Abstract
Membrane phosphatidylcholine homeostasis is maintained in part by a sensing device in the key regulatory enzyme, CTP:phosphocholine cytidylyltransferase (CCT). CCT responds to decreases in membrane phosphatidylcholine content by reversible membrane binding and activation. Two prominent isoforms, CCTα and -β2, have nearly identical catalytic domains and very similar membrane binding amphipathic helical (M) domains but have divergent and structurally disordered N-terminal (N) and C-terminal phosphorylation (P) regions. We found that the binding affinity of purified CCTβ2 for anionic membranes was weaker than CCTα by more than an order of magnitude. Using chimeric CCTs, insertion/deletion mutants, and truncated CCTs, we show that the stronger affinity of CCTα can be attributed in large part to the electrostatic membrane binding function of the polybasic nuclear localization signal (NLS) motif, present in the unstructured N-terminal segment of CCTα but lacking in CCTβ2. The membrane partitioning of CCTβ2 in cells enriched with the lipid activator, oleic acid, was also weaker than that of CCTα and was elevated by incorporation of the NLS motif. Thus, the polybasic NLS can function as a secondary membrane binding motif not only in vitro but in the context of cell membranes. A comparison of phosphorylated, dephosphorylated, and region P-truncated forms showed that the in vitro membrane affinity of CCTβ2 is more sensitive than CCTα to phosphorylation status, which antagonizes membrane binding of both isoforms. These data provide a model wherein the primary membrane binding motif, an amphipathic helical domain, works in collaboration with other intrinsically disordered segments that modulate membrane binding strength. The NLS reinforces, whereas the phosphorylated tail antagonizes the attraction of domain M for anionic membranes.
Keywords: Phospholipid Metabolism, Phospholipid Vesicle, Protein Folding, Protein Phosphorylation, Ultracentrifugation, Amphipathic Helix, Intrinsically Disordered, Lipid-Protein Interactions, Nuclear Localization Signal, Polybasic Motif
Introduction
Most regulatory enzymes come in different forms while performing the same enzymatic reaction within the cell. This may provide functional redundancy to safeguard against catastrophic consequences of a mutation in one form. Alternatively, it may provide a means of strict and specialized regulation of cellular metabolism and physiology. The contribution of non-redundant isoforms is abundantly exemplified in the realm of lipid metabolic enzymes, where differential tissue expression and subcellular localization is thought to establish spatially distinct lipid pools for signaling and binding events (1).
CTP:phosphocholine cytidylyltransferase (CCT)2 is the key regulatory enzyme in the biosynthesis of phosphatidylcholine (PC), the most abundant phospholipid component in most eukaryotic biomembranes. CCT responds to variation in the membrane content of PC relative to other phospholipids by reversible membrane binding and subsequent activation, which provides a mechanism for modulating its activity and maintaining PC homeostasis (2). In mammals there are two CCT genes encoding CCTα and CCTβ (3). There are three variations of CCTβ (1–3) as a result of alternative splicing of the gene and an alternate start site (4). Northern analysis of murine and human tissue showed that whereas CCTα is expressed ubiquitously, CCTβ isoforms are expressed very weakly relative to CCTα, with the exception of the adult brain and reproductive organs, and embryonic tissue (3–5). CCTα is essential for cell proliferation and survival (6, 7). However, targeted disruption of the CCTα gene in mouse macrophages causes no deleterious phenotype as CCTβ2 is up-regulated (8). This suggests that CCTβ2 may serve as a redundant, precautionary backup to CCTα. On the other hand, a critical role for CCTβ2 in specific tissues is evidenced by reduced fertility and gonadal defects in mice as a consequence of disruption of the CCTβ gene (9). As well, CCTβ2 is specifically up-regulated and activated in growing neuronal cells in culture in response to neuronal growth factor (10) and plays a critical role in neurite outgrowth and branching (11). Lastly, although CCTα is typically localized in cell nuclei, CCTβ2 is confined to the cytoplasm (5). These results suggest that CCTα and CCTβ2 may perform specialized functions in some cells.
Of the β isoforms, CCTβ2 most closely resembles CCTα, with a very similar domain structure (Fig. 1A): the N-terminal segment (region N), the catalytic domain (domain C), the membrane binding domain (domain M), and the C-terminal phosphorylation segment (region P). The structure, regulation, and membrane binding properties of CCTα have been well studied, but these areas remain relatively unexplored for CCTβ. Domain C and domain M are well conserved between these CCT isoforms, both having 98% sequence similarity (Fig. 1A). In contrast, the amino acid sequences of the N and P regions differ, having only 54 and 75% sequence similarity, respectively. How these divergent regions affect the function of the two CCT isoforms was unknown. We hypothesize that CCTα and -β2 have evolved similar but distinct modes of regulation to allow for varied control of PC synthesis in different subcellular localizations and cell types.
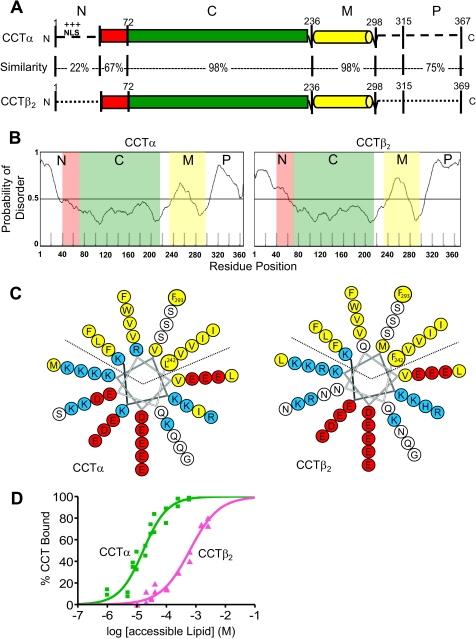
FIGURE 1.
CCTα and -β2 have highly similar C and M domains but divergent N and P domains and different membrane affinities. A, domain structure and sequence conservation are shown. Rat CCTα and -β2 sequence alignment and percent similarity were calculated using ClustalW. Domain boundaries are approximate. Structured regions shown as rectangles or cylinders and disordered regions as dashed lines. B, prediction of disordered/ordered segments using the server RONN (79). Outputs from other prediction programs appear in supplemental Fig. S2. C, domain M comparison is shown. Amphipathic α-helical M domains of the two CCT isoforms (residues 242–293) are represented as 11/3 helical wheel diagrams (80). D, binding analysis of His-CCTα and His-CCTβ2 to SLVs composed of PC/PG (3:2) at 20 °C is shown. Data were compiled from two independent experiments and were fit using GraphPad Prism 4 to the equation % Bound = 100Kp [L]/(1 + Kp[L]) (42), where [L] is the concentration of accessible lipid (½ of total lipid). Top and bottom values were constrained to 100 and 0%, respectively.
The N region of CCTα contains a nuclear localization signal (NLS) sequence at residues 12–16 (RKRRK), and this isoform is found in the nucleus in many (5, 12, 13) but not all cells (14–17). The N region of CCTβ2 lacks this sequence and is expressed in the cytoplasm, where it binds the endoplasmic reticulum (5, 11). The NLS is necessary and sufficient for nuclear localization of CCTα, as shown by deletion and transfer mutagenesis (18) and the impedance of nuclear import when the NLS is masked by proximal mono-ubiquitination (19). The recently solved structure of the first 236 residues of CCTα revealed a catalytic domain fold with strong structural homology to the CTP:glycerol-3-phosphate cytidylyltransferase family (20). It also showed that residues 40–72 of the N-terminal region are in fact part of the catalytic domain fold (20). This portion of CCT is partially conserved (67% similar) between the CCTα and -β2 isoforms.
Domain M functions as a negative regulator of CCTα activity when CCTα is in its soluble form (21). Membrane binding serves to alleviate that inhibition. The association of domain M with membranes is governed by electrostatic interactions, hydrophobic interactions, and bilayer curvature strain, which are dependent on the lipid content of the membrane (22–25). When membranes are enriched in anionic lipids or lipids that promote negative curvature strain, domain M converts into a long amphipathic helix and intercalates into one leaflet of the membrane bilayer (26–28). The structure of domain M when CCTα is soluble is unknown, but circular dichroism data suggest a mix of conformers (29). CCTβ1 also showed evidence of activity dependent on anionic lipids (3).
In addition to its regulation by membrane composition, CCTα activity is also regulated by reversible phosphorylation, which antagonizes membrane binding and activation (23, 30–33). CCTα is phosphorylated on up to 16 serine residues in region P (34, 35), which are in a Pro-rich milieu. CCTβ2 is a phosphoprotein (5), but the extent of phosphorylation and its affect on membrane binding had not been investigated when we began this study. Rat CCTβ2 has 18 serine residues as well as 3 threonine residues in the Pro-rich P region, all of which may be phosphorylated.
CCTα functions as a homodimer (36) and can cross-bridge two anionic lipid vesicles in vitro (37, 38). Although we originally proposed that tethering is mediated by each domain M engaging a separate vesicle (38), we later found that a CCT heterodimer containing only one domain M tethered anionic lipid vesicles just as well as the wild type CCTα homodimer (37). This suggested that another membrane binding motif pairs with the M domain in CCTα. Deletion of the polybasic NLS (12RKRRK16) from the CCTα dimer abolished vesicle cross-bridging (37); thus, a secondary role for the NLS as a membrane tether was proposed.
