mRNA in strains with the indicated mutation. Unspliced HAC1u and spliced (induced) HAC1i are indicated; lower bands are splicing intermediates. C, control and arv1Δ strains expressing GFP driven by an UPRE. D, random spore analysis of a diploid strain heterozygous for deletions of ARV1 and IRE1 on media selecting for the mutant genotypes (arv1Δ, (G418), ire1Δ (clonNAT), or arv1Δ ire1Δ (G418 and clonNAT)).
We then went on to assess the status of the UPR directly by an IRE1-mediated HAC1 splicing assay. ARV1 mutants exhibit constitutive HAC1 splicing, independent of DTT treatment (Fig. 3B). Furthermore, arv1Δ strains expressing a GFP reporter driven by an UPRE (36) exhibit constitutive UPR activation, as demonstrated by UPRE-GFP fluorescence (Fig. 3C).
The key role of UPR induction in the physiology of ARV1-deficient cells was further confirmed when we investigated the genetic interaction between ARV1 and IRE1, the major ER stress transducer in yeast (5). Random spore analysis of a diploid strain heterozygous for deletions of ARV1 and IRE1 demonstrated that arv1Δ ire1Δ is a lethal combination (Fig. 3D). Furthermore, in a tetrad manipulation analysis of 70 individual meiotic events of the same diploid, we could not recover arv1Δ ire1Δ double mutants (although single mutants were recovered at Mendelian frequencies). Thus, ARV1 deficiency renders cells dependent on the UPR for viability.
UPR Induction and Membrane Lipid Homeostasis
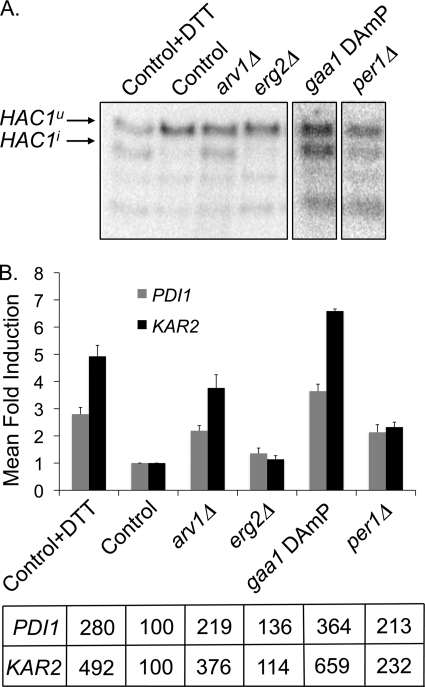
Arv1p is required for lipid egress from the ER in yeast. Furthermore, its loss results in induction of the UPR. We questioned whether this was a general response to disruption of ER lipid homeostasis. We screened 76 mutant strains known to be deficient in sterol, fatty acid, and sphingolipid metabolism by a HAC1 splicing assay (supplemental Table S4). Deletions in non-essential genes or hypomorphic (DAmP (37)) alleles of essential genes were assessed for UPR induction by comparing the ratio of spliced HAC1 (HAC1i) to total HAC1 by densitometry of Northern blots. Strains exhibiting a HAC1i/total HAC1 ratio of 3 or greater were then subjected to real-time expression analyses of canonical UPR target genes. The fold-induction of PDI1 and KAR2 was expressed as a fraction of the control strain. Of the strains examined, only the arv1Δ strain exhibited a PDI1 and KAR2 induction of 2-fold or more (Fig. 4, A and B). Thus, arv1Δ strains are unusual within the set of lipid mutants screened in terms of severity of UPR induction.
FIGURE 4.
UPR induction in arv1Δ strains is distinct from strains exhibiting defects in lipid metabolism and GPI anchor biosynthesis. A, Northern blot analysis of HAC1 mRNA in strains defective in sterol, sphingolipid, fatty acid, and GPI anchor metabolism. Unspliced HAC1u and spliced (induced) HAC1i are indicated; lower bands are splicing intermediates. B, real-time PCR profiles of UPR target genes PDI1 and KAR2 in strains with defective lipid or GPI anchor metabolism. Mean -fold induction of PDI1 and KAR2 is expressed as a percentage of the control strain (bottom). In contrast to DTT treatment of control cells, the majority of mutants (e.g. erg2Δ) were negative by these criteria. Error bars, S.D.
