Abstract
Conformational change during protein–ligand binding may significantly affect both the binding mechanism and the rate constant. Most earlier theories and simulations treated conformational change as stochastic gating with transition rates between reactive and nonreactive conformations uncoupled to ligand binding. Recently, we introduced a dual-transition-rates model in which the transition rates between reactive and nonreactive conformations depend on the protein–ligand distance [H.-X. Zhou, Biophys. J. 98, L15 (2010)]. Analytical results of that model showed that the apparent binding mechanism switches from conformational selection to induced fit, when the rates of conformational transitions increase from being much slower than the diffusional approach of the protein–ligand pair to being much faster. The conformational-selection limit (kCS) and the induced-fit limit (kIF) provide lower and upper bounds, respectively, for the binding rate constant. Here we introduce a general model in which the energy surface of the protein in conformational space is coupled to ligand binding, and present a method for calculating the binding rate constant from Brownian dynamics simulations. Analytical and simulation results show that, for an energy surface that switches from favoring the nonreactive conformation while the ligand is away to favoring the reactive conformation while the ligand is near, kCS and kIF become close and, thus, provide tight bounds to the binding rate constant. This finding has significant mechanistic implications and presents routes for obtaining good estimates of the rate constant at low cost.
INTRODUCTION
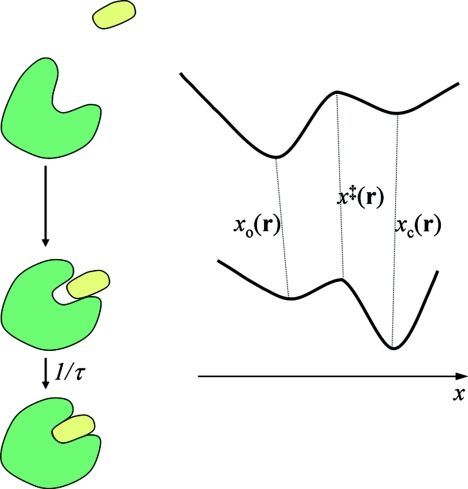
Protein–ligand binding is generally accompanied by conformational change. Both the diffusional approach of the protein–ligand pair and the conformational change can be rate-limiting steps.1 When binding is rate-limited by diffusion, only the relative translation and the overall rotation of the binding molecules need to be considered; theories and simulations have had great successes in this regime.2, 3 Outside this regime, one has to explicitly treat conformational degrees of freedom. Theories have been presented where the conformational degrees of freedom are uncoupled to the translational–rotational degrees of freedom.4, 5, 6, 7 However, one expects the energy surface in conformational space to change significantly when the protein–ligand pair moves from far apart to near contact. Indeed, a far more realistic energy surface is one that switches from favoring a “nonreactive” conformation while the protein–ligand pair is far apart to favoring a “reactive” conformation while the protein–ligand pair is near contact (Fig. 1). In this paper we deal with such a binding-coupled energy surface.
Figure 1.
A model in which conformational change and ligand binding are coupled. The energy surface of the protein depends on the protein–ligand separation r.
Szabo et al.4 introduced a model in which conformational change of the protein is treated as stochastic gating,
| (1) |
with the transition rates ω± between reactive and nonreactive conformations independent of the ligand position. Binding to the protein can occur only when the ligand is in the “reactive” region while the protein is in the reactive conformation. Such a model predicts the following relation for the binding rate constant kon:4, 7
| (2) |
where is the Laplace transform of the time-dependent rate coefficient when the protein always stays in the reactive conformation and kon0 is the long-time limit of that rate coefficient. The limits of kon, when the gating rates are infinitely slow and infinitely fast, are of particular interest. We denote these two limits as kslow and kfast. When the reactivity of the protein–ligand pair in the reaction region is small, kslow and kfast are identical:
| (3) |
where p+ = ω+∕(ω+ + ω−) is the equilibrium probability for the protein to adopt the reactive conformation. However, when the reactivity is large the two limits are very different:
| (4a) |
| (4b) |
So the slow-gating limit can be significantly less than the fast-gating limit.
We6 developed a related model, for the binding of a ligand to a buried site in a protein, which is accessible only through a gate that undergoes open–closed transitions. The open conformation allows for ligand entrance but the closed conformation blocks it. The open–closed transitions are assumed to be independent of the ligand position and described by the scheme of Eq. 1. Instead of stochastic gating, Agmon5 treated the conformational change of an enzyme as diffusion along a continuous coordinate, but here again the energy surface of the enzyme was assumed to be unaffected by substrate binding.
Wade, McCammon, and co-workers have carried out Brownian dynamics and molecular dynamics simulations to explore the conformational changes of a number of proteins in the absence of ligands.8, 9, 10, 11, 12 Equation (4) was used to estimate the ligand-binding rate constant. Similarly, the gating dynamics of an enzyme with a buried site, obtained from molecular dynamics simulations in the absence of the substrate, was used to estimate the substrate-binding rate constant.13 In another interesting study,14 an ensemble of conformations of an enzyme was generated by molecular dynamics simulations and multiple “static” snapshots were used to calculate the substrate-binding rate constant.
The most expensive and of most relevance to the present paper is a study by Wade et al.,15 in which conformational fluctuation and substrate binding to an enzyme were coupled. The diffusional motion of the substrate and conformational fluctuation of the enzyme were both modeled by Brownian dynamics simulations. To reduce the computational cost of simulating the conformational fluctuation, only a critical loop over the active site was allowed to be flexible during the simulations. We note that Kim and Lee16 have recently derived an formal expression for the binding rate coefficient for reactant molecules with internal degrees of freedom.