In the present study we found that the binding affinity of purified CCTβ2 for anionic vesicles is more than an order of magnitude weaker than that of CCTα. This was surprising because the sequences of the membrane binding amphipathic helical domain (M) are 98% similar. Mutagenesis revealed that the differential membrane affinities can be attributed to the NLS sequence acting as a secondary membrane binding motif in the α isoform. In vitro dephosphorylation and region P truncation of CCTs showed a stronger modulating influence of phosphorylation on the membrane affinity of CCTβ2, presumably due to the lack of a secondary membrane binding motif, the NLS. Importantly, these same trends were confirmed in the context of cellular membranes. These studies show that membrane binding of CCT isoforms via the amphipathic helix domain can be modulated by other regions and highlight the functional importance of CCT intrinsically disordered regions.
EXPERIMENTAL PROCEDURES
Construction of CCT Isoform Variant cDNAs
The methods for construction of plasmids containing His-tagged and untagged CCT isoform cDNAs are found in the supplemental material.
Expression and Purification of CCTs
COS-1 cells were transiently transfected as described (37) using 20 μg of plasmid DNA per 15-cm dish. The duration of transfection was 64 h, with the exception of His-CCTα-βN and His-CCTα ΔNLS. These were transfected for 48 h to limit aggregation due to overexpression. Cells were harvested and homogenized, and His-tagged proteins were purified as described (36, 37). The purification incorporated two steps with ∼10,000 molar excess Triton to eliminate any lipid bound to the CCT, and the effectiveness of this step is manifest in the low specific activity obtained in the absence of lipid (supplemental Table 2). The CCTs eluted from the nickel resin were dialyzed against 10 mm Tris, pH 7.4, 100 mm NaCl, 0.25 mm Triton X-100, and 2 mm DTT and stored at −80 °C. The His-CCTα312 obtained in the COS cell lysate was insoluble and was, therefore, denatured and re-folded before purification. The cell homogenate (in 20 mm K2HPO4, 5 mm NaH2PO4, pH 8.0, 1% Nonidet P-40, 500 mm NaCl, and 15 mm imidazole) was centrifuged at 15,000 × g for 10 min at 4 °C. The protein pellet containing His-CCTα312 was dissolved in 6 m guanidine hydrochloride and centrifuged at 15,000 × g for 15 min at 4 °C. The supernatant was then dialyzed at 4 °C in three stages: (i) 4 h against 100 mm NaCl, 20 mm NaH2PO4, pH 7.4, 0.25 mm Triton X-100, 2 mm DTT, and 3 m guanidine hydrochloride (GuHCl), (ii) 3 h against the same buffer but with 1.5 m GuHCl, and (iii) overnight against the same buffer without GuHCl. The dialyzed protein was centrifuged at 15,000 × g for 30 min at 4 °C, and the His-CCTα312 in the supernatant was purified as described above. The His tag was left intact where indicated but was cleaved with tobacco etch virus (37) for most experiments. Protein concentration was determined (39) using ovalbumin as a standard.
CCT Activity Assay
Purified CCTs were assayed for enzymatic activity as described previously (40). Briefly, 0.1 μg of purified protein was assayed in the presence of 8 mm CTP, 88 mm NaCl, 12 mm MgCl2, 20 mm Tris, pH 7.4, 10 mm DTT, and 1 mm [14C]phosphocholine (Amersham Biosciences; specific activity, 1 mCi/mmol). Activating vesicles in the form of SUVs (22) composed of egg PC/PG (3:2) were added to a final concentration of 0.2 mm. For the enzyme kinetic analysis, the phosphocholine dependence (0–5 mm) was determined in the presence of 16 mm CTP, and the CTP dependence (0–20 mm) was determined in the presence of 1.5 mm phosphocholine. Data analysis of primary plots (velocity versus concentration) using Prism 4 provided Km and Vmax values. In experiments comparing the activation of CCTα and -β2, the maximum activities in the presence of PC/PG (3/2) LUVs were extrapolated from plots of specific activity versus lipid concentration using GraphPad Prism 4. This apparent Vmax was used to normalize the activity of each CCT isoform. The normalized activation data were used to determine values for L at ½ maximal activation (EC50) using GraphPad Prism 4.
Chemical Cross-linking with BS3 or Copper Phenanthroline
His-tagged proteins (0.4 μm final) in 20 mm K2HPO4, pH 7.4, 100 mm NaCl, 0.15 mm Triton X-100, 2 mm DTT were equilibrated for 3 min in a 37 °C shaking water bath in the absence or presence of 2 mm PG sonicated vesicles. Bis(sulfosuccinimidyl)suberate (BS3) was added to 1 mm, and the samples were incubated for a further 20 min. The reaction was stopped by the addition of glycine (0.1 m final). In prequenched samples, glycine was added before the addition of BS3. Samples were then analyzed by SDS-PAGE and silver stain (41). For copper phenanthroline reactions His-tagged CCTs were first reduced with 1 mm DTT for 10 min in a 37 °C shaking water bath. Reduced His-CCTs were placed on ice and were diluted to a protein concentration of 0.34 μm and a DTT concentration of 60 μm with 20 mm K2HPO4, pH 7.4, 100 mm NaCl in the presence of 0–2 mm PG-sonicated vesicles. Samples were equilibrated in a 37 °C shaking water bath for 3 min. Stocks of CuSO4 and phenanthroline were mixed and added to initiate the reaction (final concentration of 0.4 mm CuSO4 and 1.2 mm phenanthroline). After 10 min of shaking at 37 °C, samples were analyzed by non-reducing SDS-PAGE and silver stain.
Vesicle Aggregation Assay
CCT-induced aggregation of anionic lipid vesicles was monitored by measuring the apparent absorbance at 400 nm (38). Varying amounts of CCT (0 to ∼900 nm) were added to sonicated PG vesicles (0.1 mm PG SUVs, 10 mm Tris, pH 7.4, 2 mm DTT, 130 mm NaCl, 0.65 mm EDTA). Triton X-100 was present in the purified protein stocks and was, therefore, present in the reaction mixture at a concentration of 0.05–0.08 mm, which was kept constant in all samples. Upon the addition of CCT to the mixture, the increase in absorbance at 400 nm was recorded every 30 s for 3 min. A clear plateau value was reached during this time, and this value was plotted as a function of dimeric CCT concentration.
In Vitro Dephosphorylation
CCTs (7.5 μm) were dephosphorylated by the catalytic subunit of protein phosphatase 1 in the presence of 0.2 mm MnCl2, 10 mm Tris, pH 7.4, 100 mm NaCl, 2 mm DTT, and 0.25 mm Triton X-100. EDTA was added to 2 mm and K2HPO4 to 20 mm, and MnCl2 was omitted in reactions where protein phosphatase 1 catalytic subunit was prequenched. Protein phosphatase 1 catalytic subunit was prepared according the manufacturer's instructions, and 2 units/μg CCT were used. Samples were incubated for 45 min in a 37 °C shaking water bath. The reactions were not quenched at the end but were used immediately in membrane binding assays or stored at −80 °C for later use. The phosphatase was not removed from the untagged CCT samples before vesicle binding analysis; thus, as a control we examined the binding of CCTα and CCTα with prequenched phosphatase. The binding curves were not statistically different (p = 0.115; supplemental Fig. S1); thus, the presence of the phosphatase did not interfere in these binding assays. Dephosphorylation was confirmed by monitoring a band shift via SDS-PAGE and silver stain as well as by whole protein MALDI mass spectrometry using an Applied Biosystems Voyager-DE STR (Framingham, MA) mass spectrometer. LC/MS was used to investigate the phosphorylation status of CCTβ313 using an Applied Biosystems MDS-SCIEX API QSTAR Pulsar (Sciex, Thornhill, ON).
Sucrose-loaded Vesicle Binding Assay
Binding of various CCT constructs was measured and analyzed as described (37, 42) with a few minor modifications. We used 0.5 μm CCT and varying amounts of PC/PG (3:2) sucrose-loaded vesicles (SLVs). The protein-SLV mixtures were sedimented at 100,000 × g for 30 min at 20 °C. The intensity of the pellet band was corrected for contamination by supernatant as well as protein that sedimented in the absence of SLVs (37). The latter was 27% ± 6 (mean ± S.D. for all constructs; n = 42). One outlier was His-CCTα-βN, where lipid-independent sedimentation was ∼40%; therefore, this construct was precentrifuged at 100,000 × g for 30 min at 20 °C, and the supernatant was used in binding assays.
Cell Membrane Partitioning Assay
CCT partitioning into cell membranes enriched with oleic acid was performed as described (28). Cells were transiently transfected with 5 μg of plasmid DNA per 10-cm dish for various durations to obtain similar expression levels (∼100 units CCT activity/mg of lysate protein): CCTα, 20 h; CCTα312, CCTα ΔNLS, and CCTα312 ΔNLS, 24 h; CCTβ+NLS and CCTβ313+NLS, 30 h; CCTβ2 and CCTβ313, 36 h. Medium containing 1 mm sodium oleate and 0.25–20 mg/ml fatty acid-free bovine serum albumin (BSA) (molar ratios OA:BSA from 266 to 3.3) was used to enrich cell membranes. Cells were harvested in 1.5 ml of hypotonic buffer (10 mm Tris, pH 7.4, 1 mm EDTA, 2 mm PMSF, 2 mm DTT) and lysed by sonication 2 × 30 s on ice. NaCl was added to 100 mm, and the samples were centrifuged at 100,000 × g for 1 h at 4 °C. The supernatant was removed (cytosol), and the pellet was resuspended by sonication in 1.5 ml of 10 mm Tris, pH 7.4, 100 mm NaCl, 1 mm EDTA, 2 mm PMSF, 2 mm DTT, and 1% Triton X-100. The sample was centrifuged again to separate the membrane and Triton-insoluble (particulate) fractions. The units of CCT activity of each fraction were determined as described above in the presence of saturating levels of the activating lipid (250 μm sonicated PG vesicles). CCT units in each fraction were directly proportional to the quantity of cell sample up to ≥6 μl; 6 μl were assayed. Thus, the mass of CCT in each fraction is faithfully and accurately represented by the units of enzyme activity using conditions that are limited only by the amount of CCT in the cell fraction. This protocol routinely yielded ≤20% of the total WT CCTα or β2 in the membrane fraction of untreated cells. The percent in the Triton-insoluble particulate fraction varied somewhat among CCT constructs. It ranged from a low value of ∼1% for CCTβ2, CCTβ313, and CCTα312ΔNLS to a high value of 5–7% for CCTα312 and CCTβ313+NLS.
