Abstract
Ribosome-inactivating proteins (RIPs) are toxic N-glycosidases that depurinate the universally conserved α-sarcin loop of large rRNAs. They have received attention in biological and biomedical research because of their unique biological activities toward animals and human cells as cell-killing agents. A better understanding of the depurination mechanism of RIPs could allow us to develop potent neutralizing antibodies and to design efficient immunotoxins for clinical use. Among these RIPs, ricin exhibited remarkable efficacy in depurination activity and highly conserved tertiary structure with other RIPs. It can be considered as a prototype to investigate the depurination mechanism of RIPs. In the present study, we successfully identified a novel functional domain responsible for controlling the depurination activity of ricin, which is located far from the enzymatic active site reported previously. Our study indicated that ricin A-chain mAbs binding to this domain (an α-helix comprising the residues 99–106) exhibited an unusual potent neutralizing ability against ricin in vivo. To further investigate the potential role of the α-helix in regulating the catalytic activity of ricin, ricin A-chain variants with different flexibility of the α-helix were rationally designed. Our data clearly demonstrated that the flexibility of the α-helix is responsible for controlling the depurination activity of ricin and determining the extent of protein synthesis inhibition, suggesting that the conserved α-helix might be considered as a potential target for the prevention and treatment of RIP poisoning.
Keywords: Antibodies, Enzyme Catalysis, Molecular Dynamics, Ribosomal RNA (rRNA), RIP
Introduction
Ribosome-inactivating proteins (RIPs)3 are depurinating rRNA N-glycosidases (E.C. 3.2.2.22) that cleave a single bond between a specific adenine and ribose of rRNA in eukaryotes (1). They are generally divided into two classes (1–3). Type I RIPs such as trichosanthin and gelonin are monomeric enzymes of ∼30 kDa. Type II RIPs are heterodimeric proteins with an approximate molecular mass of 60 kDa, in which one polypeptide with RIP activity (A-chain) is linked by a disulfide bridge to a galactose-binding lectin (B-chain). The B-chain is able to bind to a galactose-containing receptor on the surface of sensitive cells and mediate transport of the A-chain through the secretory pathways into the cytoplasm.
Ricin (a type II RIP), which has emerged as a powerful catalyst for mammalian ribosomes, is a good prototype to investigate the N-glycosidase mechanism of RIPs. Ricin A-chain (RTA) is the catalytic subunit of ricin, which catalyzes the depurination of an invariant adenosine residue, A4324, within the GA4324GA tetraloop motif of the highly conserved sarcin-ricin loop of eukaryotic 28 S rRNA (4). Most previous studies have focused on the role of the active-site residues that are crucial for catalytic activity of RIPs. Day et al. (5) have presented the crystal structure of ricin, indicating that RTA has a prominent cleft able to recognize the target rRNA stem-loop. Site-directed mutagenesis, as well as analysis by systematic deletion of amino acids, strongly supports that the pronounced cleft is the enzymatic active site (6–8). Recently, the transition state analogues in structures of ricin and saporin ribosome-inactivating proteins were studied. The data confirmed that the invariant residues of RIPs in the catalytic active site of ricin were essential for the efficient catalysis by RTA (9). Although the biochemical properties of RIPs have been extensively studied, the enzymatic mechanism of RIPs is still elusive.
Deep understanding of the catalytic mechanism of RIPs could help us develop potent neutralizing antibodies for protecting against ricin, a potential weapon of bioterrorism, and to design more effective therapeutic immunotoxins. Most of previous studies have demonstrated that antibodies can be utilized as a powerful tool to investigate the structural and functional relationship of target proteins (10, 11). In the present study, we firstly employed antibodies obtained from individual mice immunized with RTA to study the relationship between the antibody recognition site on RTA and the neutralizing capacity of these RTA antibodies. In line with previous studies, we found that the antibodies specifically recognizing the enzymatic active site of RTA displayed substantial protective efficacy in vitro. One of the most striking findings in our present study is that the RTA mAbs 6C2 and 6G3, whose combining sites are distant from the catalytic active site of ricin reported previously (5, 7, 12, 13), exhibit more marked neutralizing ability than the Abs binding to the enzymatic active site of RTA. The computational and experimental data strongly indicated that the flexibility of an α-helix, which is recognized by 6C2 and 6G3, plays an important role in regulating the enzymatic activity of ricin, suggesting that the α-helix can serve as an attractive target for protection and rescue from ricin toxicity.