To further investigate how conformational change that is coupled to ligand binding affects binding mechanism and rate constant, we17 extended the model of Szabo et al.4 to include two sets of transition rates, one when the ligand is far away from the protein and the other when the ligand is near. The transition rates were chosen to favor the nonreactive conformation when the ligand is away but the reactive conformation when the ligand is near. In order to obtain analytical results, the geometry of the model considered was very simple. The protein was modeled as a sphere with a uniformly reactive surface. It was found that, when the rates of conformational transitions are low, binding proceeds via an apparently conformational-selection (CS) mechanism; in contrast, when the rates of conformational transitions are high, binding proceeds via an apparently induced-fit (IF) mechanism. Moreover, the rate constants in these limiting situations, kCS and kIF, provide lower and upper bounds, respectively.
Here we increase the realism of this line of development by restricting the reactive region to a localized site on the protein surface. We describe a method for calculating the binding rate coefficient from Brownian dynamics simulations. Our simulation results on a model system showed that the CS and IF limits become closer and closer as the reactive region becomes more and more localized. This finding prompted us to derive general relations for kCS and kIF. They confirm that, for an energy surface that switches from favoring the nonreactive conformation while the ligand is away to favoring the reactive conformation while the ligand is near, kCS and kIF are close. Therefore, these two limiting values provide tight bounds on the binding rate constant. No simulation of conformational change is necessary in the calculation of either kCS or kIF; hence these limits provide good estimates of kon at low computational cost. The closeness of kCS and kIF under conditions that realistically model protein–ligand binding has significant mechanistic implications.
The rest of the paper is organized as follows. In Sec. 2, we present a general formulation of the time-dependent binding rate coefficient when the energy surface of the protein is coupled to ligand binding. There we also present a method for calculating the rate coefficient from Brownian dynamics simulations. In Sec. 3, we present analytical results for the binding rate constant and its CS and IF limits. In Sec. 4, we illustrate our Brownian dynamics method for calculating the rate constant on a model system. Details of the implementation are described. In Sec. 5, we present simulation results and compare them against available analytical results. Finally, in Sec. 4 we discuss the implications and further extensions of the present study.
FORMULATION OF THE GENERAL PROBLEM
Here, we present a general formulation of the time-dependent binding rate coefficient when the energy surface of the protein is coupled to ligand binding. A method to calculate the rate coefficient from Brownian dynamics simulations is described. For simplicity, we treat a pointlike ligand and describe the conformational space by a single coordinate x. Our basic formulation can be easily extended to a more general case involving both translational and rotational diffusion of the protein and the ligand as well as a higher-dimensional conformational space. The motion along x is first treated as continuous diffusion and then as transitions between two discrete conformations.
Continuous diffusion in conformational space
As illustrated in Fig. 1, in the system that we consider, the protein with a conformation space specified by a one-dimensional variable x binds with a pointlike ligand. For example, x may represent the opening size of the ligand-binding pocket on the protein surface. We note that the mathematics is identical if the protein is modeled as rigid and the ligand, such as a peptide, is modeled as flexible; in that case x represents the conformational degree of freedom of the ligand. Let the protein–ligand separation be a three-dimensional vector r. We treat the motions in r and along x both as diffusion, with diffusion constants D and D1, respectively. Let the potential of mean force in r and x be U(r, x). We envision that, for a given r, U(r, x) has a double-well shape along x (Fig. 1); for example, one well may correspond to a closed conformation and the other well an open conformation. When the ligand is far away, the Boltzmann weight of the open conformation is much greater than that of the closed form, so that the protein mostly adopts the open conformation. We denote the potential as U∞(x) when the protein and ligand are far apart. When the ligand is near or inside the binding pocket, due to protein–ligand interactions, the Boltzmann weights of the open and closed conformations are reversed to favor the latter. Within this loosely-bound state, the protein–ligand pair can form the native complex. We treat the last step as a rate process with a rate constant 1∕τ.
In the Smoluchowski theory for diffusion-influenced bimolecular reactions, the rate coefficient, k(t), for protein–ligand binding is calculated from the pair distribution function, P(r, x, t), where t denotes time.7, 16, 18 The pair distribution function satisfies the Smoluchowski equation augmented by a “sink” term to account for the formation of the native complex:
| (5) |
where β = (kBT)−1 and H(r, x) is either 1 if the ligand is in the “reactive” region (forming the loosely-bound state with the protein in the closed conformation) or 0 otherwise. At infinite protein–ligand separation the pair distribution function is
| (6) |
The inner boundary is reflecting, since the protein and ligand cannot interpenetrate. The initial distribution is assumed to be an equilibrium distribution:
| (7) |
The rate coefficient is then
| (8) |
This result generalizes the usual formulation of k(t) by including the coordinate x for modeling conformational change. Using Eq. 7, the initial value k(0) is found as
| (9) |
If diffusions (both in r and along x) were infinitely fast (or equivalently, reactions were infinitely slow, i.e., τ → ∞), the protein–ligand pair would stay in the equilibrium distribution and the rate coefficient would stay at k(0). In general, k(t) would decay over time.
We have previously developed an algorithm for calculating k(t) from Brownian dynamics simulations.19 The algorithm is based on the relationship between the pair distribution function P(r, x, t) and the survival probability S(t|r, x):
| (10) |
The survival probability is the probability that a protein–ligand pair, started at time t = 0 from separation r and conformation x, has not formed the native complex by time t. Using Eq. 10 in Eq. 8, we have
| (11) |
Using Eq. 9, we find the ratio k(t)∕k(0) as
| (12) |
The last result is the basis of our Brownian dynamics algorithm for calculating the binding rate coefficient. When Brownian trajectories are started at t = 0 from the reactive region with a distribution proportional to Peq(r, x), the average survival probability at time t is k(t)∕k(0). Multiplied by k(0) calculated according to Eq. 9, we obtain k(t).