Immunoblot of Cell Fractions
Equivalent proportions of soluble and membrane fractions from COS cells expressing various CCT constructs and variably treated with oleic acid were electrophoresed, transferred, and blotted with a 1/10,000 dilution of an antibody against the conserved catalytic domain (36). This antibody was the generous gift of Dr. Rama Mallampalli, and it reacts similarly with both CCTα and CCTβ. Immunoreactivity was assessed via chemiluminescence and film.
Statistical Analysis
We used the f-test in Prism 4 to compare the statistical differences between models imposed on data sets that appeared to be similar. The significance threshold, α, was set to 0.05. p values are reported in the results only for data sets that are not obviously different. The error reported for membrane partition coefficients (Kp) is the 95% confidence interval with respect to the best fit.
RESULTS
The N- and C-terminal Regions of CCTα and -β2 Are Intrinsically Disordered
The first and last 40–50 residues of CCTα and -β2 are predicted to be intrinsically disordered regions by multiple algorithms (Fig. 1B; supplemental Fig. S2). In support of this prediction the N-terminal 39 residues of CCTα (residues 1- 236) were unresolved in the crystal (20). As well, both this region and the entire P region are highly susceptible to proteolysis (34, 43), suggesting flexibility. Domain M in both isoforms is predicted to house both ordered and disordered subregions (Fig. 1B; supplemental Fig. S2).
CCTα and CCTβ2 Have Different Anionic Membrane Binding Affinities in Vitro
As a first step in determining how the divergent regions among CCT isoforms may influence membrane binding and subsequent activation, we characterized the lipid vesicle binding of CCTβ2 and CCTα in parallel. His-tagged proteins were purified, and their binding to SLVs composed of PC and phosphatidylglycerol (PG), a molar ratio of 3/2, was measured. We found that His-CCTβ2 had a significantly weaker membrane binding affinity than His-CCTα, requiring more than an order of magnitude higher lipid concentration to achieve the same percent binding (Fig. 1D). Partition coefficients (Kp) were calculated from these curves (Table 1). The Kp for His-CCTα was ∼40-fold higher than that of His-CCTβ2.
TABLE 1.
Partition coefficients and phosphorylation state for CCT constructs
The average number of phosphate groups on full-length CCT isoforms and constructs and CCTβ313 was determined by mass spectrometry of whole proteins. Partition coefficients were calculated from the binding curves in Fig. 3 where Kp = 1/[accessible lipid] when protein is 50% bound (37, 42). The error reported is the ±95% confidence interval with respect to the best fit Kp value. ND, not determined and/or no published data.
| Set | Construct | Phosphorylation state | Partition coefficient | r2 Value for curve fit |
|---|---|---|---|---|
| ×103, m−1 | ||||
| 1 | His-CCTα | ND | 60 ± 13 | 0.94 |
| His-CCTβ2 | ND | 1.6 ± 0.3 | 0.97 | |
| His-CCTα-βN | ND | 3.0 ± 0.5 | 0.98 | |
| His-CCTβ-αN | ND | 10 ± 2 | 0.97 | |
| 2 | CCTα | 6 | 24 ± 6 | 0.95 |
| CCTβ2 | 7 | 1.1 ± 0.3 | 0.94 | |
| CCTα ΔNLS | 2 | 3.2 ± 0.9 | 0.90 | |
| CCTβ+NLS | 14 | 2.8 ± 0.8 | 0.87 | |
| 3 | Dephos-CCTα | 0 | 43 ± 5 | 0.97 |
| Dephos-CCTβ2 | 0 | 7 ± 2 | 0.91 | |
| Dephos-CCTα ΔNLS | 0 | 15 ± 3 | 0.93 | |
| Dephos-CCTβ+NLS | 0 | 87 ± 25 | 0.92 | |
| 4 | CCTα312 | 0a | 133 ± 17 | 0.97 |
| CCTβ313 | 0 | 12 ± 3 | 0.90 | |
| CCTα312 ΔNLS | ND | 12 ± 3 | 0.94 |
a Phosphorylation status of rat CCTα312 is based on previous analyses (47).
What could account for this difference in binding affinity for anionic membranes? NMR and CD analyses indicated that residues 242–293 contribute to the amphipathic helix responsible for membrane binding of CCTα (29, 44). This region of CCTα and CCTβ2 has several amino acid substitutions that are semiconservative but only one non-conservative change when analyzed by ClustalW (Ile-272 in CCTα to His-272 in CCTβ2; Fig. 1C), and this occurs outside the non-polar face of the helix. Both isoforms have 0 net charge over residues 242–293. The peak hydrophobic moment and total hydropathies of the non-polar and polar faces of this region of domain M are also very similar between CCT isoforms (supplemental Table 1). The subtle differences in M domains seemed unlikely to account for the significantly different lipid response between CCT isoforms. This prediction was borne out by comparative membrane binding analyses of purified constructs containing only the M domain plus unphosphorylated P region of the two isoforms. We monitored the induction of α-helical content by PC/PG (3:2)-sonicated vesicles (supplemental Fig. S3). Lipid-induced helix acquisition in domain M coincides with other measures of membrane binding (28). There was no significant difference in the Kp values calculated from these curves (Kp for CCTβ2 = 11,200 ± 1,300 versus 12,200 ± 650 m−1 for CCTα).
We hypothesized that the divergent and disordered regions N and P influence the membrane binding and subsequent activation of CCT isoforms. Region N is the most divergent segment between CCT isoforms and analysis of overexpressed constructs in crude cell lysates suggested that region N of CCTβ1 negatively influences catalytic activity (3). We, therefore, focused our initial attention on the role of region N in the lipid response of CCT isoforms by creating region N-swapped chimeric enzymes.
CCTβ2 Is a Dimer, and Region N Participates in Forming the Dimer Interface
Before creating CCTα/CCTβ chimeras, we needed to ensure that the N regions of CCTα and CCTβ2 perform analogous structural roles. If so, this would alleviate concerns about folding disruption in the chimeric enzymes. Region N of CCTα participates in forming the dimer interface and interacts intimately with domain C as revealed in the solved structure of CCTα (residues 1–236 (20) and by chemical cross-linking (36).
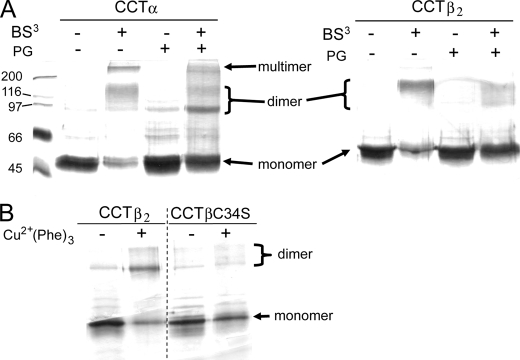
We determined the quaternary interactions of CCTβ2 using the chemical cross-linker BS3. We found that, like CCTα (36), a homodimeric ∼90-kDa species of CCTβ2 is trapped in the presence of the cross-linker (Fig. 2A). The addition of activating lipids such as PG SUVs reduced the amount of dimer trapped. This feature is shared with CCTα (Fig. 2A and Ref. 36) and suggests that upon membrane binding a conformational change occurs that results in an alteration of the dimer interface that abrogates cross-linking. These experiments confirm that CCTβ2 is a dimer and that the dimer interface rearranges upon membrane binding, like that of CCTα, but did not determine whether region N of CCTβ2, like that of CCTα, participates in forming the dimer interface.
FIGURE 2.
Both CCTβ2 and -α are dimers with intersubunit contacts involving region N. A, BS3 reactions are shown. Purified His-tagged CCTα or β2 (0.4 μm) was reacted with BS3 in the presence or absence of 2 mm PG SUVs. Reactions were prequenched (−) or quenched after 20 min at 37 °C (+). B, copper phenanthroline reactions are shown. Purified His-tagged CCTβ2 or CCTβ2 C34S (0.34 μm) were either added to 0.4 mm CuSO4 and 1.2 mm phenanthroline (+) or untreated (−). The samples were electrophoresed on 10% polyacrylamide gels and stained with silver.
CCTα has a pair of cysteines located in region N that are in the region that forms the dimer interface. Although there is no evidence that these cysteines form a disulfide bond under physiological conditions, a disulfide bond can be formed when CCTα is oxidized in vitro with copper phenanthroline (Cu(Phe)3), thereby trapping CCTα as a covalently linked dimer (36). Mutation of Cys-37 in CCTα to serine severely reduced disulfide-trapped dimers (36). The CCTβ2 dimer has a pair of cysteines at position 34. To test whether this pair is homologous to the CCTα Cys-37 pair, we mutated Cys-34 to serine. This switch did not affect activity or lipid dependence (supplemental Table 2). The Cu(Phe)3-treated His-CCTβ C34S did not show an increase in disulfide-trapped dimers after oxidation (Fig. 2B), similar to that of CCTα C37S (36). These results indicate very similar roles for region N in the quaternary structure of the two CCT isoforms.