EXPERIMENTAL PROCEDURES
A detailed description of all of the procedures is provided under supplemental Methods. Briefly, recombinant RTA was prokaryotically expressed in our laboratory. Then, RTA Abs were generated by immunizing Balb/c mice. Hybridomas secreting RTA mAbs were characterized by ELISA using isotope-specific reagents (Sigma) and purified from ascites fluid using protein A (GE Healthcare). The relative binding avidity of each mAb was assessed by ELISA using RTA protein as the target antigen (14). The inhibition of the enzymatic activity of ricin by RTA mAbs was measured in a cell-free in vitro translation assay using rabbit reticulocyte lysates (Promega) as both the source of mRNA and ribosomes (14, 15). Then, a standard 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide dye reduction assay was performed to evaluate the neutralization of ricin-mediated cytotoxicity. The in vivo mouse protection assays were done to evaluate the in vivo protective efficacy of RTA mAbs. The mice were randomized into several groups (n = 10 mice/group) and intraperitoneally injected with ricin holotoxin diluted in 0.2 ml of PBS (50 μg/kg). Subsequently, the mice were immediately administered RTA mAbs (0.5 μg or 2.5 μg). For passive transfer experiments, we gave ricin-challenged mice a single dose of RTA mAbs at indicated time points (1 h, 2 h, 4 h, or 8 h). The survival of mice was monitored until the experiment was terminated.
The RTA variants with different flexibility of the α-helix were designed based on inspection of the crystal structure of ricin. Then, the flexibility of the α-helix was evaluated by using the molecular dynamic method. The dynamic simulations were performed with the AMBER 9.0 suite of programs (16). The detailed procedure can be seen under supplemental Methods.
RESULTS
RIPs with Very Low Similarity in Primary Structure Are Highly Conserved in Tertiary Structures
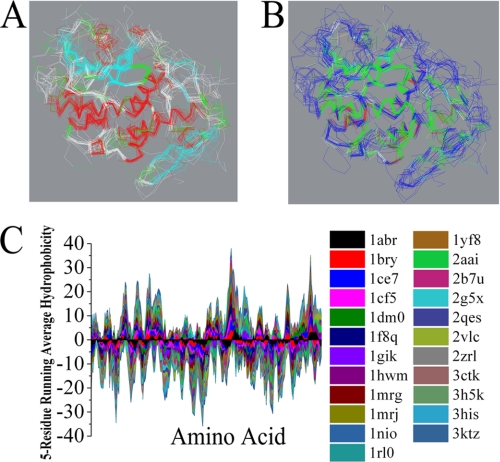
To date, the structures of 15 type I RIPs and eight type II RIPs have been solved by x-ray crystallography (supplemental Table 1). First, multiple protein sequences were aligned by using the Align123 algorithm, a progressive pairwise alignment algorithm modified from the CLUSTAL W program (17). Supplemental Fig. 1 shows the best alignment of sequences of these 23 RIPs (15 type I RIPs and eight A-chains of type II RIPs). Because the lengths of these proteins differ, some insertions or deletions were required for optimal alignment. Our data showed that the sequence identity among these RIPs was very low (2.3%), and the sequence similarity was only 7.9%. However, further study revealed that all of these type I RIPs and the A-chain of the type II RIPs have a very similar pattern in the secondary and tertiary structures (Fig. 1A). When each is compared with RTA, the root mean squared distance between corresponding Cα positions is generally less than 3.5 Å (supplemental Table 1). As we know, the solvent accessibility is much more related to the tertiary interactions between residues far apart in the sequence but close in three-dimensional space. Fig. 1B indicates that all of these RIPs exhibited a similar solvent accessibility. Moreover, the interesting feature in this alignment is the significant matching of hydrophobic amino acid residues in all 23 RIPs (supplemental Fig. 1). Hydrophobicity plots were designed to display the distribution of hydrophobic and hydrophilic residues along a protein sequence and are useful for identifying both local and global properties for a protein sequence (18, 19). In this study, the analysis of primary sequences of these RIPs by using a Kyte-Doolittle hydrophobicity plot indicated that these RIPs have a similar hydrophobic and hydrophilic pattern (Fig. 1C). These data revealed that RIPs are highly conserved in the secondary and tertiary structures, although the similarity of the primary structures is very low. These findings may suggest that the evolutionary conserved tertiary structure may be essential for unique catalysis and rigid regulation of N-glycosidases.