ARV1 has also been implicated in GPI anchor maturation by a proposed GPI-flippase activity (13). To determine if activation of the UPR in arv1Δ strains was distinct from other strains defective in GPI-anchor biosynthesis, we screened 22 mutations in the yeast GPI biosynthetic pathway (38) for UPR induction (supplemental Table S4). Of the GPI mutants screened, only the ARV1 and PER1 deletions and the gaa1 hypomorph exhibited HAC1 splicing and a PDI1-KAR2 induction that was 2 times greater than control cells (Fig. 4, A and B). Gaa1p is a component of the GPI transamidase complex, and, similar to ARV1 mutants, the GAA1 mutant is defective in the synthesis of complex sphingolipids and sensitive to aureobasidin A (13). Per1p is responsible for remodeling lipid moieties in the GPI anchor such that GPI-anchored proteins are unable to associate with detergent-resistant lipid rafts in PER1 mutants (39). ARV1 deficiency is also unusual in its severity of UPR induction compared with the remainder of the genome. Of ∼4500 nonessential gene deletions investigated, the arv1Δ strain exhibits the second strongest induction of the UPR (40).
Interaction of Protein Misfolding and Lipid Dysregulation in arv1Δ Strains
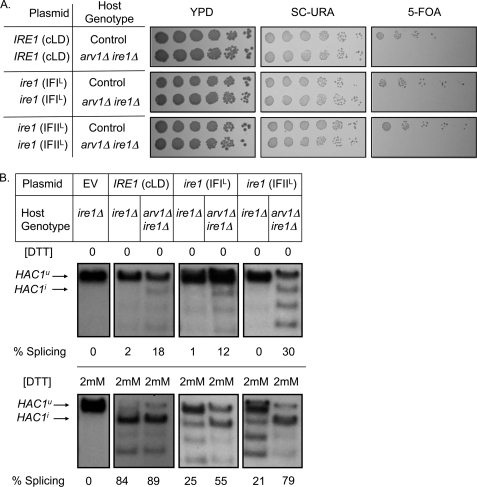
We hypothesize that the unusual severity of UPR induction in arv1Δ strains reflects the synergism of lipid accumulation and protein misfolding in the ER. Structural studies of Ire1p demonstrate that the Ire1p core luminal domain (cLD) aligns with other cLDs in a head-to-head (interface I (IFI)) and tail-to-tail (interface II (IFII)) fashion. Ire1p cLD constructs in which the interfaces are mutated (IFIL and IFIIL), disrupt Ire1p oligomerization and the response to unfolded proteins (19). We transformed arv1Δ ire1Δ heterozygous diploids with plasmids expressing the control and mutant cLDs and sporulated the strains. Haploid arv1Δ ire1Δ progeny were not obtained from diploids containing the vector control. By contrast, expression of control and mutant cLDs rescued arv1Δ ire1Δ lethality (Fig. 5A). Rescue was lost upon selection against the URA3-based cLD plasmid by growth on 5-fluoroortic acid, which is converted into a toxic substance in the presence of the URA3 gene product. The mutant cLDs were sufficient to induce the UPR signaling cascade in an arv1Δ background (Fig. 5B, top). We then assayed UPR induction in ARV1 mutants treated with DTT to induce protein misfolding. HAC1 splicing mediated by the Ire1p variants was further elevated in an arv1Δ strain under these conditions (Fig. 5B, bottom), indicative of an independent and additive interaction between ARV1 function and protein misfolding.
FIGURE 5.
UPR induction in arv1Δ mutants is synergistic with the detection of unfolded proteins in the ER lumen. A, plasmids mutated at interface 1 (IFIL) or interface 2 (IFIIL) of the Ire1p core luminal domain rescue arv1Δ ire1Δ synthetic lethality, as represented by growth on a URA-based selection medium. Counterselection for the plasmid by growth on 5-fluoroortic acid (5-FOA) restores the synthetic lethal phenotype. The empty vector control (pRS416) in the arv1Δ ire1Δ strains was inviable. Control genotype refers to yeast strains in the same genetic background as the double mutants. B, Northern blot analysis of HAC1 mRNA in ire1Δ and arv1Δ ire1Δ strains expressing control Ire1p, IFIL Ire1p, or IFIIL Ire1p with or without 2 mm DTT treatment. EV, empty vector control. Unspliced HAC1u and spliced (induced) HAC1i are indicated; lower bands are splicing intermediates. HAC1 splicing was calculated as HAC1i/(HAC1i + HAC1u) based on densitometry.