Region N Distinguishes the Membrane Binding Affinity of CCT Isoforms in Vitro
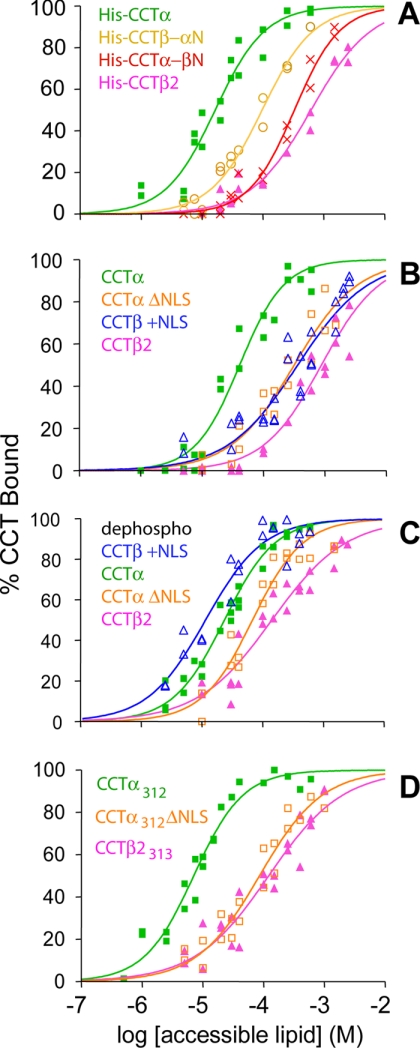
Having demonstrated analogous roles of region N in the quaternary structure of CCTα and -β2, we constructed region N chimeras, swapping the first 83 residues of the two CCT isoforms. The purified region N swapped mutants were active and lipid-dependent (supplemental Table 2). Binding of these constructs to PG/PC (2:3) SLVs was measured, and partition coefficients (Kp) were calculated. Exchange of region N of CCTα with that of CCTβ2 (His-CCTα-βN) resulted in a right shift of the binding curve (Fig. 3A), translating to a 20-fold reduction in Kp (Table 1). The reciprocal mutation of CCTβ2 (His-CCTβ-αN) resulted in a left shift of the binding curve (Fig. 3A), translating to a >6-fold increase in Kp value (Table 1).
FIGURE 3.
The NLS is responsible for the differential membrane binding affinity of CCT isoforms. Binding analysis to SLVs composed of PC/PG (3:2) at 20 °C. A, His-tagged CCT isoforms and region N-swapped chimeras are shown. B, untagged CCT isoforms and NLS insertion/deletion mutants are shown. C, in vitro dephosphorylated, untagged CCT isoforms, and NLS insertion/deletion mutants are shown. D, region P truncation mutants are shown. Data were compiled from at least two independent experiments and were fit using GraphPad Prism 4 to the equation given in the legend to Fig. 1D. Top and bottom values were constrained to 100 and 0%, respectively.
The Polybasic NLS Distinguishes the Membrane Binding Affinity of CCT Isoforms in Vitro
We have shown that the NLS sequence in region N of CCTα cooperates with the M domain to facilitate cross-bridging (tethering) of anionic lipid vesicles, with domain M serving as one membrane anchor and the NLS as the other (37). We hypothesized that the NLS serves to elevate the membrane binding affinity of CCTα and, as it is absent in CCTβ2, may explain the large difference in lipid response between CCT isoforms.
To probe this idea, a CCTα mutant lacking the NLS (CCTα ΔNLS) and a CCTβ2 mutant with the NLS sequence from CCTα (CCTβ+NLS) were created. The exact sequences of their N termini are provided in the supplemental Methods. The polyhistidine tag used in purifying CCT can contribute to membrane binding when paired with the polybasic NLS (37). The His tag was, therefore, cleaved. CCT constructs were active and lipid-dependent (supplemental Table 2). The binding affinity for PC/PG (3:2) SLVs of untagged CCTαΔNLS and CCTβ+NLS compared with untagged CCTα and CCTβ2 was determined. Deletion of the NLS reduced the binding affinity of CCTα (Fig. 3B), and the resultant Kp was similar to that of the chimeric His-CCTα-βN (Table 1). The addition of the NLS to CCTβ2 increased its binding affinity (Fig. 3B) as reflected by an ∼3-fold increase in Kp (Table 1).
Membrane Binding of the NLS Contributes to CCTα Activation by Highly Anionic Vesicles
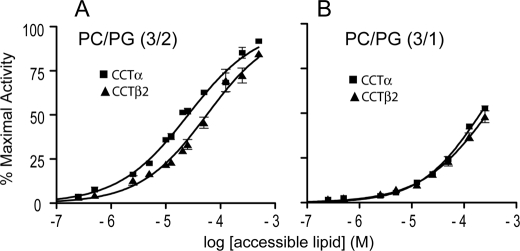
We explored whether the additional binding strength afforded by the NLS motif would translate into more efficient lipid activation of CCTα versus CCTβ2. Before comparing the anionic lipid requirement for activation of the two isoforms, we determined the substrate Km values so that the analyses would use conditions saturating for substrate. The Km values for CTP were approximately the same for the two CCTs (CCTα = 1.3 ± 0.4 mm, CCTβ2 = 0.9 ± 0.4 mm), and the Km values for phosphocholine were 0.4 ± 0.06 mm for CCTα and 0.25 ± 0.05 mm for CCTβ2. These values are in the range of those reported elsewhere for CCTα (21, 45, 46). The maximal activity of CCT β2 is lower than CCTα (10.1 ± 1.4 versus 14.6 ± 1.1 μmol/min/mg in the plots shown in Fig. 4; see also supplemental Table 2). Fig. 4 plots enzyme activation by lipids normalized to the maximal activity of each isoform. In the presence of 40 mol % PG LUVs, there was a 2.2-fold stronger response to the lipids on the part of CCTα. EC50 values extracted from the curves in Fig. 4A were 2.6 ± 0.3 for CCTα and 5.6 ± 0.9 for CCTβ2; p < 0.0001. Previous work showed that NLS binding to membranes requires in excess of 25 mol % anionic lipid (38). In keeping with this, the lipid activation of CCTα and -β2 were nearly coincident when we used LUVs containing 25 mol % PG (Fig. 4B), reflecting a dependence solely on their highly similar M domains for activation. Thus, when the lipid vesicles are strongly anionic, the binding reinforcement via the NLS motif impacts on the lipid activation of CCTα.
FIGURE 4.
CCTβ2 has a weaker anionic lipid activation response than CCTα. The specific activities (nmol of CDP-choline/min/μg of CCT) of purified untagged CCT isoforms were measured in parallel as a function of increasing concentrations of the indicated LUVs. The data were normalized to the maximal activities obtained in the presence of 3:2 PC/PG LUVs, (CCTα, 14.6 ± 1.1 nmol/min/μg; CCTβ2, 10.1 ± 1.4 nmol/min/μg). Data represent the mean ± S.D. of four independent determinations. ■, CCTα; ▴, CCTβ2.
The NLS Distinguishes the Vesicle Tethering Activity of CCT Isoforms
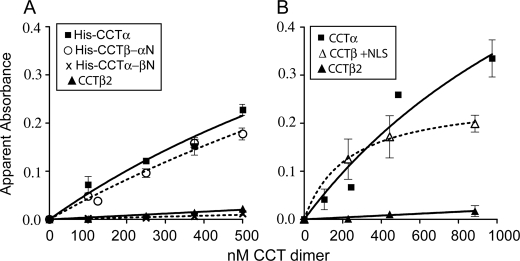
We hypothesized that because CCTβ2 contains no NLS motif, it would lack the tethering activity that has been shown for CCTα (37). We compared the anionic vesicle tethering activity of CCT constructs by monitoring the increased turbidity due to aggregated anionic lipid vesicles (37, 38). Fig. 5A shows that CCTβ2 is completely deficient in vesicle tethering function. Swapping the N regions between the α and β2 isoforms results in loss of function from CCTα and gain of function in CCTβ2. Fig. 5B shows that the CCTβ2 construct carrying the NLS causes a gain-of-function, as it is nearly as effective as CCTα. These data confirm that the NLS can function as a membrane binding motif and demonstrate that the NLS accounts for the difference in tethering activity between CCTα and -β2 isoforms.
FIGURE 5.
CCTβ2 cannot tether lipid vesicles because it lacks a NLS. The increase in apparent absorbance (400 nm) of PG SUVs due to the addition of CCT was monitored for 3 min. The apparent absorbance due to vesicles alone was subtracted from the plateau value of each CCT concentration. The data represent the means ± S.E. or range for at least two independent determinations. A, His-tagged CCT isoforms and region N-swapped chimeras are shown. B, untagged CCTβ2 and CCTβ+NLS are shown. Untagged CCTα was included as a positive control.
Dephosphorylation of CCTs Amplifies the Effect of the NLS on Membrane Binding
The NLS, acting in its traditional capacity, targets proteins to the nucleus. Deletion of the NLS from CCTα and the addition of the NLS to CCTβ2 would presumably result in their mislocalization during expression in COS-1 cells. As both isoforms are phosphoproteins, these mislocalized proteins could have alternative phosphorylation states as compared with their wild type counterparts, which could influence Kp and potentially mask the true contribution of the NLS to membrane binding affinity.