Let us briefly outline how our formulation of k(t) can be extended to the more general case involving both translational and rotational diffusion of the protein and the ligand as well as a higher-dimensional conformational space. Now r represents the protein–ligand relative translational and overall rotational degrees of freedom and x represents coordinates in the higher-dimensional conformational space. The function H(r, x) still defines the reaction criterion: it has a value of 1 only if the protein–ligand pair is separated and orientated such that it is in the loosely-bound state with the two molecules correctly facing each other and in their respective reactive conformations. The protein–ligand pair is started from the region in configurational–conformaitonal space where H(r, x) = 1 and then propagated. Whenever it is found in the region where H(r, x) = 1, the protein–ligand pair is allowed to be transformed, with a mean lifetime τ, into the native complex. Again, the survival probability at time t gives k(t)∕k(0).
Transitions between two discrete conformations
For the model shown in Fig. 1, at each protein–ligand separation r, the conformation of the protein is either open or closed. Let the location of the energy barrier separating the two conformations be x = x‡(r). The open and closed conformations are specified by −∞ < x < x‡(r) and x‡(r) < x < ∞, respectively. If the energy barrier is high, then equilibration within each well is fast and transitions between the two wells can be treated as rate processes:
| (13) |
According to Kramers,20 the transition rate constants are given by
| (14a) |
| (14b) |
where xo(r) and xc(r) are the values of x at the bottoms of the open and closed energy wells, respectively. In line with fast equilibration within both energy wells, we define conformation-specific potential energies, Uo(r) and Uc(r), for protein–ligand interaction by
| (15a) |
| (15b) |
where is the location of the energy barrier separating the open and closed conformations when the ligand is far away. The denominators in Eq. (15) ensure that Uo(r) and Uc(r) go to zero at infinite protein–ligand separation. Correspondingly we denote the pair distribution as Po(r, t) and Pc(r, t) when the protein is in the open and closed conformations, respectively,
| (16a) |
| (16b) |
The initial values of the pair distribution function are
| (17a) |
where g = o or c, and the equilibrium probability, p∞g, of conformation g at infinite protein–ligand separation is
| (17b) |
With the transitions between the two conformations treated as rate processes, the pair distribution now satisfies (see Appendix A)
| (18a) |
| (18b) |
where Hc(r) is given by Eq. A8. Note that the ratio of the transition rates between the two conformations is
| (19a) |
Denoting the transition rate constants at infinite separation as ω∞±, we have
| (19b) |
and Eq. 19a can be expressed as
| (19c) |
This result has been written down based on detailed balance.17 The gating model of Shoup et al.4 has ω+(r) = ω∞+ and ω−(r) = ω∞−; correspondingly Uc(r) = Uo(r) in their model.
The binding rate coefficient is now given by
| (20) |
In the present case, the pair distribution function Pg(r, t) relates to the survival probability S(t|r, g) via
| (21) |
Using the last result, we find
| (22) |
| (23) |
The last result shows that k(t)∕k(0) is given by the average survival probability at time t when Brownian trajectories are started with the ligand in the reactive region specified by the condition Hc(r) = 1, with position following the Boltzmann distribution exp[−βUc(r)], and with the protein in the closed, i.e., reactive conformation.
Asymptotic behavior of k(t)
While the Brownian dynamics algorithm outlined above gives the full time dependence of the binding rate coefficient, the limit of k(t) at t → ∞, i.e., the steady-state value, is of particular interest. We refer to this steady-state value the rate constant and denote it as kon. In previous work,7 we have shown that k(t) has the following asymptotic behavior at long times:
| (24) |
Even though Brownian dynamics simulations must have a finite cutoff time (tcut), yielding k(t) up to t = tcut, by fitting the long-time portion of k(t) to Eq. 24 we can obtain the steady-state value kon.19 The fitting function is linear when the independent variable is chosen as (πDt)−1∕2. The intercept of the linear fit is kon and the slope is kon2∕4πD which can be calculated from the intercept. That the slope is not a free parameter but can be calculated from the intercept provides a convenient way to validate the simulation results and determine whether tcut is sufficiently long to ensure asymptotic behavior.
ANALYTICAL RESULTS FOR RATE CONSTANTS
The preceding section outlines a Brownian dynamics algorithm for calculating the binding rate coefficient when protein–ligand binding is coupled to conformational change. The conformational change is either treated as continuous diffusion along a one-dimensional coordinate or as rate processes between two discrete conformations. The latter treatment will be the focus for the remainder of this paper. We now present some analytical results for the rate constant kon, i.e., the steady-state value of k(t). Both explicit kon results for model systems and general relations for the CS and IF limits of kon will be given. The analytical results obtained on model systems can serve an important role in validating simulation algorithms (see Sec. 5).
Dual-transition-rates model: Spherical symmetry
In earlier work we introduced the dual-transition-rates model.17 The model has spherical symmetry in r. The transition rates between the open and closed conformations have two sets of values, ω± in the loosely-bound state, where R < r < R + Δ ≡ R1, and ω∞±, when r > R1. The conformation-specific interaction potentials are 0 when r > R1 and constants Uo and Uc, respectively, for the open and closed conformations when R < r < R1. According to Eq. 19c, the two sets of transition rates are constrained by
| (25) |
The reactive region is a very thin shell in R < r < R + ɛ. When ɛ → 0 and τ → 0 but ɛ∕τ has a finite value κ, the sink term is equivalent to a radiation boundary condition19
| (26) |
The rate constant kon for this model is given by17
| (27) |
where
| (28a) |
| (28b) |
| (28c) |
| (28d) |
| (28e) |
| (28f) |
| (28g) |
In the dual-transition-rates model, conformational selection and induced fit are manifest at two extremes of the transition rates. When the transition rates are extremely small, conformational selection appears as the binding mechanism and the rate constant is
| (29) |
where kon0 is the rate constant when the protein stays in the closed (i.e., reactive) conformation, given by
| (30) |
In the opposite extreme, where the transition rates are large, induced fit appears as the binding mechanism, and the rate constant is given by
| (31) |
Note that the last result is equivalent to the rate constant when the protein stays in the reactive conformation but with the reactivity κ replaced by pcκ and the potential Uc replaced by Ueff. The CS and IF limits are lower and upper bounds of kon, respectively.
