Abstract
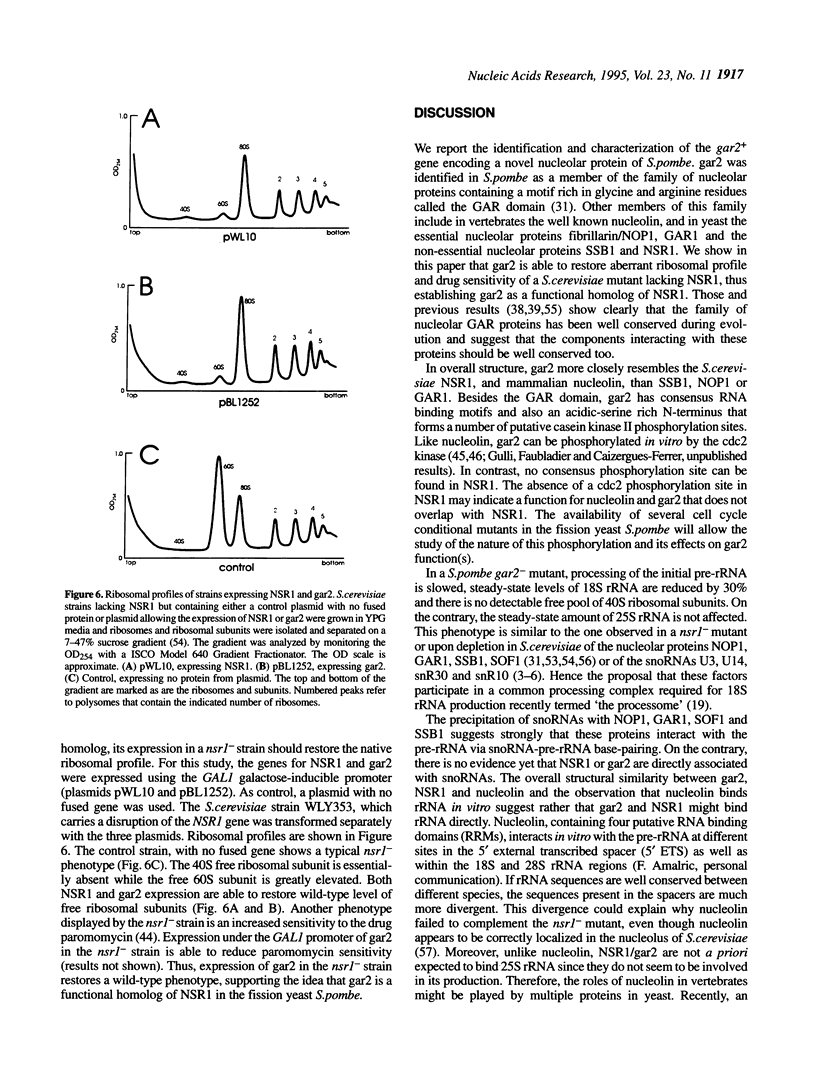
Several nucleolar proteins, such as nucleolin, NOP1/fibrillarin, SSB1, NSR1 and GAR1 share a common glycine and arginine rich structural motif called the GAR domain. To identify novel nucleolar proteins from fission yeast we screened Schizosaccharomyces pombe genomic DNA libraries with a probe encompassing the GAR structural motif. Here we report the identification and characterization of a S.pombe gene coding for a novel nucleolar protein, designated gar2. The structure of the fission yeast gar2 is reminiscent of that of nucleolin from vertebrates and NSR1 from Saccharomyces cerevisiae. In addition, like these proteins, gar2 has a nucleolar localisation. The disruption of the gar2+ gene affects normal cell growth, leads to an accumulation of 35S pre-rRNA and a decrease of mature 18S rRNA steady state levels. Moreover, ribosomal profiles of the mutant show an increase of free 60S ribosomal subunits and an absence of free 40S ribosomal subunits. gar2 is able to rescue a S.cerevisiae mutant lacking NSR1, thus establishing gar2 as a functional homolog of NSR1. We propose that gar2 helps the assembly of pre-ribosomal particles containing 18S rRNA.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alderuccio F., Chan E. K., Tan E. M. Molecular characterization of an autoantigen of PM-Scl in the polymyositis/scleroderma overlap syndrome: a unique and complete human cDNA encoding an apparent 75-kD acidic protein of the nucleolar complex. J Exp Med. 1991 Apr 1;173(4):941–952. doi: 10.1084/jem.173.4.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aris J. P., Blobel G. cDNA cloning and sequencing of human fibrillarin, a conserved nucleolar protein recognized by autoimmune antisera. Proc Natl Acad Sci U S A. 1991 Feb 1;88(3):931–935. doi: 10.1073/pnas.88.3.931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belenguer P., Caizergues-Ferrer M., Labbé J. C., Dorée M., Amalric F. Mitosis-specific phosphorylation of nucleolin by p34cdc2 protein kinase. Mol Cell Biol. 1990 Jul;10(7):3607–3618. doi: 10.1128/mcb.10.7.3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltrame M., Tollervey D. Identification and functional analysis of two U3 binding sites on yeast pre-ribosomal RNA. EMBO J. 1992 Apr;11(4):1531–1542. doi: 10.1002/j.1460-2075.1992.tb05198.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergès T., Petfalski E., Tollervey D., Hurt E. C. Synthetic lethality with fibrillarin identifies NOP77p, a nucleolar protein required for pre-rRNA processing and modification. EMBO J. 1994 Jul 1;13(13):3136–3148. doi: 10.1002/j.1460-2075.1994.tb06612.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caizergues-Ferrer M., Belenguer P., Lapeyre B., Amalric F., Wallace M. O., Olson M. O. Phosphorylation of nucleolin by a nucleolar type NII protein kinase. Biochemistry. 1987 Dec 1;26(24):7876–7883. doi: 10.1021/bi00398a051. [DOI] [PubMed] [Google Scholar]
- Caizergues-Ferrer M., Mariottini P., Curie C., Lapeyre B., Gas N., Amalric F., Amaldi F. Nucleolin from Xenopus laevis: cDNA cloning and expression during development. Genes Dev. 1989 Mar;3(3):324–333. doi: 10.1101/gad.3.3.324. [DOI] [PubMed] [Google Scholar]