To clarify the contribution of the NLS to membrane binding affinity without the complication of variable and undefined phosphorylation states, purified CCT constructs were dephosphorylated in vitro. CCT constructs were incubated with the catalytic subunit of protein phosphatase I, which has been successfully used to dephosphorylate CCTα (23). Dephosphorylation of CCTs was confirmed by mass spectrometry of whole proteins and by monitoring the resultant band shift on SDS-PAGE (supplemental Fig. S4). Mass spectrometry analysis gave an average number of six phosphates for CCTα, in agreement with previously published work (34, 35), whereas CCTβ2 had seven. CCTα ΔNLS had an average of 2 phosphate groups, whereas CCTβ+NLS had a remarkable 14. This high degree of phosphorylation could explain why the addition of the NLS to CCTβ2 was associated with only a 3-fold increase in membrane binding affinity.
Binding of in vitro dephosphorylated CCTα, CCTβ2, CCTα ΔNLS, and CCTβ+NLS to PC/PG (3:2) SLVs was measured, and Kp values were calculated from the binding curves. Differences in membrane binding affinities between dephosphorylated CCT isoforms are clearly evident (Fig. 3C; Table 1, compare Set 2 with Set 3). The Kp value of dephosphorylated CCTα is 6-fold higher than that of dephosphorylated CCTβ2. Deletion of the NLS from CCTα reduced binding affinity ∼3-fold, and the addition of the NLS to CCTβ2 increased binding affinity more than 10-fold (Table 1). Thus, when phosphorylation was eliminated as a variable, the effect of the NLS on membrane binding affinity became clearer. Moreover, we observed that for constructs lacking an NLS (CCTβ2 and CCTα ΔNLS), membrane affinity was more profoundly affected by phosphorylation status. For example, removal of 6 phosphate groups from CCTα increased its Kp value just 2-fold, whereas removal of 7 phosphates from CCTβ2 resulted in a 7-fold increase in Kp value, and loss of only 2 phosphate groups from CCTα ΔNLS resulted in a 5-fold increase in Kp (Table 1, compare Sets 2 and 3). These data suggest an antagonism between membrane attraction by the NLS and membrane repulsion by the phosphorylated P region.
Region P Antagonizes Membrane Binding via Its Phosphorylation Status
Can region P also influence the affinity of domain M for membranes via a mechanism unrelated to its phosphorylation status? To answer this question we prepared CCTs missing the entire region P. ProfSec secondary structure predictions suggested that region P deletion would not affect the integrity of domain M. Furthermore, this deletion in CCTα does not affect its activity (47–49); supplemental Table S2). It is known that phosphorylation is restricted to residues 315–367 in CCTα, but the sites of phosphorylation in CCTβ2 were unknown. To determine whether phosphorylation is restricted to the C-terminal 55 residues in CCTβ2, we compared the mass of purified CCTβ313 before and after dephosphorylation with PP1α using conditions that resulted in loss of seven phosphates in full-length CCTβ2. Unlike full-length CCTβ2, phosphatase treatment produced no band shift on gels (supplemental Fig. S4) and no change in mass as determined by mass spectrometry (untreated, 38,334 Da; phosphatase-treated, 38,334 Da; which agrees with the theoretical mass within an error of 1 Da). Thus, residues 314–369 constitute the phosphorylation region of CCTβ2.
We measured the binding to PC/PG (3:2) SLVs of the CCTα, CCTα ΔNLS, CCTβ2, and CCTβ+NLS constructs truncated before region P (Fig. 3D). For CCTβ313 and CCTα312 ΔNLS there was very little difference in the Kp values when comparing dephosphorylated full-length versus region P-truncated constructs (Table 1; Set 3 versus Set 4), in support of the notion that region P modulates membrane binding solely via its phosphorylation status. As well, there was no significant difference in the Kp values of these two region P truncated constructs (p > 0.05). CCTβ313+NLS, although active, formed protein aggregates, leading to sedimentation of ∼50% of the protein in the absence of lipid. The partitioning of the soluble component of this construct into the vesicles, determined after subtracting the insoluble component, yielded a Kp value of 71 ± 17 × 103 m−1, not significantly different from that of the dephosphorylated CCTβ2+NLS construct. However, given its solubility problems, we are hesitant to make conclusions based on results with this construct. The partitioning of CCTα312 was ∼3 times higher than that of dephosphorylated full-length CCTα. This suggests a negative contribution of region P to binding, i.e. region P stabilizes the soluble form of the enzyme especially when the NLS is present. In keeping with this idea, like CCTβ313+NLS, the CCTα312 construct was more prone to aggregation and losses during purification than the other constructs. Most importantly, the role of the NLS is clearly revealed in the region P truncated CCTs (Table 1, Set 4).
In summary, the in vitro analyses indicate that CCTβ2 has a weaker binding response to lipids than CCTα and that although differential phosphorylation among isoforms does influence membrane binding affinity, the NLS is largely responsible for this difference. Next we sought to validate these in vitro findings in the context of cellular membranes. Can the NLS function as a secondary membrane binding motif in cells to distinguish the membrane partitioning of CCTα and -β2?
CCTβ2 Has a Weaker Membrane Binding Affinity in Cells Because of the Absence of the NLS
CCTα translocation to COS cell membranes can be promoted by enrichment of membranes with the anionic lipid, OA, which can be delivered exogenously from a complex with BSA (28, 32, 50). This method was used to determine the relative binding affinity of CCTβ2, CCTβ+NLS, and CCTα ΔNLS for OA-enriched cell membranes compared with CCTα. These constructs were expressed in COS cells to approximately equivalent levels, as assessed by activity (40–50-fold above endogenous levels). The membrane partitioning was analyzed as a function of the OA/BSA molar ratio, which we previously showed results in an increase in fatty acid content up to ∼25 mol % of total phospholipids (28). Our protocol to analyze membrane bound versus soluble CCT was designed so that ≤20% of wild type CCTα would appear in the membrane fraction in untreated cells. The ratio of membrane-bound/free CCT is a function of the volume of lysis buffer, which was constant in these analyses. This ratio does not represent how much CCT is membrane-bound in the intact cell but enables measurement of the changes in membrane partitioning due to OA enrichment and a comparison of the responses of the different CCT constructs.
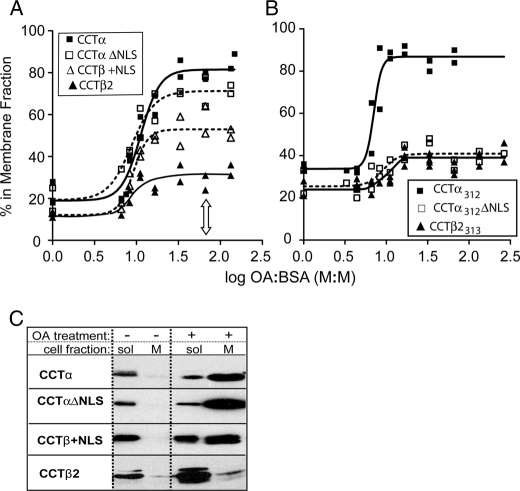
CCTβ2 displayed weaker membrane partitioning than CCTα in both untreated and OA-enriched cells (Fig. 6A). Induction of partitioning into the membrane fraction occurred at approximately the same OA/BSA ratio for CCTα, β2, and the NLS insertion/deletion mutants. This suggests that they have similar negative charge-sensing mechanisms. On the other hand, the partitioning at saturating OA contents varied among the four CCT constructs. The ratio of bound/free CCTβ2 in OA-saturating conditions was ∼10-fold lower than that of CCTα (Table 2), and the addition of the NLS to CCTβ2 increased this ratio from 0.43 to 1.2. These results are consistent with the in vitro membrane binding data. However, deletion of the NLS from CCTα appeared to have only a small (<2-fold) but significant reduction (p < 0.05) in the membrane partitioning (Fig. 6A). In the analysis shown in Fig. 6A, we monitored units of CCT enzyme activity in each cell fraction using assay conditions limiting only for CCT protein (substrates and lipid activating vesicles were saturating). As a complementary approach, we assessed CCT mass by immunoblot of cell fractions from untreated cells or cells treated with a saturating concentration of OA (Fig. 6C). The results parallel the activity analysis, showing dramatic differences in the soluble versus membrane distribution of CCTα and -β2 constructs in OA-treated cells and a large increase in membrane partitioning upon the addition of the NLS sequence to CCTβ2 (CCTβ+NLS) but minimal effect of NLS deletion from CCTα.
FIGURE 6.
Differential membrane partitioning of CCT isoforms to oleic acid-enriched cell membranes. COS-1 cells expressing CCT constructs at approximately equivalent levels were treated with OA/BSA in molar ratios of 3.3/1 to 266/1 for 1 h at 37 °C. Cells were harvested and fractionated, and the CCT activity units in each fraction were determined. The proportion of CCT in the membrane fraction versus the total CCT in the cytosol, membrane, and particulate fraction for each OA/BSA ratio was plotted. The individual data points from at least two independent determinations were fit to curves by GraphPad Prism 4 or fitted manually in the case of CCTβ2. A, untagged full-length CCT isoforms and NLS insertion/deletion mutants are shown. B, region P truncated CCT isoforms and NLS insertion/deletion mutants are shown. C, equivalent proportions of the soluble (sol) and membrane (M) fractions of cells transfected with the indicated full-length CCTs were analyzed by immunoblot with an antibody against the conserved catalytic domain. Cells were untreated or treated with OA/BSA at a molar ratio of 66; this concentration is indicated in panel A with an arrow.
TABLE 2.