Dual-transition-rates model: Reactive patch
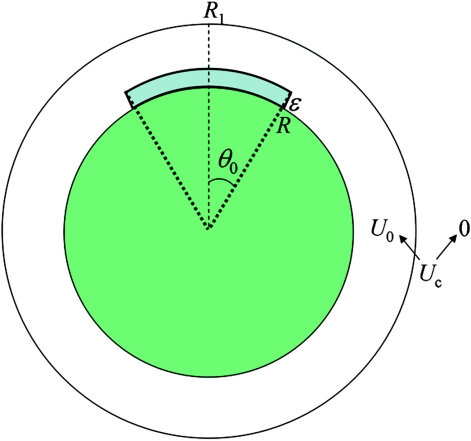
The spherically symmetric dual-transition-rates model can be made more realistic when the reactive region is restricted to a patch, with the polar angle θ limited to the range from 0 to θ0 (Fig. 2). We have not been able to find a general expression for the rate constant of this model. However, the CS and IF limits can be obtained. These results are based on the rate constant for the situation where the protein always stays in the reactive conformation. The latter, denoted as kon0(θ0, κ, U) to signify the dependence on the patch angle θ0, the reactivity κ at r = R, and the potential U in R < r < R1, is given by21
| (32) |
where
| (33a) |
| (33b) |
| (33c) |
with Pl(x) denoting Legendre polynomials. The first term on the right hand side of Eq. 32 gives the rate constant when the reactivity is small, i.e., κR∕D → 0, whereas the second term gives the rate constant when the reactivity is large, i.e., κR∕D → ∞. A similar interpretation applies to the two terms of Eq. 30 or 31.
Figure 2.
The model system for which analytical and simulation results are obtained in the present study.
When θ0 = 180°, Eq. 32 reduces to Eq. 30 with U = Uc. However, for other values of θ0, the derivation of Eq. 32 relies on the so-called constant-flux approximation.22 As θ0 → 0, Eq. 33a is known to underestimate kD(θ0, U) by a factor 32∕3π2 ≈ 1.08. Following our earlier work,21 we scale up this kD(θ0, U) by 32∕3π2 to calculate kon0(θ0, κ, U) when comparison is made against simulation results for small reactive patches (i.e., θ0 ≤ 15°).
In the limit that conformational transitions are extremely slow, the protein cannot change its conformation during the binding process. If the protein started out in the closed (i.e., reactive) conformation, the binding rate constant would be kon0(θ0, κ, Uc). If the protein started out in the open (i.e., nonreactive) conformation, it would not be able to bind with the ligand at all and the binding rate constant would be 0. Now the probability for the protein to start in the closed conformation while the ligand is far away is p∞c. Therefore the binding rate constant is the average over the two starting conformations:
| (34) |
In the limit that conformational transitions are extremely fast, the ligand always experiences an average effective potential, given by Eq. 28d. In the reactive region, the ligand likewise experiences an average reactivity, given by pcκ. The binding rate constant is, thus,
| (35) |
When κR∕D → 0, kCS becomes
| (36) |
which is the initial value k(0) of the time-dependent rate coefficient [see Eq. 22]. In fact, when κR∕D → 0, the rate constant should always be k(0) irrespective of the timescale of the conformational transitions. It can be easily verified that that kIF approaches k(0) when κR∕D → 0.
How do kCS and kIF compare when κR∕D → ∞? For notational simplicity we denote kon0(θ0, κ, U) as kD(θ0, U) when κR∕D → ∞. For a small reactive patch (i.e., θ0 → 0), which models stereospecific ligand binding, an approximate dependence of kD(θ0, U) on U,21, 23
| (37) |
is very useful here. The accuracy of this approximation is also affected by the range (Δ in the present case) of the interaction potential U, improving as Δ increases. Assuming that Eq. 37 holds, then when κR∕D → ∞, kCS becomes
| (38) |
and kIF becomes
| (39) |
Note that the second term of Eq. 39 is the same as kCS of Eq. 38. According to Eq. 25, the ratio of the second term to the first term on the right hand side of Eq. 39 is pc∕po. For systems of biological interest, this ratio should be much greater than 1. Therefore, Eq. 39 is dominated by the second term. We can thus conclude that kCS and kIF are close when κR∕D → ∞. Since kCS and kIF are actually identical when κR∕D → 0, we can further conclude that kCS and kIF are close irrespective of the value of κR∕D, as long as the reactive patch is small.
Stereospecific binding in general
The closeness of kCS and kIF derived for the model with a small reactive patch actually holds in general, as long as the reactive region is small relative to the range of the interaction potential. This is a highly significant result. Because kCS and kIF provide lower and upper bounds of kon, if they are close in value then either of them can provide a good estimate for kon. Below we outline a derivation for the closeness of kCS and kIF, making clear the approximation involved. The derivation given here is for the case where conformational change is treated as rate processes between two discrete conformations. The case where conformational change is treated as continuous diffusion along a one-dimensional coordinate is presented in Appendix B.
When the reactivity is small, both kCS and kIF are identical to k(0). The difference between kCS and kIF is the greatest when the reactivity is big. In that situation, when conformational change is treated as rate processes between two discrete conformations, we have
| (40) |
where kD[Uc(r)] is the rate constant when the protein does not undergo conformational change, the interaction potential is Uc(r), and the reactivity is infinite. Correspondingly kIF approaches
| (41) |
where Ueff(r) is the conformation-averaged effective potential
| (42a) |
| (42b) |
When the reactive region is small relative to the range of the interaction potential U(r), our previous work21, 23 suggests an approximate dependence of kD[U(r)] on U(r):
| (43) |
where kD0 denotes the counterpart of kD[U(r)] when the interaction potential U(r) is turned off, and rRR is a representative point in the reactive region. This approximation holds when the range of the potential U(r) is significantly greater than the size of the reactive region. Applying this approximation, we have
| (44) |
| (45) |
The second term of Eq. 45 is the same as Eq. 44. In addition, we also expect the second term of Eq. 45 to dominate over its first term. The preceding derivation thus shows that, even when the reactivity increases to infinity, where the difference between kCS and kIF is the greatest, these two bounds of kon are close as long as Uo(r) and Uc(r) vary slowly over the reactive region and beyond.