FIGURE 1.
Structure analysis of RIPs. Stereo view of the superimposed backbone atoms for the structures of RIPs. A, secondary structure elements are color coded: helix (red), strand (cyan), and coil (white). B, hydrophobic and hydrophilic residues are colored green and blue, respectively. C, Kyte-Doolittle hydropathy analysis using a window of five amino acids. Hydrophobicity is shown as the vertical axis, with the hydrophobic side of the plot having a positive value. The horizontal axis shows the amino acid residue number along the sequence.
Generation and Characterization of Monoclonal Antibodies against the Ricin A-Chain
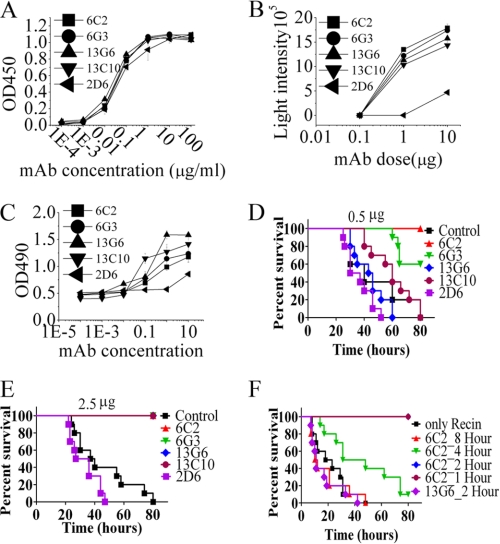
Because of exhibiting a highly conserved tertiary structure with other RIPs and near perfect performance in depurination of the invariant adenosine residue from eukaryotic ribosome, ricin is an ideal prototype to understand the mechanism of toxicity of RIPs and to develop potent neutralizing antibodies for protecting against RIP toxins. Antibodies specific for different epitopes of RTA can be used as valuable tools for elucidating the structure-function relationship of RTA. Therefore, we first produced a panel of mAbs that bind determinants on the ricin A-chain. Seventeen mAbs specific for RTA have been identified. As shown in supplemental Table 2, Ig subclass and the protective effects of the supernatant were evaluated. Among these anti-RTA mAbs, 6C2, 6G3, 13C10, 13G6, and 2D6 showed protective effects against toxicity of ricin from Vero cells (supplemental Table 2). An enzyme-linked immunosorbent assay was subsequently performed to measure the relative binding avidity of the RTA antibodies. As shown in Fig. 2A, our data indicated that these five RTA mAbs had similar binding avidities. Then, these Abs were tested for their ability to inhibit ricin enzymatic activity, which was measured as the inhibition of protein synthesis in a cell-free system using rabbit reticulocyte lysates. In line with our results described above, 6C2, 6G3, 13C10, and 13G6 showed the best inhibition of ricin activity, with 2D6 exhibiting moderately inhibitory activity (Fig. 2B). Next, the protective effects of these RTA mAbs were investigated by evaluating the viability of Vero cells after incubation with ricin (6.4 ng/ml) and the indicated concentrations of Abs (Fig. 2C). In accordance with the protein inhibition experiment, the best neutralizing activity was seen with 6C2, 6G3, 13C10, and 13G6 (Fig. 2, B and C). To evaluate the correlation between in vitro neutralization and in vivo protection, we assessed the therapeutic activity of different neutralizing mAbs by in vivo mouse protection assay. Mice were injected intraperitoneally with ricin at a dose of 50 μg/kg and immediately administered a single dose of mAbs. Notably, 0.5 μg of the neutralizing mAbs 6C2 and 6G3 provided potent protection efficacy (Fig. 2D). In contrast, 13C10 and 13G6 only exhibited moderate protection against ricin. Our data indicate that even a single administration of 2.5 μg of 2D6 gave slightly better protection than that of negative control (Fig. 2E).
FIGURE 2.