In vivo membrane partitioning of CCT constructs
Basal and maximum % membrane partitioning was determined as the mean value ± S.E. of the base line and plateau of the curves in Fig. 6 using GraphPad Prism 4. Bound/free is the ratio of CCT units in the membrane fraction to the CCT units in the soluble fraction.
| Set | Construct | Basal |
Maximum |
||
|---|---|---|---|---|---|
| % Partitioning | Bound/free | % Partitioning | Bound/free | ||
| 1 | CCTα | 19 ± 4 | 0.24 | 82 ± 3 | 4.85 |
| CCTβ2 | 12 ± 3 | 0.14 | 30 ± 1 | 0.43 | |
| CCTα ΔNLS | 19 ± 4 | 0.24 | 70 ± 2 | 2.80 | |
| CCTβ+NLS | 12 ± 3 | 0.14 | 53 ± 2 | 1.22 | |
| 2 | CCTα312 | 34 ± 4 | 0.55 | 87 ± 3 | 14.50 |
| CCTβ313 | 27 ± 1 | 0.38 | 38 ± 1 | 0.63 | |
| CCTα312 ΔNLS | 26 ± 3 | 0.35 | 41 ± 2 | 0.72 | |
We considered that differential phosphorylation status of these constructs could again mask the impact of the NLS on membrane binding affinity, especially for CCTαΔNLS. The complete set of kinases responsible for the phosphorylation of the 16 sites in CCTα has not been identified (47, 51–54), and the kinases other than Cdk5 (11) that act on CCTβ2 are unknown. In vitro SLV binding analysis suggested that region P truncation has a similar effect on membrane binding affinity as dephosphorylation. Rather than attempting to inhibit all kinases acting on CCTs, we deleted region P from CCTα and -β2 and the NLS mutants and expressed these in COS cells to ∼equal levels, as measured by activity analysis of lysate fractions (data not shown). Because of its poor folding properties (see above), the membrane partitioning of CCTβ313+NLS is not included in our results. The partitioning results for the other CCT constructs lacking the P region clearly reveal a positive role for the NLS in membrane binding in cells (Fig. 6B). The membrane partitioning (bound/free) of CCTβ313 in cells saturated with OA was more than an order of magnitude lower than that of CCTα312 (Table 2). Deletion of the NLS from CCTα312 resulted in a large reduction in the membrane partitioning to mimic that of CCTβ2. These effects of NLS addition and deletion on the membrane partitioning of region P-truncated CCTs measured in cells were very similar to the effects of the same mutations analyzed in vitro (Tables 1 and 2). These results provide evidence that the NLS is a membrane binding motif in the context of cellular membranes and that the NLS is largely responsible for the difference in membrane binding affinity between CCT isoforms.
DISCUSSION
Characterization of CCTβ2 Structure, Activity, and Membrane Affinity
This work represents the first biochemical characterization of purified CCTβ2. We have shown that CCTβ2, like CCTα, is a homodimer and that the N-terminal region in the vicinity of C34 participates in subunit interaction. Also like CCTα, the N-terminal 40 residues and the C-terminal ∼65 residues are strongly predicted to be intrinsically disordered. Previous analyses of CCTβ2 in cells or partially purified cell extracts showed that it was a phosphoprotein (5, 11). We found that, like CCTα (34, 35, 47), the phosphorylation region of CCTβ2 purified from a mammalian cell is confined to the C-terminal residues 314–369. We found that rat CCTβ2 has 30–40% lower Vmax than rat CCTα even though their catalytic domains are 90% identical. This difference was maintained despite NLS deletions/additions, dephosphorylation, and/or region P truncations. However, when the entire N regions were swapped, the Vmax values also switched; i.e. the Vmax for CCTα-βN was 35% lower than that of CCTβ-αN. This suggests that a portion of the N region can influence CCT catalytic function. Lykidis et al. (3) also observed a depression of CCTα activity in a CCTα-βN domain chimera, assayed in cell lysates. Because residues 40–72 of the N region crown the catalytic domain and contribute many contacts to the catalytic dimer (20), it may be that this segment, which is just 67% similar between isoforms, functions less effectively as a dimer stabilizer in the β versus α isoform. This intriguing idea warrants further investigation.
Lykidis et al. (3) showed that CCTβ1, analyzed in lysates from transfected COS-7 cells, is activated by anionic lipids. In that study neither the activity nor the membrane binding of CCTβ was compared with that of CCTα in side-by-side experiments. We have shown here that although CCTβ2 does bind to anionic lipid vesicles, the binding is weaker than that of CCTα. Insertion of the CCTα NLS sequence into CCTβ2 caused an increase in membrane affinity in vitro and in cells, and this was especially marked when CCTβ2 was dephosphorylated, indicating that the NLS distinguishes the membrane affinities of the two isoforms. Dephosphorylation or deletion of the C-terminal 55 residues from CCTβ2 resulted in a 7–10-fold increase in membrane binding affinity, indicating that the phosphorylation region antagonizes membrane binding, as it does for CCTα. Thus, although the structure and regulatory mechanisms of CCTβ2 resemble those of CCTα, there may be differences in the relative impact of regulatory processes.
Dual Functions of the NLS of CCTα in Nuclear Import and Membrane Binding/Tethering
We previously identified the polybasic NLS in CCTα as a secondary membrane binding/tethering motif in vitro (37). The membrane affinity of the NLS motif on its own is too weak for accurate measurement using the SLV sedimentation assay (37), but its effect on partitioning is clearly seen when paired with the domain M amphipathic helix in the absence of a phosphorylated tail, an ∼10-fold enhancement (see Tables 1 and 2).
Recent studies with several other proteins also highlight a dual function for NLS sequences; that is, nuclear import and membrane binding. The polybasic segment in PLCζ that contains a nuclear-importing NLS sequence can also bind to anionic lipid vesicles in vitro (55). A polybasic plasma membrane-targeting motif in Rit or Rin GTPases could be converted into a nuclear targeting signal by replacing a single tryptophan with alanine (56). The results of mutagenesis and mis-localization with four other proteins (the yeast proteins Mid1p, Ste5, Opi1p, and mammalian lipin1β) have suggested competition between a nuclear import and membrane binding role for the NLS that could regulate the function of these proteins (57–60).
Our work extends this body of evidence in that we have quantified the membrane partitioning effect of an NLS binding motif in concert with an amphipathic helix motif (domain M) and showed that the NLS can enhance membrane partitioning of an amphitropic protein in cells. We envision that in the case of CCTα, the two functions of the NLS may not necessarily compete with each other. Rather, once CCTα has been imported into the nucleus and has dissociated from importin-α, its free basic NLS motif can bind electrostatically to sites of high negative charge density on the inner nuclear membrane. Alternatively, in cells where CCTα is found predominantly in the cytoplasm (14–17, 61), membrane binding of the NLS may serve to block binding to importin-α.
Role of Region P Phosphorylation on the Membrane Affinity of CCT Isoforms
The binding affinities of all constructs tested in vitro were increased by dephosphorylation or by region P truncation, suggesting that region P antagonizes the binding of domain M and that this effect is related to its phosphorylation state. The negative effect of region P was also observed in the context of cell membranes. CCT partitioning into OA-enriched cell membranes was higher when constructs were missing region P, with the exception of CCTα312ΔNLS. Cumulatively, these findings suggest that region P antagonizes the binding of domain M in both CCTα and CCTβ2 and that this effect is related to its phosphorylation state.
Antagonism of membrane binding by phosphorylation of region P is supported by three decades of studies on CCTα, starting with the discovery that treatment of cells with conditions inhibitory to kinases resulted in increased CCT activity, membrane binding, and PC synthesis and that phosphatase inhibitors had the opposite effect (62, 63). Many agents that stimulate PC synthesis are associated with CCT dephosphorylation (64–66) and increased membrane association (30, 32, 67, 68). Similar to work presented here, region P truncation (33) or dephosphorylation (23) increases CCTα affinity for anionic lipid vesicles. Dephosphorylation of CCTα during the cell cycle is associated with increased CCT activity and PC synthesis (17, 69). Conversely, in murine lung epithelial cells ERK1/2-dependent phosphorylation of region P in CCTα was associated with inhibition of activity, although the effect of this phosphorylation on the membrane affinity of the CCT was not explored (52).
The mechanism whereby the phosphorylated region P antagonizes membrane binding remains unresolved. Possible mechanisms include electrostatic repulsion between the phospho-serines proximal to domain M and the negatively charged membrane surface, neutralization of the positive charges in neighboring domain M (23), or enhancement of disorder in the entire tail (M+P) segment.
The NLS motif mitigates the antagonistic effects of region P phosphorylation on CCT membrane affinity. The membrane affinities of the CCTs missing the NLS motif (CCTβ2 and CCTαΔNLS) were more strongly affected by phosphorylation changes than was CCTα. The NLS motif may dampen the effects of phosphorylation by providing an additional membrane anchor.
The Different Intrinsic Membrane Affinities of CCT Isoforms May Reflect Their Distinct Cellular Localizations
Why would CCTβ2 have evolved to bind membranes more weakly than the α-isoform and to rely more on phosphorylation signals to modulate its membrane association and activity? For CCTβ2, its very low membrane affinity, even for the 40% anionic vesicles or cell membranes highly enriched in OA used in this study, suggest that this isoform would require nearly complete dephosphorylation for membrane translocation. Yet despite its weak membrane binding affinity, cells can function with only CCTβ2 to provide CDP-choline for PC synthesis (5, 8). The different membrane binding affinities of CCT isoforms may have evolved as a consequence of their subcellular location. CCTβ2 is a cytoplasmic enzyme that shuttles on and off the endoplasmic reticulum membrane, whereas CCTα is typically found translocating on and off the inner nuclear membrane. The endoplasmic reticulum composes ∼50% of the total membrane area within a cell, whereas the inner nuclear membrane composes only ∼0.2% (70). In effect, the local concentration of target membrane for CCTβ2 is much higher than for CCTα, and therefore, CCTβ2 would not require as high intrinsic affinity to achieve the same level of membrane binding as CCTα. There is some experimental support for the idea that cytoplasmic/endoplasmic reticulum-localized CCT experiences a higher density of membrane than does nuclear CCT, which could compensate for lower intrinsic Kp. Deletion of the NLS from CCTα, while inducing a large shift in subcellular localization from the nucleus to the cytoplasm, does not have a deleterious effect on the rate of PC synthesis (18, 71). Thus, we predict that, provided equivalent expression, the cytoplasmically localized CCTβ2 would generate PC synthesis at rates similar to that of nuclear CCTα.