ILLUSTRATION OF THE BROWNIAN DYNAMICS ALGORITHM
In Sec. 2 we described an algorithm for calculating k(t) from Brownian dynamics simulations. This algorithm is adapted from our earlier work19 but now accounts for protein conformational change that is coupled to ligand binding. It consists of the following steps. (i) At the start of each Brownian trajectory, the protein is in the closed, i.e., reactive conformation and the ligand is in the reactive region. The starting position of the ligand follows the Boltzmann distribution exp[−βUc(r)]. (ii) The trajectory is then propagated (conformational transition for the protein and translational diffusion for the ligand). (iii) Whenever the ligand moves into the reactive region while the protein is in the closed conformation, they can react with the rate constant 1∕τ to form the native complex. If the reaction occurs, the trajectory is terminated; otherwise it is propagated to a preset cutoff time tcut. (iv) Finally, after all the trajectories (totaling Ntraj) are finished, the fraction of surviving trajectories at time t < tcut is calculated. That fraction is equal to k(t)∕k(0). Multiplying by k(0) [calculated according to Eq. 22], we obtain k(t). Below we further explain steps (ii) and (iii) of our algorithm and provide details of its implementation on a model system.
Trajectory propagation
Our simulations consist of simultaneous transition of the protein in conformational space and translational diffusion of the ligand in the three-dimensional space exterior to the protein. The translational diffusion follows the Ermak–McCammon algorithm:24
| (46) |
where r0 is the current position, r is the position after timestep Δt, F0 = −∇Ug0(r0) is the force calculated for the current ligand position r0 and the current protein conformation g0, and R is a random vector with components generated from a normal distribution.
The conformational transition of the protein is implemented as follows. Suppose that the current conformation of the protein is open. The probability that, within the timestep Δt, the protein switches to the closed conformation is exp[−(ω+(r0) + ω+(r))Δt∕2]. This probability is compared with a random number uniformly distributed between 0 and 1. If the random number is smaller, then the conformational switch takes place; otherwise the protein stays in the open conformation. The case where the current conformation is closed can be similarly treated.
The reflecting protein surface has to be dealt with. We follow a very simple recipe.19 If the new position of the ligand penetrates inside the protein surface, the ligand is “reflected” back to its current position and, at the same time, the protein is restored to its current conformation. The lifetime is incremented by the timestep, just like in a regular move.
Determination of whether reaction occurs
The probability that, within the timestep Δt, the protein and ligand react to form the native complex is exp[−(i0 + i)Δt∕2τ], where i0 = 1 if the current ligand position is inside the reactive region and the current protein conformation is closed and = 0 otherwise; and i is the counterpart of i0 after the timestep. This probability is compared with a random number uniformly distributed between 0 and 1. If the random number is smaller, then the reaction takes place and the trajectory is terminated; otherwise the trajectory is continued.
Implementation on a model system
As illustration, we implemented the Brownian dynamics algorithm for calculating k(t) on the dual-transition-rates model with a reaction patch, introduced in Subsection III.B (see Fig. 2). The interaction potential Uo for the open conformation is 0 for any r > R. The interaction potential Uc for the closed conformation has the form
| (47) |
which, when L∕Δ → 0, approaches the step-function potential, having value U0 for R < r < R1 and 0 for r > R1, considered in the analytical solutions of Subsections III A and III B. This smooth potential function is much easier to deal with in the Brownian dynamics simulations than the step-function potential. We chose R = 20 Å, R1 = 22 Å, and L = 0.1 Å. When r − R1 ≫ L, Uc(r) is practically 0. To minimize force calculations, we turned off Uc(r) when r > R2 = 23 Å.
We set ω − = ω∞−. There was no particular reason for this choice except to reduce the number of parameters. Then Eq. 19c gives
| (48) |
We set ω∞+∕ω∞− to 0.1, reflecting the fact that the closed, i.e., reactive conformation is disfavored when the ligand is away. The value of ω∞− was varied to explore the full timescale range of the conformational transitions, approaching the CS limit at small ω∞− and the IF limit at large ω∞−. The CS and IF limits were also studied directly. In these simulations, the protein was kept in the closed (i.e., reactive) conformation. The interaction potential was Uc(r) for the CS limit and the effective potential Ueff(r) for the IF limit. For the present model system, exp[−βUeff(r)] = p∞cexp[−βUc(r)] + p∞o. The reactivity was κ and pcκ, respectively, in the CS and IF simulations. The rate constant from the former simulations, after multiplication by p∞c, yields kCS, while the rate constant from the latter simulations directly gives kIF.
The ligand diffusion constant D was chosen to be 10 Å2∕ns. The timestep was Δti = 10−6 ns for r < R2. For r > R2, the timestep was Δti + 10−3(r − R2)2∕2D. The cutoff time for all the simulations was 104 ns.
The reactive region was defined by R < r < R + ɛ and θ < θ0. Corresponding to a reactivity κ, the rate constant for forming the native constant is 1∕τ = κ∕ɛ. We fixed ɛ at 2 Å and studied a range of κ values. When the reactive region covered the full surface (θ0 = 180°), κ was 0.5 Å∕ns; 2 × 104 trajectories were run. For small reactive patches, κ was 5, 50, and 500 Å∕ns; 2 × 104, 4 × 104, and 105 trajectories, respectively, were run.