Characterization and evaluation of mAbs against RTA. A, relative binding avidity of RTA mAbs. RTA mAbs were purified on protein G. Purified mAbs were titrated in ELISA for binding to RTA (100 ng/well). B, inhibition of ricin enzymatic activities by RTA mAbs. A fixed concentration of ricin holotoxin was mixed with varying concentrations of RTA mAb. The ability of mAbs to block the inhibition of cell-free protein synthesis was measured. C, Ab-mediated inhibition of ricin cytotoxicity. Vero cells were incubated with ricin (1 μg) and the indicated Ab. After 40 h, cell viability was assayed by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide dye reduction. D–F, therapeutic efficacy of RTA mAbs. After an intraperitoneal injection of ricin into the left side on the basis of weight (50μ/kg), mice were administered a single dose of 0.5 μg (D) or 2.5 μg (E) of RTA mAb. Data reflect ∼10 mice per condition. F, efficacy of RTA-specific mAb therapy at different time points. A single dose (5 μg) of mAb 6C2 was administered either 1 h, 2 h, 4 h, or 8 h after the ricin injection. Data are mean ± S.D. of at least three experiments.
Because mAb 6C2 and 6G3 exhibited unusually strong protection against ricin both in vitro and in vivo, we further investigated the protective effects of these two mAbs by ascertaining the therapeutic window for post-exposure treatment using the BALB/c mouse model. As shown in Fig. 2F, when ricin-challenged mice were administrated 6C2 mAb at 2 h post-challenge, all animals were fully protected (100% survival). When 6C2 mAb treatment was delayed to 4 h, median survival in the 6C2-treated group was extended to 39.9 h, with statistically significant survival extension in this model by log-rank analysis (p < 0.05 compared with the untreated group). In line with these results, our data indicate that 6G3 showed a similar high level of protection against ricin in the post-exposure therapy (supplemental Fig. 2). However, the protective effect of 13G6 could not be detected although the mice were administered 13G6 at 2 h post-challenge (Fig. 2F).
Mapping of Neutralizing Monoclonal Antibodies to RTA
To further investigate the reason of the variation in the neutralizing capacity of these anti-RTA mAbs, the immunological characterization of mAbs was carried out. As a preliminary form of epitope mapping, antibody cross-inhibition assays were performed among the anti-RTA mAbs, in which the binding of an horseradish peroxidase-conjugated anti-RTA mAb was inhibited by unconjugated Abs (Fig. 3, A and B). Our data showed that RTA mAbs 6C2 and 6G3 appeared to have similar epitope specificity, which was inhibited by unconjugated 6C2 in a dose-dependent manner. Similarly, 13G6 and 13C10 had comparable patterns of inhibition, indicating that they blocked the binding of each other. Then, the epitopes of RTA mAbs were subsequently mapped with random peptide phage display libraries. The 6C2 mAb binds to the amino acid sequence EXITH, which corresponds exactly to residues 102–106 of RTA. Phages selected by the 13G6 mAb have a common motif of YYYSXXXT (supplemental Table 3). Overlaying this sequence on the three-dimensional structure of RTA (13, 20) provides a best fit with residues 150–157 of RTA, which fold into part of the enzyme active site (Fig. 3C) (5). Mapping the 2D6 mAb with a random peptide library yielded a consensus sequence of IPXLPXRV. The successfully characterized epitopes of these RTA mAbs were plotted onto the crystal structure of ricin (Fig. 3C). It is interesting to find that, although the RTA mAbs 6C2 and 6G3 display an unusually strong neutralizing ability, the binding sites of these two mAbs are situated on an α-helix (residues 99–106 of RTA) far from the catalytic active site of ricin. However, the RTA mAb 2D6, whose binding site is located close to the enzymatic cleavage site of ricin, has a very limited ability to protect against ricin toxicity both in vitro and in vivo.
FIGURE 3.
Epitope Identification for RTA mAbs. A and B, microtiter plates were coated with 50 ng/well of RTA. Unconjugated RTA mAbs (5 μg/ml), indicated on the horizontal axis, were added to the coated microtiter wells and incubated for 60 min. The HRP-conjugated Ab indicated at the top of the graph was added, and the mixture was incubated for 60 min. The plates were washed, and colorimetric substrate was added. Antibodies were tested in up to four different experiments, of which this is representative. C, epitope mapping of RTA mAbs. Epitopes defined by mapping using peptide display phage libraries were plotted onto the three-dimensional structure of RTA. Red indicates the epitope of RTA mAbs 6C2 and 6G3, green the epitope of mAb 2D6, yellow the enzyme active site, and blue the residue present in the active site and bound by RTA mAbs 13G6 and 13C10. The graphs are representative of at least three experiments, each of which showed similar results.