Decreased ARV1 Expression in Mammalian Cells Induces ER Stress and Apoptosis
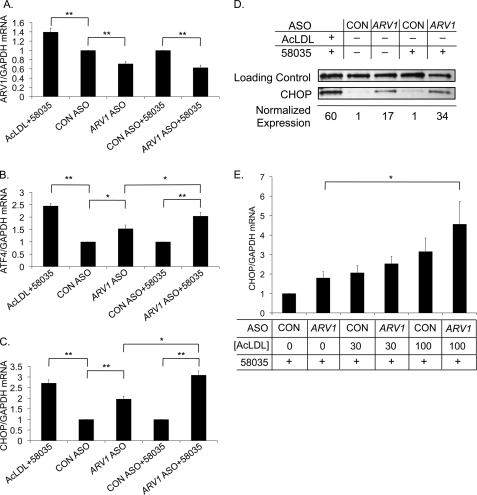
We questioned whether lipid accumulation in the ER due to decreased ARV1 expression could activate the UPR in mammalian cells. ASOs to ARV1 are effective in modulating ARV1 expression (11). Because organs replete with macrophages (e.g. spleen) were not impacted in these studies, we used the ASOs in primary murine macrophages in vitro and observed a significant decrease in ARV1 expression (Fig. 6A). In order to determine if the UPR is induced in ASO-treated macrophages, we probed for activation of the three major branches of the UPR. Mammalian IRE1 splices XBP1 transcripts (analogous to HAC1 transcripts in yeast) to form an active transcription factor (41). Surprisingly, ARV1 ASO-treated macrophages did not exhibit increased XBP1 splicing relative to controls (data not shown). Furthermore, induction of the UPR via processing of ATF6, another branch of the UPR in mammalian cells, was also unchanged (data not shown). However, ATF4 levels were significantly increased upon ARV1 knockdown, reflecting activation of PERK-mediated UPR (Fig. 6B). Prolonged ER stress induces apoptosis mediated by CHOP (42), another target of PERK-mediated UPR that is selectively activated by ATF4. ARV1 knockdown significantly increased the levels of CHOP transcripts (Fig. 6C). To exacerbate cholesterol accumulation at the ER, we treated macrophages with the ACAT inhibitor 58035 in conjunction with the ASOs. We observed a significant increase in ATF4 and CHOP expression in comparison with ARV1 ASO treatment alone (Fig. 6, B and C). ARV1 ASO treatment also increased CHOP expression at the protein level, which was further elevated by the addition of the ACAT inhibitor (Fig. 6D).
FIGURE 6.
ARV1 knockdown in murine macrophages induces PERK-mediated UPR induction. ARV1 (A), ATF4 (B), and CHOP (C) mRNA expression in macrophages treated with control (CON) or ARV1 ASO with or without the ACAT inhibitor, 58035. Asterisks show statistically significant (*, p < 0.05; **, p < 0.005) differences of the mean -fold induction (ΔΔCt) ± S.E.; n = 6. D, CHOP protein levels in macrophages treated with control or ARV1 ASO with or without 58035. CHOP expression was calculated by densitometry and normalized to a nonspecific loading control. E, CHOP mRNA expression after treatment with control or ARV1 ASO for 48 h with increasing concentrations of AcLDL (0–100 μg/ml) and 58035 for 5 h; n = 5. *, statistically significant (p < 0.05) difference of the mean -fold induction ± S.E. (error bars).
To determine if the effect of ARV1 knockdown was synergistic with exogenous cholesterol loading, we treated ASO-transfected macrophages with increasing concentrations of AcLDL and 10 μg/ml ACAT inhibitor 58035 (2). Using CHOP transcript levels as a marker for UPR induction, we demonstrate that cholesterol loading increased the UPR in ARV1 knockdown cells in a statistically significant, dose-dependent manner (Fig. 6E).