SIMULATION RESULTS
Here we present simulation results for the rate coefficient of the model system shown in Fig. 2 and compare them with available analytical results. We pay particular attention to how close the CS and IF limits of the rate constant are.
Asymptotic behavior of k(t)
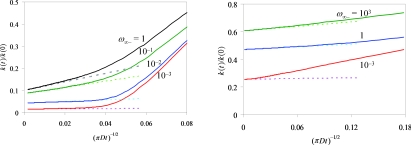
At long times, k(t) is predicted to follow the asymptotic behavior given by Eq. 24 Specifically, k(t) is a linear function of (πDt)−1∕2. Furthermore, the slope is determined by the intercept. All our simulation results conform to this asymptotic behavior. For illustration, in Fig. 3 we show two sets of results, one for the case where the reactive region covers the whole surface of the protein with κ = 0.5 Å∕ns and exp(−βU0) = 100; the other for the case where the reactive region spans up to θ = 10° with κ = 5 Å∕ns and exp(−βU0) = 10. The long-time portion of each k(t)∕k(0) curve can be fitted well to a linear function in (πDt)−1∕2, with the slope m related to the intercept b via m = b2k(0)∕4πD, exactly as Eq. 24 predicts. Form here on we focus on the steady-state rate constant kon, which is given by bk(0).
Figure 3.
The asymptotic behavior of the rate coefficient. The solid curves show simulation results for k(t)∕k(0); the dashed line show fits to a linear function of (πDt)−1∕2. The slope of the linear function is determined from the intercept according to Eq. 24. The values of ω∞−, in units of ns−1, are shown in the figure. Results for two sets of model parameters are given: (a) θ0 = 180°, κ = 0.5 Å∕ns, and exp(−βU0) = 100; (b) θ0 = 10°, κ = 5 Å∕ns, and exp(−βU0) = 10.
Dual-transition-rates model: Spherical symmetry
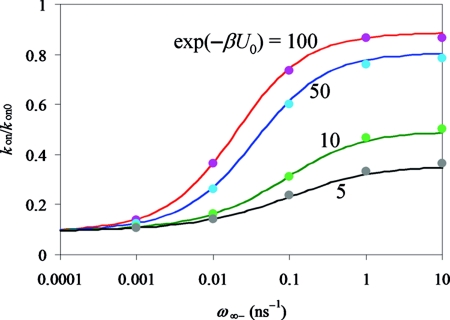
For the case where the reactive region covers the whole surface of the protein, the analytical result for kon is given by Eq. 27. In Fig. 4 we compare the simulation results for kon against the analytical theory. In the simulations, the reactive region has a finite thickness ɛ, but in the theory the thickness approaches 0, while keeping the same reactivity κ. To reduce the effect of this difference for treating the reactive region, we scale both the simulation and analytical results by their respective results for kon0. Figure 4 shows that the simulation results for kon∕kon0 over the full range of ω∞− and a range of U0 are in excellent agreement with the analytical theory.
Figure 4.
Comparison of simulation (symbols) and analytical (curves) results for kon∕kon0. The model parameters are: θ0 = 180°, κ = 0.5 Å∕ns, with values of exp(−βU0) shown in the figure.
At a given U0, as ω∞− increases from 0 to ∞, kon increases from the CS limit to the IF limit. Note that kCS∕kon0 = p∞c. That is why the kon∕kon0 curves for different U0 values all start from the same ω∞− → 0 limit. However, the ω∞− → ∞ limit, kIF∕kon0, increases with increasing |U0|.
Dual-transition-rates model: Reactive patch
We now turn attention to the model with a small reactive patch. In Table 1 we compare our simulation and analytical results on kCS and kIF for patch angles at 3°, 5°, 8°, 10°, and 15°. It can be seen that there is close agreement between simulation and theory. The simulation results do seem to be slightly larger than the analytical results. We attribute these small discrepancies to the different treatments of the reaction region in the simulations and in the theory. The effect of the different treatments is magnified when the reactive patch becomes very small.
Table 1.
Comparison of simulation and theoretical results on kCS and kIF for a range of reactive patch angles.
|
kCS∕k(0) |
kIF∕k(0) |
|||||
|---|---|---|---|---|---|---|
| θ0 ° | exp(−βU0) | κ | Simulation | Theory | Simulation | Theory |
| 3 | 5 | 5 | 0.73 | 0.665 | 0.93 | 0.881 |
| 3 | 10 | 5 | 0.69 | 0.634 | 0.89 | 0.826 |
| 5 | 5 | 5 | 0.55 | 0.516 | 0.87 | 0.817 |
| 5 | 10 | 5 | 0.49 | 0.468 | 0.79 | 0.735 |
| 8 | 5 | 5 | 0.38 | 0.370 | 0.78 | 0.738 |
| 8 | 10 | 5 | 0.31 | 0.314 | 0.67 | 0.629 |
| 10 | 5 | 5 | 0.31 | 0.306 | 0.72 | 0.693 |
| 10 | 10 | 5 | 0.24 | 0.250 | 0.61 | 0.573 |
| 15 | 5 | 5 | 0.20 | 0.208 | 0.62 | 0.605 |
| 15 | 10 | 5 | 0.15 | 0.158 | 0.49 | 0.469 |
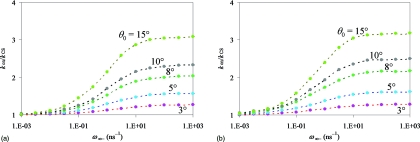
To show the transition of kon from the CS limit to the IF limit and the difference in magnitude between kIF and kCS, in Figs. 56 we display kon∕kCS as a function of ω∞− for a variety of model parameters. Figure 5a presents the results for exp(−βU0) = 5 and κ = 5 Å∕ns. As the patch angle decreases from 15° to 3°, kIF∕kCS reduces from 3.1 to 1.3. This is the main finding of the present study: kIF and kCS becomes close (i.e., kIF∕kCS ≈ 1) as the reactive patch becomes smaller. Figure 5b shows that the closeness of kIF and kCS is affected by the magnitude of the interaction potential. At exp(−βU0) = 10, kIF∕kCS increases slightly to 3.2 for θ0 = 15° and remains at 1.3 for θ0 = 3°.