The Ideal Recognition Site for RTA Neutralizing Antibody Is Not Located at the Enzymatic Active Site of RTA
To investigate whether the significant protective effect of these RTA antibodies accounts for blocking of the interaction between RTA and substrate RNA, biacore binding studies were performed. We firstly synthesized the biotin-labeled stem-loop RNA (slRNA), which contains the required substrate motif for RTA. Then, RTA at the concentration of 60 μg/ml was allowed to bind to slRNA, and a typical binding curve was obtained (supplemental Fig. 3A). However, when we pre-incubated RTA with the excess amount (1:4 molar ratio) of 13G6, the binding signal could not be detected (supplemental Fig. 3B). However, the binding signal could be observed after incubating RTA with an excessive amount of 6C2 or 2D6 (supplemental Fig. 3, C and D). We further investigated the binding affinity for the substrate with or without 6C2 and 6G3. The significant difference was not observed (supplemental Table 4). Our data clearly demonstrate that the antibodies 6C2 and 2D6 are unable to block the interaction between RTA and slRNA. However, 13G6 could completely block the binding of RTA to slRNA when pre-incubated with RTA. Then, the extent of inhibition of RNA hydrolysis by RTA antibody was measured by HPLC determination of the amount of adenine released. As shown in supplemental Fig. 3E, when we incubated an excessive amount of mAbs 6C2 or 13G6 with RTA, more than 80% of adenine released from ribosome could be inhibited. The noncompetitive inhibition constants (Ki) calculated for 6C2 and 6G3 are shown in supplemental Fig. 4. Our data indicate that 6C2 displays a similar inhibition constant to that of 6G3. The results revealed that 6C2 and 6G3 have a similar increase of neutralizing activity for ribosome. As shown in supplemental Fig. 4, 6C2 and 6G3 exhibited a similar dose-dependent inhibitory effect, with 21, 43, and 66% of inhibition at antibody concentrations of 0.033, 0.100, and 0.200 nm, respectively. All these results indicated that the substantial protective effect of 6C2 is not attributable to a direct block of ricin-slRNA interaction but to disruption of the depurination activity of RTA.
Rational Design of RTA Variants to Investigate the Functional Role of the α-helix in Regulating the Enzymatic Activity of Ricin
A considerable amount of research has been done to study the role of flexibility in enzyme action, indicating that enzyme flexibility plays an important role in regulating enzymatic activity (21, 22). We suspect that the flexibility of the α-helix may play an important role in regulating the enzymatic activity of ricin. To investigate this issue, we rationally designed RTA variants with different levels of α-helix flexibility and evaluated the differences of these RTA variants in the depurination activity and protein synthesis inhibition. First, experimental variants were designed based on inspection of the crystal structure of ricin. As shown in Fig. 4A, Ala was substituted for functional or biophysical reasons: substitution of Glu and Asp at positions 99 and 100 of Ala was expected to disrupt the salt bridge formation with Arg-48 and Arg-56. Ala was substituted for His and Leu at positions 106 and 107 to avoid hydrophobic interactions with Ile-53 and Phe-57. Substitution of Asp-96 was performed to prevent the formation of a weak salt bridge between Asp-96 and Arg-125. To investigate the role of the different point mutations we introduced in RTA in modulating the flexibility of the α-helix (residues 99–106 of RTA), we carried out molecular dynamics simulations of RTA and its variants, and analyzed the structural fluctuations throughout the Molecular dynamics trajectories.
FIGURE 4.
Rational design of RTA variants. A, stereo diagram showing the overall fold of the RTA. The epitope of mAb 6C2 is colored in blue. B and C, the average fluctuation per residue calculated over the last 40 ns of the trajectory. Root mean square fluctuation values were calculated relative to the starting structure at 300 K (B) and 500 K (C).