- Chooi W. Y., Leiby K. R. An electron microscopic method for localization of ribosomal proteins during transcription of ribosomal DNA: a method for studying protein assembly. Proc Natl Acad Sci U S A. 1981 Aug;78(8):4823–4827. doi: 10.1073/pnas.78.8.4823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark M. W., Yip M. L., Campbell J., Abelson J. SSB-1 of the yeast Saccharomyces cerevisiae is a nucleolar-specific, silver-binding protein that is associated with the snR10 and snR11 small nuclear RNAs. J Cell Biol. 1990 Nov;111(5 Pt 1):1741–1751. doi: 10.1083/jcb.111.5.1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fournier M. J., Maxwell E. S. The nucleolar snRNAs: catching up with the spliceosomal snRNAs. Trends Biochem Sci. 1993 Apr;18(4):131–135. doi: 10.1016/0968-0004(93)90020-n. [DOI] [PubMed] [Google Scholar]
- Ghisolfi L., Joseph G., Amalric F., Erard M. The glycine-rich domain of nucleolin has an unusual supersecondary structure responsible for its RNA-helix-destabilizing properties. J Biol Chem. 1992 Feb 15;267(5):2955–2959. [PubMed] [Google Scholar]
- Girard J. P., Caizergues-Ferrer M., Lapeyre B. The SpGAR1 gene of Schizosaccharomyces pombe encodes the functional homologue of the snoRNP protein GAR1 of Saccharomyces cerevisiae. Nucleic Acids Res. 1993 May 11;21(9):2149–2155. doi: 10.1093/nar/21.9.2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard J. P., Feliu J., Caizergues-Ferrer M., Lapeyre B. Study of multiple fibrillarin mRNAs reveals that 3' end formation in Schizosaccharomyces pombe is sensitive to cold shock. Nucleic Acids Res. 1993 Apr 25;21(8):1881–1887. doi: 10.1093/nar/21.8.1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard J. P., Lehtonen H., Caizergues-Ferrer M., Amalric F., Tollervey D., Lapeyre B. GAR1 is an essential small nucleolar RNP protein required for pre-rRNA processing in yeast. EMBO J. 1992 Feb;11(2):673–682. doi: 10.1002/j.1460-2075.1992.tb05099.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagan I. M., Hyams J. S. The use of cell division cycle mutants to investigate the control of microtubule distribution in the fission yeast Schizosaccharomyces pombe. J Cell Sci. 1988 Mar;89(Pt 3):343–357. doi: 10.1242/jcs.89.3.343. [DOI] [PubMed] [Google Scholar]
- Henríquez R., Blobel G., Aris J. P. Isolation and sequencing of NOP1. A yeast gene encoding a nucleolar protein homologous to a human autoimmune antigen. J Biol Chem. 1990 Feb 5;265(4):2209–2215. [PubMed] [Google Scholar]
- Hirano T., Konoha G., Toda T., Yanagida M. Essential roles of the RNA polymerase I largest subunit and DNA topoisomerases in the formation of fission yeast nucleolus. J Cell Biol. 1989 Feb;108(2):243–253. doi: 10.1083/jcb.108.2.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes J. M., Ares M., Jr Depletion of U3 small nucleolar RNA inhibits cleavage in the 5' external transcribed spacer of yeast pre-ribosomal RNA and impairs formation of 18S ribosomal RNA. EMBO J. 1991 Dec;10(13):4231–4239. doi: 10.1002/j.1460-2075.1991.tb05001.