Figure 5.
Dependence of kon∕kCS on ω∞− for a range of reactive patch angles (shown in the figure). Results for two values of exp(−βU0): (a) 5 and (b) 10 are shown to illustrate that the difference between the CS and IF limits of kon increases as the magnitude of the interaction potential increases. κ = 5 Å∕ns. Data for the same patch angle are connected by dash.
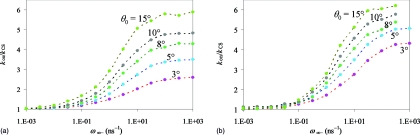
Figure 6.
Dependence of kon∕kCS on ω∞− for a range of reactive patch angles. Results for two values of κ: (a) 50 and (b) 500 Å∕ns are shown to illustrate that the difference between the CS and IF limits of kon increases as the reactivity increases. exp(−βU0) = 5.
Figure 6 further shows that, with exp(−βU0) fixed at 5, kIF∕kCS increases as the reactivity κ increases. At κ = 50, kIF∕kCS changes from 5.8 to 2.6 as the patch angle decreases from 15° to 3°. In comparison, at κ = 500, kIF∕kCS changes from 6.3 to 4.3 as the patch angle decreases from 15° to 3°.
The simulation results presented here are for the extreme case where the range of the interaction potential is the same as the thickness of the reactive region. The analytical results of Subsections III B and III C show that the approximation kIF∕kCS ≈ 1 improves when the range of the interaction potential widens beyond the reactive region.
DISCUSSION
In the present study we have introduced a general model for protein–ligand binding in which the energy surface of the protein in conformational space is coupled to the binding of its ligand. Two common features of most protein–ligand systems are that the energy surface switches from favoring nonreactive conformations while the ligand is away to favoring reactive conformations while the ligand is near and the reactive region is highly localized.25, 26 We have also presented a method for calculating the binding rate constant from Brownian dynamics simulations.
For a given energy surface, the timescale of the conformational transitions may affect the binding mechanism and the binding rate constant. The binding mechanism appears as conformational selection when conformational transitions are slow relative to the time required for the diffusional approach of the protein–ligand pair, and gradually shifts to manifestly induced fit when the rate of conformational transition increases. The CS and IF limits provide lower and upper bounds, respectively, for the binding rate constant. The difference between kIF and kCS increases as the reactivity increases. At infinite reactivity, we have derived the general result,
| (49) |
where the reactive region is assumed to be small relative to the range of the interaction potential, and pc(rRR) is the equilibrium probability of the closed (i.e., reactive) conformation in the reactive region. For systems of actual interest, when the ligand is in the reactive region, the equilibrium probability for the reactive conformation is much higher than that for the nonreactive conformation, i.e., pc(rRR) → 1. Hence kIF∕kCS is close to 1. Our simulation results on a model with a small reactive patch confirm (in fact, motivated the analytical derivation of) the closeness of kIF and kCS.
The closeness of kIF and kCS in our model is in contrast to the relation between the limits of the rate constant under fast and slow gating in the model of Szabo et al.4 In their model the energy surface of the protein is unaffected by ligand binding. At infinite reactivity, their model predicts kfast∕kslow → 1∕p∞c, where p∞c is the equilibrium probability for the reactive conformation when the ligand is away. For systems of actual interest, one expects that p∞c → 0; hence kfast ≫ kslow in the model of Szabo et al. Our model in fact assumes a low equilibrium probability (i.e., p∞c → 0) for the protein being in the reactive conformation when the ligand is away. However, when near the protein, intermolecular interactions place the ligand on the Uc(r) potential, which facilitates the diffusion of the ligand toward the binding site and therefore enhances the binding rate constant. Consequently, the difference between the limits (kCS and kIF) of the rate constant under slow and fast conformational transitions is significantly reduced.
Because kCS and kIF are lower and upper bounds of the binding rate constant kon, when they are close they provide good estimates of kon. This is significant because the calculation of kon involves expensive simulations of protein conformational fluctuation while running the Brownian dynamics simulations of the ligand translational motion. In contrast, for the calculations of both kCS and kIF, the ligand experiences a fixed interaction potential and hence no simulation of protein conformational fluctuation is necessary. For kCS, the interaction potential is Uc(r) as the protein is fixed in the reactive conformation. For kIF, the interaction potential is Ueff(r) as it is assumed that, for each give ligand position r, the protein instantaneously adopts an equilibrium distribution in conformational space. The closeness of kCS and kIF and how good estimates they provide for kon will be investigated in the future with realistic representations of protein–ligand systems.
If conformational selection and induced fit can achieve similar rate constants, is it still significant to distinguish the two binding mechanisms? If so, what kinetic experiments can ascertain the binding mechanism to be one but not the other? These mechanistic questions remain to be addressed.
Finally, we note that the formulation of k(t) depends on a single protein–ligand pair. As we pointed out in Subsection II A, k(t) is unaffected whether the conformational coordinate x refers to the protein or the ligand. That is, the same k(t) is obtained whether it is the protein or the ligand that undergoes conformational change. However, in experiments, rate coefficients can be measured only when many protein–ligand pairs are present. Consider the case where one species is in excess (with concentration C) so that the binding kinetics is pseudo-first-order. If the ligand is the species that is both in excess and undergoing conformational change, then the many ligand molecules around a single protein molecule are independent. The probability that the protein escapes binding with all the ligand molecules is27
| (50) |
However, when the ligand is in excess but the protein undergoes conformational change (or vice versa), then Eq. 50 is no longer valid, except in the limit of fast conformational change. That is because now the ligand molecules are no longer independent: they experience the same change in reaction condition at the same time whenever the protein goes through a conformational transition. Therefore one has to deal with a many-body problem. For the situation where conformational change is uncoupled to ligand binding, Zhou and Szabo27 have derived an approximate approach for calculating the many-body S(t), with k(t) from the pair problem as input. When conformational change is coupled to ligand binding we anticipate that the many-body binding kinetics will have much richer features. In particular, using analyses based on ordinary chemical kinetics, Hammes et al.28 proposed that increasing ligand concentration can shift the binding mechanism at the many-body level from conformational selection to induced fit. We plan to investigate the many-body problem in the future.