The values of root mean square fluctuations reveal the extent of motion of an atom around its equilibrium position, providing information on flexibility or rigidity of molecules or parts or them (23, 24). As shown in Fig. 4B, the single point mutation D96A in RTA presented an increase in flexibility in the α-helix. When the temperature was elevated up to 500 K, the mutations D96A or H106A/L107A could significantly increase the flexibility of the α-helix. But the double mutation E99A/D100A substantially reduced the flexibility of the α-helix, even under the high-temperature conditions (Fig. 4, B and C). The conformational change and dynamics of the α-helix were illustrated by showing root mean square deviation and by superimposing the coordinates of RTA and RTA variants. In line with the RMSF analysis, our data showed that a large fluctuation was produced by introducing the D96A mutation after 33 ns of dynamic simulation (Fig. 5, A and C). The double mutation H106A/L107A could markedly increase the fluctuation of the α-helix at the high temperature, suggesting that the double mutation H106A/L107A has the potential to make the α-helix more flexible at room temperature. On the other hand, the double mutation E99A/D100A significantly decreased the flexibility of the α-helix even at the high-temperature conditions (Fig. 5B). Previous studies have demonstrated that there are four amino acids (Tyr-80, Tyr-123, Glu-177, and Arg-180) critical for the depurination activity of ricin (6, 9, 12, 25). To investigate how the flexibility of the α-helix affects the enzyme activity of ricin, we performed a dihedral angle analysis. Our results indicated that the flexibility of the α-helix could affect the side chain orientation of Glu-177 (supplemental Fig. 5).
FIGURE 5.
Characterization and comparison of RTA variants. A and B, RMS deviations for the α-helix (residues 99–106 of RTA). Molecular dynamic simulation of RTA variants were performed at 300 K (A) and 500 K (B). The last 40 ns of each trajectory were used for analysis. C, the average structures of each RTA variant over the last 10 ns of simulation at 300 K are superimposed and shown. RTA is colored in gray, RTA_D96A in red, RTA_E99A/D100A in blue, and RTA_H106A/L107A in green. D, quantity of adenine released from 80 S rabbit reticulocyte ribosomes. The extent of ribosome depurination was quantified by using HPLC. E, the extent of protein synthesis inhibition by RTA variants was evaluated in cell-free assays. The different concentrations of RTA variants were incubated with a cell-free in vitro translation mixture. Luciferase activity reported in relative light intensity is representative of protein translation and measured after 90 min at 30 °C. Data are mean ± S.D. of at least three experiments.
After these RTA variants were successfully constructed and expressed, the amount of adenine released and subsequent inhibition of protein synthesis produced by RTA variants were studied. As shown in Fig. 5, D and E, introducing the point mutation at position 96 (RTA_D96A) or at positions 106 and 107 (RTA_H106A/L107A) led to a substantial increase in the toxicity of RTA, but the double mutation RTA_E99A/D100A can substantially decrease the amount of adenine released and protein synthesis inhibition. In line with our expectations of the rational design, these data clearly demonstrate that the flexibility of the α-helix is responsible for modulating the depurination activity of RTA and, ultimately, the degree of inhibition of protein synthesis.
DISCUSSION
In this study, we first collected a set of crystal structures of RIPs from the Protein Data Bank. Then, bioinformatic methods for analyzing the sequence-structure relationship of RIPs were performed. It is striking to note that, although all these RIPs have a very low sequence similarity, the secondary and tertiary structures are highly conserved. Many previous investigations have revealed that RIPs depurinate RNA in ribosomes at a specific universally conserved position in the rat 28 S rRNA(1–3), suggesting that the highly conserved tertiary structure may be responsible for unique catalytic properties of RIPs. Further analysis indicated that all these RIPs had a similar pattern in the distribution of hydrophobic and hydrophilic residues along the protein sequence. Previous studies have collectively demonstrated that the hydrophobic effect is the dominant force in protein folding and protein stability (26–29). In agreement with these results, our data suggest that the hydrophobic distribution pattern might play a significant role in determining the secondary and tertiary structures of RIPs, which are highly conserved in ribosome depurination.