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen R. P., Hurt E. C., Kern H., Lehtonen H., Carmo-Fonseca M., Lapeyre B., Tollervey D. Evolutionary conservation of the human nucleolar protein fibrillarin and its functional expression in yeast. J Cell Biol. 1991 May;113(4):715–729. doi: 10.1083/jcb.113.4.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen R., Tollervey D., Hurt E. C. A U3 snoRNP protein with homology to splicing factor PRP4 and G beta domains is required for ribosomal RNA processing. EMBO J. 1993 Jun;12(6):2549–2558. doi: 10.1002/j.1460-2075.1993.tb05910.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jong A. Y., Clark M. W., Gilbert M., Oehm A., Campbell J. L. Saccharomyces cerevisiae SSB1 protein and its relationship to nucleolar RNA-binding proteins. Mol Cell Biol. 1987 Aug;7(8):2947–2955. doi: 10.1128/mcb.7.8.2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kass S., Tyc K., Steitz J. A., Sollner-Webb B. The U3 small nucleolar ribonucleoprotein functions in the first step of preribosomal RNA processing. Cell. 1990 Mar 23;60(6):897–908. doi: 10.1016/0092-8674(90)90338-f. [DOI] [PubMed] [Google Scholar]
- Kenan D. J., Query C. C., Keene J. D. RNA recognition: towards identifying determinants of specificity. Trends Biochem Sci. 1991 Jun;16(6):214–220. doi: 10.1016/0968-0004(91)90088-d. [DOI] [PubMed] [Google Scholar]
- Lapeyre B., Bourbon H., Amalric F. Nucleolin, the major nucleolar protein of growing eukaryotic cells: an unusual protein structure revealed by the nucleotide sequence. Proc Natl Acad Sci U S A. 1987 Mar;84(6):1472–1476. doi: 10.1073/pnas.84.6.1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapeyre B., Mariottini P., Mathieu C., Ferrer P., Amaldi F., Amalric F., Caizergues-Ferrer M. Molecular cloning of Xenopus fibrillarin, a conserved U3 small nuclear ribonucleoprotein recognized by antisera from humans with autoimmune disease. Mol Cell Biol. 1990 Jan;10(1):430–434. doi: 10.1128/mcb.10.1.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapeyre B., Michot B., Feliu J., Bachellerie J. P. Nucleotide sequence of the Schizosaccharomyces pombe 25S ribosomal RNA and its phylogenetic implications. Nucleic Acids Res. 1993 Jul 11;21(14):3322–3322. doi: 10.1093/nar/21.14.3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W. C., Xue Z. X., Mélèse T. The NSR1 gene encodes a protein that specifically binds nuclear localization sequences and has two RNA recognition motifs. J Cell Biol. 1991 Apr;113(1):1–12. doi: 10.1083/jcb.113.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W. C., Zabetakis D., Mélèse T. NSR1 is required for pre-rRNA processing and for the proper maintenance of steady-state levels of ribosomal subunits. Mol Cell Biol. 1992 Sep;12(9):3865–3871. doi: 10.1128/mcb.12.9.3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. D., Zagorski J., Fournier M. J. Depletion of U14 small nuclear RNA (snR128) disrupts production of 18S rRNA in Saccharomyces cerevisiae. Mol Cell Biol. 1990 Mar;10(3):1145–1152. doi: 10.1128/mcb.10.3.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lygerou Z., Mitchell P., Petfalski E., Séraphin B., Tollervey D. The POP1 gene encodes a protein component common to the RNase MRP and RNase P ribonucleoproteins. Genes Dev. 1994 Jun 15;8(12):1423–1433. doi: 10.1101/gad.8.12.1423. [DOI] [PubMed] [Google Scholar]