Detection of a membrane binding/tethering interaction of the NLS in CCTα in vitro requires between 25–33 mol % anionic lipid content (38). In the more concentrated cellular milieu the anionic lipid requirement may be less. In fact we reliably detected a difference between CCT312 constructs with and without an NLS motif in the cell membrane partitioning trials even without OA enrichment. Recent lipid compositional analyses of membranes derived from the nucleus show very high anionic phospholipid content (72), adding credence for a membrane binding function of the NLS of nuclear CCTα.
Functional and Evolutionary Significance of Intrinsically Disordered (ID) Regulatory Domains in CCT
The ID regions N and P contribute to the activation of CCT by modulating the membrane binding affinity of domain M. Moreover, in the soluble form of CCT, domain M is quasi-unstructured (29); see Fig. 1B and supplemental Fig. S2), and in this state it functions to silence the active site.3 Membrane binding of domain M alleviates its inhibition of catalysis (21) and is responsible for CCT activation. Why would such important functions be contained within structural disorder? According to the “fly-casting model” (73, 74), the disordered N region and the partially disordered M domain would have relatively large capture radii for sampling membranes. For domain M the coupling of membrane binding to folding into an α-helix would decrease the entropic penalty associated with folding (74, 75). Contact with a membrane site enriched in activating lipids might nucleate helical structure in segments of domain M. Propagation of the amphipathic helix to form a strong hydrophobic face for membrane insertion may be facilitated by binding of the NLS, residing at the tip of an ID segment, as this would increase the time that domain M is resident on the membrane. The M domain of CCTβ2 would have an identical search radius to that of CCTα, but its interaction with the membrane would not be reinforced by a second membrane binding motif.
Although our data show that the NLS can increase CCT binding to 40 mol % of PG vesicles by an order of magnitude, this translated into only a modest 2.2-fold increase in activation efficiency of CCTα compared with CCTβ2 by the same vesicles. As well, CCT partitioning in cells showed that all constructs, regardless of NLS content, initiated membrane partitioning at approximately the same range of oleic acid enrichment. Thus, in vitro and in cells, it is the charge-sensing function of domain M that primarily determines the response to anionic lipids and the activation status of CCT. We surmise that domain M initiates binding and the NLS serves to reinforce this binding as is reflected in the higher maximal partitioning of constructs possessing NLS motifs. In its cross-bridging mode the NLS might not have this consequence, but there are no data suggesting that the NLS, residing at the tip of a long disordered chain, cannot also bind to the same membrane as the M domain. In terms of activation, it is only when domain M is membrane-bound that the active site inhibition is de-repressed. The NLS can assist the de-repression associated with membrane binding of domain M by increasing the lifetime of membrane-bound M domain.
The structural disorder of region P may serve a different purpose than that of region N. Phosphorylated regions tend also to be areas of intrinsic disorder (76). The flexibility of region P may allow better access of kinases and phosphatases to enable the fine-tuning of the membrane binding affinity of domain M. In CCTα there appears to be an antagonistic competition between two ID domains for influence on domain M.
The sequences of the N and P regions of CCTs are the least conserved. Although little is known of their special roles, we note that the CCTβ1 isoform is missing most of the disordered P region and CCTβ3 is missing most of the disordered N region. CCTs containing these ID regions enable the evolution of new functions and binding partners, because maintaining a specific fold is not necessary. It will be no surprise to find that the N-and C-terminal ID segments of CCTs across phyla have evolved novel regulatory devices to modulate the function of CCTs. The utility of intrinsically disordered regions has been elegantly described for proteins involved in regulatory interactions with other proteins, ligands, or DNA (75, 77). By comparison, the contribution of ID regions to regulation of the catalytic functions of enzymes has been neglected, although large-scale analysis of the mammalian “unfoldome” suggests that ID regions are frequently found in metabolic enzymes (78). Our study is, thus, rather unique in documenting how two disordered regions impact on the membrane binding function of the enzyme that controls PC synthesis.
Acknowledgments
We thank Joseph Lee and Ziwei Ding for preparation of constructs composed of the M+P regions of CCT and Jaeyong Lee for assistance with some of the figures. We are grateful to Suzanne Perry, Agnes Yuen, and Wendy Cheng at the Michael Smith Laboratory-Proteomics Core Facility, University of British Columbia for the mass spectrometry analyses. We thank E. C. Young and Rob Kay for critiquing our manuscript.
This work was supported by a Canadian Institutes for Health Research operating grant (to R. B. C.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables 1 and 2 and Figs. S1–S4.
H. K. Huang, Z. Ding, and R. B. Cornell, unpublished data.
- CCT
- CTP:phosphocholine cytidylyltransferase
- PC
- phosphatidylcholine
- NLS
- nuclear localization signal
- PG
- phosphatidylglycerol
- Kp
- molar partition coefficient
- OA
- oleic acid
- SLV
- sucrose-loaded vesicle
- SUV
- small unilamellar vesicle
- LUV
- large unilamellar vesicle
- ID
- intrinsically disordered.
REFERENCES
- 1. Vance J. E. (1998) Trends Biochem. Sci. 23, 423–428 [DOI] [PubMed] [Google Scholar]
- 2. Cornell R. B., Northwood I. C. (2000) Trends Biochem. Sci. 25, 441–447 [DOI] [PubMed] [Google Scholar]
- 3. Lykidis A., Murti K. G., Jackowski S. (1998) J. Biol. Chem. 273, 14022–14029 [DOI] [PubMed] [Google Scholar]
- 4. Karim M., Jackson P., Jackowski S. (2003) Biochim. Biophys. Acta 1633, 1–12 [DOI] [PubMed] [Google Scholar]
- 5. Lykidis A., Baburina I., Jackowski S. (1999) J. Biol. Chem. 274, 26992–27001 [DOI] [PubMed] [Google Scholar]
- 6. Wang L., Magdaleno S., Tabas I., Jackowski S. (2005) Mol. Cell. Biol. 25, 3357–3363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cui Z., Houweling M., Chen M. H., Record M., Chap H., Vance D. E., Tercé F. (1996) J. Biol. Chem. 271, 14668–14671 [DOI] [PubMed] [Google Scholar]
- 8. Zhang D., Tang W., Yao P. M., Yang C., Xie B., Jackowski S., Tabas I. (2000) J. Biol. Chem. 275, 35368–35376 [DOI] [PubMed] [Google Scholar]
- 9. Jackowski S., Rehg J. E., Zhang Y. M., Wang J., Miller K., Jackson P., Karim M. A. (2004) Mol. Cell. Biol. 24, 4720–4733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Carter J. M., Waite K. A., Campenot R. B., Vance J. E., Vance D. E. (2003) J. Biol. Chem. 278, 44988–44994 [DOI] [PubMed] [Google Scholar]
- 11. Carter J. M., Demizieux L., Campenot R. B., Vance D. E., Vance J. E. (2008) J. Biol. Chem. 283, 202–212 [DOI] [PubMed] [Google Scholar]
- 12. Wang Y., Sweitzer T. D., Weinhold P. A., Kent C. (1993) J. Biol. Chem. 268, 5899–5904 [PubMed] [Google Scholar]
- 13. Watkins J. D., Kent C. (1992) J. Biol. Chem. 267, 5686–5692 [PubMed] [Google Scholar]
- 14. Houweling M., Cui Z., Anfuso C. D., Bussière M., Chen M. H., Vance D. E. (1996) Eur. J. Cell. Biol. 69, 55–63 [PubMed] [Google Scholar]
- 15. Fagone P., Sriburi R., Ward-Chapman C., Frank M., Wang J., Gunter C., Brewer J. W., Jackowski S. (2007) J. Biol. Chem. 282, 7591–7605 [DOI] [PubMed] [Google Scholar]
- 16. Ridsdale R., Tseu I., Wang J., Post M. (2001) J. Biol. Chem. 276, 49148–49155 [DOI] [PubMed] [Google Scholar]
- 17. Tseu I., Ridsdale R., Liu J., Wang J., Post M. (2002) Am. J. Respir. Cell Mol. Biol. 26, 506–515 [DOI] [PubMed] [Google Scholar]
- 18. Wang Y., MacDonald J. I., Kent C. (1995) J. Biol. Chem. 270, 354–360 [DOI] [PubMed] [Google Scholar]