Previous studies have demonstrated that the catalysis of the 80 S ribosome by RTA approached the diffusion rate limit for enzymatic reactions with a catalytic efficiency of 2.6 × 108 m−1s−1 for the discontinuous assay conditions (15, 30). Among these RIPs, ricin has evolved to become a near perfect catalyst for mammalian ribosomes. It can be seen as an ideal prototype to investigate the N-glycosidase mechanism of RIPs. In addition, epitope-specific mAbs have been widely used as a powerful tool to clarify the relation between the structure and the function of the macromolecule (31, 32). Therefore, we generated a panel of mAbs raised against the ricin A-chain, the catalytic subunit of ricin, to study the structure-function relationship of ricin. Both in vitro and in vivo assays were performed to evaluate the neutralizing ability of RTA mAbs. Although 6C2 and 13G6 had a similar protective efficacy in our in vitro experiments, our in vivo protection data clearly showed that the mAbs 6C2 and 6G3, whose binding sites are far from the enzymatic active site, had more potent neutralizing activity against ricin than the RTA mAb 13G6, recognizing the catalytic active site of ricin. Our biacore data revealed that the exceptional neutralization potency of 6C2 is not attributed to the block of ricin-slRNA interaction but the direct inhibition of depurination activity of ricin. Because the combining site of mAbs 6C2 and 6G3 is located far from the catalytic active site of ricin, the ricin-antibody complex formation cannot be blocked by the substrate. Thus, the mAbs 6C2 and 6G3 could continuously play a functional role in inhibition of depurination activity of ricin in vivo, which may be the reason why 6C2 and 6G3 exhibited more potent in vivo protective efficacy than 13G6.
The data also indicate that the α-helix recognized by 6C2 may play a significant role in controlling the depurination activity of ricin. To further investigate the role of the α-helix in regulating the enzymatic activity of ricin, the molecular dynamic method was employed to rationally design RTA variants with differences in the flexibility of the α-helix. Our subsequent experimental data clearly revealed that increasing the flexibility of the α-helix could substantially enhance the depurination activity of ricin and the inhibition of protein synthesis. On the other hand, the marked decrease in the flexibility of the α-helix has significantly reduced the enzymatic activity of ricin. As we known, binding by antibody could substantially decrease the flexibility of the targeted region. Thus, binding by 6C2 could substantially reduce the flexibility of the α-helix and substantially decrease the depurination activity of ricin. These data suggest that the flexibility of the α-helix plays a critical role in regulating the toxicity of ricin. Furthermore, we performed a dihedral angle analysis to study the relationship of the α-helix and catalytic active site of ricin. We found that the flexibility of the α-helix could affect the side chain orientation of Glu-177 (supplemental Fig. 5). Previous studies have identified that Glu-177 is critical for the depurination activity of ricin (5, 6, 9, 12). Taken together, these data suggested that the α-helix may modulate the enzymatic activity of ricin by controlling the side chain orientation of Glu-177.
In summary, our data clearly demonstrate that the α-helix recognized by 6C2 is responsible for modulation of ricin toxicity, suggesting that the α-helix might be used as an ideal target for the prevention and treatment of ricin toxicosis. Additionally, our bioinformatic analysis revealed that RIPs are highly conserved in the secondary and tertiary structures, and the conserved α-helix may be critical for the depurination activity of other RIPs.
Acknowledgment
We thank the Shanghai Supercomputing Center for allocation of computing time.
This work was supported by Program Project Grants 973 and 863 from the National Natural Science Foundation of China, Ministry of Science and Technology of China, by Grant B905 from the National Key project for New Drug Creation and Manufacture, Shanghai Commission of Science and Technology, Shanghai Leading Academic Discipline Project, and by the Program for Pujiang Scholar and Innovative Research Team in University.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables 1–4, Figs. 1–5, methods, and references.
- RIP
- ribosome-inactivating protein
- RTA
- ricin A-chain
- Ab
- antibody
- slRNA
- stem-loop RNA.