- Maser R. L., Calvet J. P. U3 small nuclear RNA can be psoralen-cross-linked in vivo to the 5' external transcribed spacer of pre-ribosomal-RNA. Proc Natl Acad Sci U S A. 1989 Sep;86(17):6523–6527. doi: 10.1073/pnas.86.17.6523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattaj I. W. RNA recognition: a family matter? Cell. 1993 Jun 4;73(5):837–840. doi: 10.1016/0092-8674(93)90265-r. [DOI] [PubMed] [Google Scholar]
- Moreno S., Klar A., Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- Morrissey J. P., Tollervey D. Yeast snR30 is a small nucleolar RNA required for 18S rRNA synthesis. Mol Cell Biol. 1993 Apr;13(4):2469–2477. doi: 10.1128/mcb.13.4.2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mougey E. B., O'Reilly M., Osheim Y., Miller O. L., Jr, Beyer A., Sollner-Webb B. The terminal balls characteristic of eukaryotic rRNA transcription units in chromatin spreads are rRNA processing complexes. Genes Dev. 1993 Aug;7(8):1609–1619. doi: 10.1101/gad.7.8.1609. [DOI] [PubMed] [Google Scholar]
- Mougey E. B., Pape L. K., Sollner-Webb B. A U3 small nuclear ribonucleoprotein-requiring processing event in the 5' external transcribed spacer of Xenopus precursor rRNA. Mol Cell Biol. 1993 Oct;13(10):5990–5998. doi: 10.1128/mcb.13.10.5990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najbauer J., Johnson B. A., Young A. L., Aswad D. W. Peptides with sequences similar to glycine, arginine-rich motifs in proteins interacting with RNA are efficiently recognized by methyltransferase(s) modifying arginine in numerous proteins. J Biol Chem. 1993 May 15;268(14):10501–10509. [PubMed] [Google Scholar]
- Peculis B. A., Steitz J. A. Disruption of U8 nucleolar snRNA inhibits 5.8S and 28S rRNA processing in the Xenopus oocyte. Cell. 1993 Jun 18;73(6):1233–1245. doi: 10.1016/0092-8674(93)90651-6. [DOI] [PubMed] [Google Scholar]
- Peter M., Nakagawa J., Dorée M., Labbé J. C., Nigg E. A. Identification of major nucleolar proteins as candidate mitotic substrates of cdc2 kinase. Cell. 1990 Mar 9;60(5):791–801. doi: 10.1016/0092-8674(90)90093-t. [DOI] [PubMed] [Google Scholar]
- Rimoldi O. J., Raghu B., Nag M. K., Eliceiri G. L. Three new small nucleolar RNAs that are psoralen cross-linked in vivo to unique regions of pre-rRNA. Mol Cell Biol. 1993 Jul;13(7):4382–4390. doi: 10.1128/mcb.13.7.4382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripmaster T. L., Vaughn G. P., Woolford J. L., Jr A putative ATP-dependent RNA helicase involved in Saccharomyces cerevisiae ribosome assembly. Proc Natl Acad Sci U S A. 1992 Dec 1;89(23):11131–11135. doi: 10.1073/pnas.89.23.11131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein R. J. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- Sachs A. B., Davis R. W. Translation initiation and ribosomal biogenesis: involvement of a putative rRNA helicase and RPL46. Science. 1990 Mar 2;247(4946):1077–1079. doi: 10.1126/science.2408148. [DOI] [PubMed] [Google Scholar]