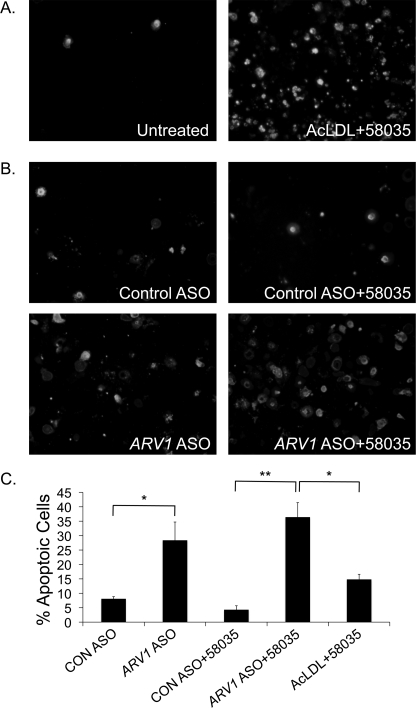
Cholesterol loading of macrophages using AcLDL and an ACAT inhibitor triggers CHOP-dependent apoptosis (Fig. 7A) (2). We treated primary macrophages with ARV1 ASOs for 24 h in the absence of AcLDL and investigated annexin V/propidium iodide cellular fluorescence to assess apoptosis. ARV1 ASO treatment of macrophages increased the number of apoptotic cells relative to control ASO treatment (Fig. 7B). This was further exacerbated when combined with ACAT inhibition, suggesting a toxic accumulation of free cholesterol at the ER. Furthermore, apoptosis in the ARV1 knockdown cells treated with the ACAT inhibitor exceeded that seen with exogenous cholesterol loading mediated by AcLDL treatment and ACAT inhibition (Fig. 7C). Thus, ARV1 knockdown results in apoptosis, probably due to elevated CHOP expression in response to ER cholesterol accumulation.
FIGURE 7.
ARV1 knockdown in murine macrophages induces apoptosis. Shown is Annexin V and PI staining of macrophages treated with AcLDL + 58035 (24 h) (A) or control or ARV1 ASO (24 h) (B) with or without 58035 for 18 h. C, the percentage of total cells stained with annexin V and PI (mean ± S.E. (error bars), four fields of cells; at least 100 cells/field). Asterisks denote statistically significant differences (*, p < 0.05; **, p < 0.005).
DISCUSSION
The endoplasmic reticulum represents a protected, sequestered environment for protein folding and lipid homeostasis. ARV1 encodes a major, previously unsuspected component in the repertoire of reactions that maintain the integrity of this compartment. As might be anticipated for such a key process, loss of ARV1 activity has many sequelae. It is growth-limiting in yeast under a variety of conditions and embryonic lethal in nematodes (43). Yeast cells and macrophages induce the UPR to survive the loss of ARV1, and this induction is apparently independent of protein misfolding, although it certainly synergizes with this additional ER burden. Ablation of ARV1 in the murine liver results in cholestasis and hypercholesterolemia (11). In addition to conservation of ARV1, the impact of its loss is also maintained throughout eukaryotic evolution.
The range of phenotypes conferred by the ARV1 null allele in yeast suggests a wide reaching role of the Arv1 protein in cell metabolism and the high impact of loss of lipid export from the ER. Our observations of sterol-loaded yeast cells (Fig. 1, C–E) indicate that sterol transport to the ER from the periphery of ARV1 mutant cells is normal but that its egress from the ER is impaired. The lipid and ultrastructural changes of ARV1 mutants are probably upstream of the UPR because constitutive induction of this cascade (e.g. in SCJ1 deletion mutants) does not phenocopy ARV1 deletion (44). Unesterified sterols, ceramide, and its derivatives optimize the fluidity and function of all eukaryotic cellular membranes. Sterol transit from the ER to the plasma membrane is rapid, efficient, and energy-requiring (45) and probably occurs via aqueous diffusion, vesicular transport, and/or carrier proteins (46). Similarly, ceramide egress from the ER to the Golgi is energy-requiring and primarily accomplished by vesicular independent carrier proteins (47). Yeast Arv1p was isolated based on its role in sterol transport from the endoplasmic reticulum to the plasma membrane (10). Subsequently, ARV1-deficient cells were found to possess lower levels of complex sphingolipids and mislocalize GPI intermediates, consistent with a general role for Arv1p in lipid mobilization at the ER (13). However, the coordinated regulation of sterol levels with other lipids (1) makes it difficult to distinguish whether ER stress seen in ARV1 mutants is a consequence of aberrant sterol transport or reflects the ARV1-mediated transport of multiple lipid species.