REFERENCES
- 1. Stirpe F., Battelli M. G. (2006) Cell Mol. Life Sci. 63, 1850–1866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hartley M. R., Lord J. M. (2004) Mini. Rev. Med. Chem. 4, 487–492 [DOI] [PubMed] [Google Scholar]
- 3. Motto M., Lupotto E. (2004) Mini. Rev. Med. Chem. 4, 493–503 [DOI] [PubMed] [Google Scholar]
- 4. Endo Y., Mitsui K., Motizuki M., Tsurugi K. (1987) J. Biol. Chem. 262, 5908–5912 [PubMed] [Google Scholar]
- 5. Day P. J., Ernst S. R., Frankel A. E., Monzingo A. F., Pascal J. M., Molina-Svinth M. C., Robertus J. D. (1996) Biochemistry 35, 11098–11103 [DOI] [PubMed] [Google Scholar]
- 6. Morris K. N., Wool I. G. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 4869–4873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kim Y., Mlsna D., Monzingo A. F., Ready M. P., Frankel A., Robertus J. D. (1992) Biochemistry 31, 3294–3296 [DOI] [PubMed] [Google Scholar]
- 8. Ready M. P., Kim Y., Robertus J. D. (1991) Proteins 10, 270–278 [DOI] [PubMed] [Google Scholar]
- 9. Ho M. C., Sturm M. B., Almo S. C., Schramm V. L. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 20276–20281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wang Y., Zhang X., Yuan L., Xu T., Rao Y., Li J., Dai H. (2008) Biochem. Biophys. Res. Commun. 372, 902–906 [DOI] [PubMed] [Google Scholar]
- 11. Conti-Fine B. M., Lei S., Mclane K. E. (1996) Annu. Rev. Biophys. Biomol. Struct. 25, 197–229 [DOI] [PubMed] [Google Scholar]
- 12. Kim Y., Robertus J. D. (1992) Protein Eng. 5, 775–779 [DOI] [PubMed] [Google Scholar]
- 13. Rutenber E., Katzin B. J., Ernst S., Collins E. J., Mlsna D., Ready M. P., Robertus J. D. (1991) Proteins 10, 240–250 [DOI] [PubMed] [Google Scholar]
- 14. Maddaloni M., Cooke C., Wilkinson R., Stout A. V., Eng L., Pincus S. H. (2004) J. Immunol. 172, 6221–6228 [DOI] [PubMed] [Google Scholar]
- 15. Sturm M. B., Schramm V. L. (2009) Anal Chem. 81, 2847–2853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Case D. A., Cheatham T. E., 3rd, Darden T., Gohlke H., Luo R., Merz K. M., Jr., Onufriev A., Simmerling C., Wang B., Woods R. J. (2005) J. Comput. Chem. 26, 1668–1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Thompson J. D., Higgins D. G., Gibson T. J. (1994) Nucleic Acids Res. 22, 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kyte J., Doolittle R. F. (1982) J. Mol. Biol. 157, 105–132 [DOI] [PubMed] [Google Scholar]
- 19. Hopp T. P., Woods K. R. (1981) Proc. Natl. Acad. Sci. U.S.A. 78, 3824–3828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Montfort W., Villafranca J. E., Monzingo A. F., Ernst S. R., Katzin B., Rutenber E., Xuong N. H., Hamlin R., Robertus J. D. (1987) J. Biol. Chem. 262, 5398–5403 [PubMed] [Google Scholar]
- 21. Závodszky P., Kardos J., Svingor, Petsko G. A. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 7406–7411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Daniel R. M., Dunn R. V., Finney J. L., Smith J. C. (2003) Annu. Rev. Biophys. Biomol. Struct. 32, 69–92 [DOI] [PubMed] [Google Scholar]
- 23. Pan Y., Nussinov R. (2010) PLoS Comput. Biol. 6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Karginov A. V., Ding F., Kota P., Dokholyan N. V., Hahn K. M. (2010) Nat. Biotechnol. 28, 743–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kitaoka Y. (1998) Eur. J. Biochem. 257, 255–262 [DOI] [PubMed] [Google Scholar]
- 26. Bowler B. E. (2007) Mol. Biosyst. 3, 88–99 [DOI] [PubMed] [Google Scholar]
- 27. Rose G. D., Fleming P. J., Banavar J. R., Maritan A. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 16623–16633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pace C. N., Shirley B. A., McNutt M., Gajiwala K. (1996) FASEB J. 10, 75–83 [DOI] [PubMed] [Google Scholar]
- 29. Pace C. N. (1995) Methods Enzymol. 259, 538–554 [DOI] [PubMed] [Google Scholar]
- 30. Olsnes S., Fernandez-Puentes C., Carrasco L., Vazquez D. (1975) Eur. J. Biochem. 60, 281–288 [DOI] [PubMed] [Google Scholar]
- 31. Chang Q., Zhong Z., Lees A., Pekna M., Pirofski L. (2002) Infect. Immun. 70, 4977–4986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zolla-Pazner S., Cardozo T. (2010) Nat. Rev. Immunol. 10, 527–535 [DOI] [PMC free article] [PubMed] [Google Scholar]