- 19. Chen B. B., Mallampalli R. K. (2009) Mol. Cell. Biol. 29, 3062–3075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lee J., Johnson J., Ding Z., Paetzel M., Cornell R. B. (2009) J. Biol. Chem. 284, 33535–33548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Friesen J. A., Campbell H. A., Kent C. (1999) J. Biol. Chem. 274, 13384–13389 [DOI] [PubMed] [Google Scholar]
- 22. Arnold R. S., Cornell R. B. (1996) Biochemistry 35, 9917–9924 [DOI] [PubMed] [Google Scholar]
- 23. Arnold R. S., DePaoli-Roach A. A., Cornell R. B. (1997) Biochemistry 36, 6149–6156 [DOI] [PubMed] [Google Scholar]
- 24. Attard G. S., Templer R. H., Smith W. S., Hunt A. N., Jackowski S. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 9032–9036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Davies S. M., Epand R. M., Kraayenhof R., Cornell R. B. (2001) Biochemistry 40, 10522–10531 [DOI] [PubMed] [Google Scholar]
- 26. Johnson J. E., Aebersold R., Cornell R. B. (1997) Biochim. Biophys. Acta 1324, 273–284 [DOI] [PubMed] [Google Scholar]
- 27. Johnson J. E., Rao N. M., Hui S. W., Cornell R. B. (1998) Biochemistry 37, 9509–9519 [DOI] [PubMed] [Google Scholar]
- 28. Johnson J. E., Xie M., Singh L. M., Edge R., Cornell R. B. (2003) J. Biol. Chem. 278, 514–522 [DOI] [PubMed] [Google Scholar]
- 29. Taneva S., Johnson J. E., Cornell R. B. (2003) Biochemistry 42, 11768–11776 [DOI] [PubMed] [Google Scholar]
- 30. Houweling M., Jamil H., Hatch G. M., Vance D. E. (1994) J. Biol. Chem. 269, 7544–7551 [PubMed] [Google Scholar]
- 31. Wang Y., Kent C. (1995) J. Biol. Chem. 270, 17843–17849 [DOI] [PubMed] [Google Scholar]
- 32. Wang Y., MacDonald J. I., Kent C. (1993) J. Biol. Chem. 268, 5512–5518 [PubMed] [Google Scholar]
- 33. Yang W., Jackowski S. (1995) J. Biol. Chem. 270, 16503–16506 [DOI] [PubMed] [Google Scholar]
- 34. Bogan M. J., Agnes G. R., Pio F., Cornell R. B. (2005) J. Biol. Chem. 280, 19613–19624 [DOI] [PubMed] [Google Scholar]
- 35. MacDonald J. I., Kent C. (1994) J. Biol. Chem. 269, 10529–10537 [PubMed] [Google Scholar]
- 36. Xie M., Smith J. L., Ding Z., Zhang D., Cornell R. B. (2004) J. Biol. Chem. 279, 28817–28825 [DOI] [PubMed] [Google Scholar]
- 37. Taneva S., Dennis M. K., Ding Z., Smith J. L., Cornell R. B. (2008) J. Biol. Chem. 283, 28137–28148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Taneva S. G., Patty P. J., Frisken B. J., Cornell R. B. (2005) Biochemistry 44, 9382–9393 [DOI] [PubMed] [Google Scholar]
- 39. Bradford M. M. (1976) Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 40. Sohal P. S., Cornell R. B. (1990) J. Biol. Chem. 265, 11746–11750 [PubMed] [Google Scholar]
- 41. Poehling H. M., Neuhoff V. (1981) Electrophoresis 2, 141–147 [Google Scholar]
- 42. Murray D., Hermida-Matsumoto L., Buser C. A., Tsang J., Sigal C. T., Ben-Tal N., Honig B., Resh M. D., McLaughlin S. (1998) Biochemistry 37, 2145–2159 [DOI] [PubMed] [Google Scholar]
- 43. Craig L., Johnson J. E., Cornell R. B. (1994) J. Biol. Chem. 269, 3311–3317 [PubMed] [Google Scholar]
- 44. Dunne S. J., Cornell R. B., Johnson J. E., Glover N. R., Tracey A. S. (1996) Biochemistry 35, 11975–11984 [DOI] [PubMed] [Google Scholar]
- 45. Veitch D. P., Gilham D., Cornell R. B. (1998) Eur. J. Biochem. 255, 227–234 [DOI] [PubMed] [Google Scholar]
- 46. Yang W., Boggs K. P., Jackowski S. (1995) J. Biol. Chem. 270, 23951–23957 [DOI] [PubMed] [Google Scholar]
- 47. Cornell R. B., Kalmar G. B., Kay R. J., Johnson M. A., Sanghera J. S., Pelech S. L. (1995) Biochem. J. 310, 699–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lykidis A., Jackson P., Jackowski S. (2001) Biochemistry 40, 494–503 [DOI] [PubMed] [Google Scholar]
- 49. Wang Y., Kent C. (1995) J. Biol. Chem. 270, 18948–18952 [DOI] [PubMed] [Google Scholar]
- 50. Lagace T. A., Storey M. K., Ridgway N. D. (2000) J. Biol. Chem. 275, 14367–14374 [DOI] [PubMed] [Google Scholar]
- 51. Sanghera J. S., Vance D. E. (1989) J. Biol. Chem. 264, 1215–1223 [PubMed] [Google Scholar]
- 52. Agassandian M., Zhou J., Tephly L. A., Ryan A. J., Carter A. B., Mallampalli R. K. (2005) J. Biol. Chem. 280, 21577–21587 [DOI] [PubMed] [Google Scholar]
- 53. Ryan A. J., Andrews M., Zhou J., Mallampalli R. K. (2006) Arch. Biochem. Biophys. 447, 23–33 [DOI] [PubMed] [Google Scholar]
- 54. Wieprecht M., Wieder T., Paul C., Geilen C. C., Orfanos C. E. (1996) J. Biol. Chem. 271, 9955–9961 [DOI] [PubMed] [Google Scholar]
- 55. Nomikos M., Mulgrew-Nesbitt A., Pallavi P., Mihalyne G., Zaitseva I., Swann K., Lai F. A., Murray D., McLaughlin S. (2007) J. Biol. Chem. 282, 16644–16653 [DOI] [PubMed] [Google Scholar]
- 56. Heo W. D., Inoue T., Park W. S., Kim M. L., Park B. O., Wandless T. J., Meyer T. (2006) Science 314, 1458–1461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Loewen C. J., Gaspar M. L., Jesch S. A., Delon C., Ktistakis N. T., Henry S. A., Levine T. P. (2004) Science 304, 1644–1657 [DOI] [PubMed] [Google Scholar]
- 58. Winters M. J., Lamson R. E., Nakanishi H., Neiman A. M., Pryciak P. M. (2005) Mol. Cell. 20, 21–32 [DOI] [PubMed] [Google Scholar]
- 59. Celton-Morizur S., Bordes N., Fraisier V., Tran P. T., Paoletti A. (2004) Mol. Cell. Biol. 24, 10621–10635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Ren H., Federico L., Huang H., Sunkara M., Drennan T., Frohman M. A., Smyth S. S., Morris A. J. (2010) Mol. Biol. Cell. 21, 3171–3181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ridsdale R., Tseu I., Wang J., Post M. (2010) Am. J. Respir. Cell Mol. Biol. 43, 74–87 [DOI] [PubMed] [Google Scholar]
- 62. Hatch G. M., Jamil H., Utal A. K., Vance D. E. (1992) J. Biol. Chem. 267, 15751–15758 [PubMed] [Google Scholar]
- 63. Pelech S. L., Vance D. E. (1982) J. Biol. Chem. 257, 14198–14202 [PubMed] [Google Scholar]
- 64. Groblewski G. E., Wang Y., Ernst S. A., Kent C., Williams J. A. (1995) J. Biol. Chem. 270, 1437–1442 [DOI] [PubMed] [Google Scholar]
- 65. Shiratori Y., Houweling M., Zha X., Tabas I. (1995) J. Biol. Chem. 270, 29894–29903 [DOI] [PubMed] [Google Scholar]
- 66. MacDonald J. I., Possmayer F. (1995) Biochem. J. 312, 425–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Watkins J. D., Kent C. (1991) J. Biol. Chem. 266, 21113–21117 [PubMed] [Google Scholar]
- 68. Northwood I. C., Tong A. H., Crawford B., Drobnies A. E., Cornell R. B. (1999) J. Biol. Chem. 274, 26240–26248 [DOI] [PubMed] [Google Scholar]
- 69. Jackowski S. (1994) J. Biol. Chem. 269, 3858–3867 [PubMed] [Google Scholar]
- 70. Weibel E. R., Stäubli W., Gnägi H. R., Hess F. A. (1969) J. Cell. Biol. 42, 68–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Gehrig K., Morton C. C., Ridgway N. D. (2009) J. Lipid. Res. 50, 966–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Garnier-Lhomme M., Byrne R. D., Hobday T. M., Gschmeissner S., Woscholski R., Poccia D. L., Dufourc E. J., Larijani B. (2009) PLoS ONE 4, e4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Shoemaker B. A., Portman J. J., Wolynes P. G. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 8868–8873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Wright P. E., Dyson H. J. (2009) Curr. Opin. Struct. Biol. 19, 31–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Smock R. G., Gierasch L. M. (2009) Science 324, 198–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Iakoucheva L. M., Radivojac P., Brown C. J., O'Connor T. R., Sikes J. G., Obradovic Z., Dunker A. K. (2004) Nucleic. Acids. Res. 32, 1037–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Uversky V. N., Dunker A. K. (2010) Biochim. Biophys. Acta 1804, 1231–1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Galea C. A., High A. A., Obenauer J. C., Mishra A., Park C. G., Punta M., Schlessinger A., Ma J., Rost B., Slaughter C. A., Kriwacki R. W. (2009) J. Proteome Res. 8, 211–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Yang Z. R., Thomson R., McNeil P., Esnouf R. M. (2005) Bioinformatics 21, 3369–3376 [DOI] [PubMed] [Google Scholar]
- 80. Jones M. K., Anantharamaiah G. M., Segrest J. P. (1992) J. Lipid. Res. 33, 287–296 [PubMed] [Google Scholar]