We also provide evidence that a component of the UPR is independent from protein misfolding. Induction of the UPR in ARV1-deficient yeast cells is synergistic with the buildup of unfolded proteins in the ER lumen, plausibly due to changes in ER lipid homeostasis. The mediators of the UPR in yeast (Ire1p) and mammals (IRE1, PERK, and ATF6) are transmembrane proteins that require activation in or release from the ER membrane to be physiologically relevant. It is not surprising that disrupting the membrane integrity of this organelle, by altering the sterol and/or sphingolipid content of the ER, impacts the UPR. Thus, the UPR appears to be a downstream and essential response to the altered lipid homeostasis in ARV1 mutants. This may be the case for many lipid metabolic disorders in humans.
It is interesting that not all UPR target genes (35) are activated upon deletion of ARV1 in yeast. Given that the UPR is constitutively active in yeast ARV1 deletions, the transcriptional changes we observed represent the impact of prolonged ER stress. Similarly, in macrophages treated with the ARV1 ASO for 48 h, it is apparent that although PERK-mediated UPR is induced (Fig. 6), not all ER stress transducers are activated. In HEK293 cells, IRE1- and ATF6-mediated UPR induction diminishes with time, whereas PERK-mediated UPR is sustained over a more prolonged period (48). Selective activation of PERK signaling is proapoptotic, impairing cell proliferation and inducing apoptosis (49), whereas activation of IRE1 is antiapoptotic. We propose that PERK-mediated UPR and cell death in ARV1-deficient cells is a consequence of prolonged ER stress due to lipid overload.
The link between disruption of cellular lipid homeostasis and ER stress is compelling. The UPR and its consequences are frequently invoked upon lipid overload of many different cell types. Strikingly, we demonstrate that both within the examined lipid mutants and also genome-wide (40), loss of ARV1 is one of the most potent inducers of the UPR. We propose that the UPR is a generalized response to ER homeostasis, resulting from independent and often synergistic perturbations in membrane structure, lipid metabolism, and protein folding. The deterioration of any of these safeguards is likely to manifest as diseases in which lipids play a role. A disease association has yet to be reported for ARV1, although variation in this gene exists in human populations (see the National Institutes of Health, NCBI, SNP Web site). Thus, mutations in ARV1 may influence diseases such as obesity, atherosclerosis, and type 2 diabetes.
Acknowledgments
We appreciate the assistance of Ying Liu, Arline Albala, and Samuel C. Silverstein.
This work was supported, in whole or in part, by National Institutes of Health Grant DK54320 (to S. L. S.). This work was also supported by the American Heart Association, the American Diabetes Association, and the Ara Parseghian Medical Research Foundation.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1–S4.
- ER
- endoplasmic reticulum
- UPR
- unfolded protein response
- ASO
- antisense oligonucleotide
- UPRE
- unfolded protein response element
- AcLDL
- acetylated low density lipoprotein
- cLD
- core luminal domain
- GPI
- glycosylphosphatidylinositol
- ACAT
- acyl-coenzyme A-cholesterol acyltransferase
- GO
- gene ontology
- NBD-cholesterol
- 22-(N-(7-nitrobenz-2-oxa-1,3-diazol-4-yl)amino)-23,24-bisnor-5-cholen-3β-ol.
REFERENCES
- 1. Gulati S., Liu Y., Munkacsi A. B., Wilcox L., Sturley S. L. (2010) Prog. Lipid Res. 49, 353–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Feng B., Yao P. M., Li Y., Devlin C. M., Zhang D., Harding H. P., Sweeney M., Rong J. X., Kuriakose G., Fisher E. A., Marks A. R., Ron D., Tabas I. (2003) Nat. Cell Biol. 5, 781–792 [DOI] [PubMed] [Google Scholar]
- 3. Sturley S. L. (2000) Biochim. Biophys. Acta 1529, 155–163 [DOI] [PubMed] [Google Scholar]