- Savino R., Gerbi S. A. In vivo disruption of Xenopus U3 snRNA affects ribosomal RNA processing. EMBO J. 1990 Jul;9(7):2299–2308. doi: 10.1002/j.1460-2075.1990.tb07401.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt M. E., Clayton D. A. Nuclear RNase MRP is required for correct processing of pre-5.8S rRNA in Saccharomyces cerevisiae. Mol Cell Biol. 1993 Dec;13(12):7935–7941. doi: 10.1128/mcb.13.12.7935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schofield P. R., McFarland K. C., Hayflick J. S., Wilcox J. N., Cho T. M., Roy S., Lee N. M., Loh H. H., Seeburg P. H. Molecular characterization of a new immunoglobulin superfamily protein with potential roles in opioid binding and cell contact. EMBO J. 1989 Feb;8(2):489–495. doi: 10.1002/j.1460-2075.1989.tb03402.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sollner-Webb B. Novel intron-encoded small nucleolar RNAs. Cell. 1993 Nov 5;75(3):403–405. doi: 10.1016/0092-8674(93)90374-y. [DOI] [PubMed] [Google Scholar]
- Stroke I. L., Weiner A. M. The 5' end of U3 snRNA can be crosslinked in vivo to the external transcribed spacer of rat ribosomal RNA precursors. J Mol Biol. 1989 Dec 5;210(3):497–512. doi: 10.1016/0022-2836(89)90126-5. [DOI] [PubMed] [Google Scholar]
- Studier F. W., Rosenberg A. H., Dunn J. J., Dubendorff J. W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- Sun C., Woolford J. L., Jr The yeast NOP4 gene product is an essential nucleolar protein required for pre-rRNA processing and accumulation of 60S ribosomal subunits. EMBO J. 1994 Jul 1;13(13):3127–3135. doi: 10.1002/j.1460-2075.1994.tb06611.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollervey D. A yeast small nuclear RNA is required for normal processing of pre-ribosomal RNA. EMBO J. 1987 Dec 20;6(13):4169–4175. doi: 10.1002/j.1460-2075.1987.tb02763.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollervey D., Lehtonen H., Carmo-Fonseca M., Hurt E. C. The small nucleolar RNP protein NOP1 (fibrillarin) is required for pre-rRNA processing in yeast. EMBO J. 1991 Mar;10(3):573–583. doi: 10.1002/j.1460-2075.1991.tb07984.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyc K., Steitz J. A. A new interaction between the mouse 5' external transcribed spacer of pre-rRNA and U3 snRNA detected by psoralen crosslinking. Nucleic Acids Res. 1992 Oct 25;20(20):5375–5382. doi: 10.1093/nar/20.20.5375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tycowski K. T., Shu M. D., Steitz J. A. Requirement for intron-encoded U22 small nucleolar RNA in 18S ribosomal RNA maturation. Science. 1994 Dec 2;266(5190):1558–1561. doi: 10.1126/science.7985025. [DOI] [PubMed] [Google Scholar]
- Xue Z., Mélèse T. Nucleolar proteins that bind NLSs: a role in nuclear import or ribosome biogenesis? Trends Cell Biol. 1994 Dec;4(12):414–417. doi: 10.1016/0962-8924(94)90095-7. [DOI] [PubMed] [Google Scholar]
- Xue Z., Shan X., Lapeyre B., Mélèse T. The amino terminus of mammalian nucleolin specifically recognizes SV40 T-antigen type nuclear localization sequences. Eur J Cell Biol. 1993 Oct;62(1):13–21. [PubMed] [Google Scholar]