- 4. Radhakrishnan A., Goldstein J. L., McDonald J. G., Brown M. S. (2008) Cell Metab. 8, 512–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sidrauski C., Walter P. (1997) Cell 90, 1031–1039 [DOI] [PubMed] [Google Scholar]
- 6. Ron D., Walter P. (2007) Nat. Rev. Mol. Cell Biol. 8, 519–529 [DOI] [PubMed] [Google Scholar]
- 7. Wang X. Z., Kuroda M., Sok J., Batchvarova N., Kimmel R., Chung P., Zinszner H., Ron D. (1998) EMBO J. 17, 3619–3630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Spassieva S. D., Mullen T. D., Townsend D. M., Obeid L. M. (2009) Biochem. J. 424, 273–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Han S., Lone M. A., Schneiter R., Chang A. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 5851–5856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tinkelenberg A. H., Liu Y., Alcantara F., Khan S., Guo Z., Bard M., Sturley S. L. (2000) J. Biol. Chem. 275, 40667–40670 [DOI] [PubMed] [Google Scholar]
- 11. Tong F., Billheimer J., Shechtman C. F., Liu Y., Crooke R., Graham M., Cohen D. E., Sturley S. L., Rader D. J. (2010) J. Biol. Chem. 285, 33632–33641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Swain E., Stukey J., McDonough V., Germann M., Liu Y., Sturley S. L., Nickels J. T., Jr. (2002) J. Biol. Chem. 277, 36152–36160 [DOI] [PubMed] [Google Scholar]
- 13. Kajiwara K., Watanabe R., Pichler H., Ihara K., Murakami S., Riezman H., Funato K. (2008) Mol. Biol. Cell 19, 2069–2082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ausubel F. M., Brent R., Kingston R. E., Moore D. D., Seidman J. G., Smith J. A., Struhl K. (1998) Current Protocols in Molecular Biology, pp. 13.0.3–13.13.7, John Wiley & Sons, Inc., New York [Google Scholar]
- 15. Tong A. H., Evangelista M., Parsons A. B., Xu H., Bader G. D., Pagé N., Robinson M., Raghibizadeh S., Hogue C. W., Bussey H., Andrews B., Tyers M., Boone C. (2001) Science 294, 2364–2368 [DOI] [PubMed] [Google Scholar]
- 16. Thomas B. J., Rothstein R. (1989) Cell 56, 619–630 [DOI] [PubMed] [Google Scholar]
- 17. Giaever G., Chu A. M., Ni L., Connelly C., Riles L., Véronneau S., Dow S., Lucau-Danila A., Anderson K., André B., Arkin A. P., Astromoff A., El-Bakkoury M., Bangham R., Benito R., Brachat S., Campanaro S., Curtiss M., Davis K., Deutschbauer A., Entian K. D., Flaherty P., Foury F., Garfinkel D. J., Gerstein M., Gotte D., Güldener U., Hegemann J. H., Hempel S., Herman Z., Jaramillo D. F., Kelly D. E., Kelly S. L., Kötter P., LaBonte D., Lamb D. C., Lan N., Liang H., Liao H., Liu L., Luo C., Lussier M., Mao R., Menard P., Ooi S. L., Revuelta J. L., Roberts C. J., Rose M., Ross-Macdonald P., Scherens B., Schimmack G., Shafer B., Shoemaker D. D., Sookhai-Mahadeo S., Storms R. K., Strathern J. N., Valle G., Voet M., Volckaert G., Wang C. Y., Ward T. R., Wilhelmy J., Winzeler E. A., Yang Y., Yen G., Youngman E., Yu K., Bussey H., Boeke J. D., Snyder M., Philippsen P., Davis R. W., Johnston M. (2002) Nature 418, 387–391 [DOI] [PubMed] [Google Scholar]
- 18. Erdeniz N., Mortensen U. H., Rothstein R. (1997) Genome Res. 7, 1174–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Credle J. J., Finer-Moore J. S., Papa F. R., Stroud R. M., Walter P. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 18773–18784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sato B. K., Hampton R. Y. (2006) Yeast 23, 1053–1064 [DOI] [PubMed] [Google Scholar]
- 21. Ito H., Fukuda Y., Murata K., Kimura A. (1983) J. Bacteriol. 153, 163–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tong A. H., Lesage G., Bader G. D., Ding H., Xu H., Xin X., Young J., Berriz G. F., Brost R. L., Chang M., Chen Y., Cheng X., Chua G., Friesen H., Goldberg D. S., Haynes J., Humphries C., He G., Hussein S., Ke L., Krogan N., Li Z., Levinson J. N., Lu H., Ménard P., Munyana C., Parsons A. B., Ryan O., Tonikian R., Roberts T., Sdicu A. M., Shapiro J., Sheikh B., Suter B., Wong S. L., Zhang L. V., Zhu H., Burd C. G., Munro S., Sander C., Rine J., Greenblatt J., Peter M., Bretscher A., Bell G., Roth F. P., Brown G. W., Andrews B., Bussey H., Boone C. (2004) Science 303, 808–813 [DOI] [PubMed] [Google Scholar]
- 23. Devries-Seimon T., Li Y., Yao P. M., Stone E., Wang Y., Davis R. J., Flavell R., Tabas I. (2005) J. Cell Biol. 171, 61–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wilcox L. J., Balderes D. A., Wharton B., Tinkelenberg A. H., Rao G., Sturley S. L. (2002) J. Biol. Chem. 277, 32466–32472 [DOI] [PubMed] [Google Scholar]
- 25. Edgar R., Domrachev M., Lash A. E. (2002) Nucleic Acids Res. 30, 207–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Livak K. J., Schmittgen T. D. (2001) Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 27. Wright R. (2000) Microscopy Res. Tech. 51, 496–510 [DOI] [PubMed] [Google Scholar]
- 28. Reynolds E. S. (1963) J. Cell Biol. 17, 208–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Malathi K., Higaki K., Tinkelenberg A. H., Balderes D. A., Almanzar-Paramio D., Wilcox L. J., Erdeniz N., Redican F., Padamsee M., Liu Y., Khan S., Alcantara F., Carstea E. D., Morris J. A., Sturley S. L. (2004) J. Cell Biol. 164, 547–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Beh C. T., Rine J. (2004) J. Cell Sci. 117, 2983–2996 [DOI] [PubMed] [Google Scholar]
- 31. Metzger M. B., Michaelis S. (2009) Mol. Biol. Cell 20, 1006–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lee E., Bussemaker H. J. (2010) Mol. Syst. Biol. 6, 412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nishimura H., Kawasaki Y., Kaneko Y., Nosaka K., Iwashima A. (1992) FEBS Lett. 297, 155–158 [DOI] [PubMed] [Google Scholar]
- 34. Pinkham J. L., Guarente L. (1985) Mol. Cell Biol. 5, 3410–3416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Travers K. J., Patil C. K., Wodicka L., Lockhart D. J., Weissman J. S., Walter P. (2000) Cell 101, 249–258 [DOI] [PubMed] [Google Scholar]
- 36. Cronin S. R., Rao R., Hampton R. Y. (2002) J. Cell Biol. 157, 1017–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Breslow D. K., Cameron D. M., Collins S. R., Schuldiner M., Stewart-Ornstein J., Newman H. W., Braun S., Madhani H. D., Krogan N. J., Weissman J. S. (2008) Nat. Methods 5, 711–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Orlean P., Menon A. K. (2007) J. Lipid Res. 48, 993–1011 [DOI] [PubMed] [Google Scholar]
- 39. Fujita M., Umemura M., Yoko-o T., Jigami Y. (2006) Mol. Biol. Cell 17, 5253–5264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Jonikas M. C., Collins S. R., Denic V., Oh E., Quan E. M., Schmid V., Weibezahn J., Schwappach B., Walter P., Weissman J. S., Schuldiner M. (2009) Science 323, 1693–1697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Calfon M., Zeng H., Urano F., Till J. H., Hubbard S. R., Harding H. P., Clark S. G., Ron D. (2002) Nature 415, 92–96 [DOI] [PubMed] [Google Scholar]
- 42. Zinszner H., Kuroda M., Wang X., Batchvarova N., Lightfoot R. T., Remotti H., Stevens J. L., Ron D. (1998) Genes Dev. 12, 982–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kamath R. S., Fraser A. G., Dong Y., Poulin G., Durbin R., Gotta M., Kanapin A., Le Bot N., Moreno S., Sohrmann M., Welchman D. P., Zipperlen P., Ahringer J. (2003) Nature 421, 231–237 [DOI] [PubMed] [Google Scholar]
- 44. Silberstein S., Schlenstedt G., Silver P. A., Gilmore R. (1998) J. Cell Biol. 143, 921–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kaplan M. R., Simoni R. D. (1985) J. Cell Biol. 101, 446–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Reinhart M. P. (1990) Experientia 46, 599–611 [DOI] [PubMed] [Google Scholar]
- 47. Hanada K., Kumagai K., Tomishige N., Kawano M. (2007) Biochim. Biophys. Acta 1771, 644–653 [DOI] [PubMed] [Google Scholar]
- 48. Lin J. H., Li H., Yasumura D., Cohen H. R., Zhang C., Panning B., Shokat K. M., Lavail M. M., Walter P. (2007) Science 318, 944–949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lin J. H., Li H., Zhang Y., Ron D., Walter P. (2009) PLoS ONE 4, e4170. [DOI] [PMC free article] [PubMed] [Google Scholar]