ACKNOWLEDGMENTS
This work was supported in part by Grant No. GM58187 from the National Institutes of Health (NIH).
APPENDIX A: GOVERNING EQUATION FOR DISCRETE CONFORMATIONS
Here we derive Eqs. 18a, 18b, the governing equation for the pair distribution function when the transitions between the two conformations of the protein are treated as rate processes. To find Po(r, t), we integrate both sides of Eq. 5 over x from −∞ to x‡(r), obtaining
| (A1) |
The third term on the right hand side is absent because H(r, x) is zero when x < x‡(r) (formation of the native complex requires that the protein be in the closed, i.e., reactive conformation). Since we assume fast equilibration within the open energy well, we have
| (A2) |
where Poeq(x|r) is the equilibrium distribution for x in x < x‡(r) with a given r:
| (A3) |
Combining the last two equations, we find
| (A4) |
The first term on the right hand side of Eq. A1 now becomes
Now the integration over x is dominated by the region around x = xo(r), and hence the precise value of the upper bound of the integration is unimportant; therefore we may switch the order of the integration over x and the gradient operator on r, leading to
To treat the second term on the right hand side of Eq. A1, we follow the well-known reduction of the diffusive barrier crossing problem to a rate-equation description. The second term is essentially the rate of change in the probability of the inactive conformation. In the rate-equation description, this is
where ω±(r) are the rate constants introduced in Eq. 14a, 14b. Combining the results for the two terms, we find
| (A5) |
The equation for the distribution function Pc(r, t) when the protein is in the closed conformation can be similarly derived. Now the integration over x is from x‡(r) to ∞ and we need to assume
| (A6) |
where
| (A7) |
In the closed conformation, formation of the native complex can occur when the ligand is in the reactive region. Correspondingly the third term on the right hand side of Eq. 5 yields
where
| (A8) |
Finally, the equation for Pc(r, t) is
| (A9) |
APPENDIX B: CLOSENESS OF kCS AND kIF FOR CONTINUOUS CONFORMATIONS
Here we show that kCS and kIF are close when conformational change is treated as continuous diffusion along a one-dimensional coordinate, provided that the reactive region is small relative to the range of the interaction potential. As in the main text, we use rRR to denote a representative point in the reactive region. When the ligand is in the reactive region, the closed conformation of the protein is specified by hc(x) = 1. We focus the calculation of kCS and kIF on an infinite reactivity, since this is when the difference between them is the greatest.
In calculating kCS, one first assumes that the protein is fixed in conformation x; the resulting rate constant is then averaged over the equilibrium distribution, Peq(x), of x. For a fixed protein conformation x, the interaction potential that the ligand experiences is ΔU(r|x) ≡ U(r, x) − U∞(x), where the subtraction by U∞(x) is made so that the interaction potential approaches 0 at infinite protein–ligand separation. When the reactivity is infinite, we have
| (B1) |
where hc(x) is inserted to indicate the fact that binding occurs only if the protein starts from a closed conformation [as specified by hc(x) = 1]; and kD[ΔU(r|x)] is the rate constant when the protein conformation is fixed at x, the interaction potential is ΔU(r|x), and the reactivity is infinite. For calculating kIF, the ligand experiences the conformation-averaged effective potential Ueff(r) given by Eq. 42a. When the reactivity is infinite,
| (B2) |
where kD[Ueff(r)] is the rate constant when the protein does not under conformational change, the interaction potential is Ueff(r), and the reactivity is infinite.
To proceed further, we now use the approximate dependence of the kD[U(r)] on the interaction potential U(r), given by Eq. 43. Using this approximation, Eq. B1 becomes
| (B3) |
where we have used the expression for Peq(x) given by Eq. 6. On the other hand, Eq. B2 becomes
| (B4) |
Note that the first term is the same as Eq. B3. In addition, we also expect the first term of Eq. B4 to dominate over its second term, since the closed (i.e., reactive) conformation should dominate over the open (i.e., nonreactive) conformation when the ligand is in the reactive region. Therefore, kCS and kIF are close even for an infinite reactivity as long as the interaction potential varies slowly over the reactive region and beyond.
References
- Alsallaq R. and Zhou H.-X., Proteins 71, 320 (2008). 10.1002/prot.21679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Northrup S. H., Allison S. A., and McCammon J. A., J. Chem. Phys. 80, 1517 (1984). 10.1063/1.446900 [DOI] [Google Scholar]
- Zhou H. X., Q. Rev. Biophys. 43, 219 (2010). 10.1017/S0033583510000120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo A., Shoup D., Northrup S. H., and McCammon J. A., J. Chem. Phys. 77, 4484 (1982). 10.1063/1.444397 [DOI] [Google Scholar]
- Agmon N., J. Theor. Biol. 113, 711 (1985). 10.1016/S0022-5193(85)80188-0 [DOI] [PubMed] [Google Scholar]
- Zhou H.-X., J. Chem. Phys. 108, 8146 (1998). 10.1063/1.476255 [DOI] [Google Scholar]
- Zhou H.-X. and Szabo A., Biophys. J. 71, 2440 (1996). 10.1016/S0006-3495(96)79437-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade R. C., Davis M. E., Luty B. A., Madura J. D., and McCammon J. A., Biophys. J. 64, 9 (1993). 10.1016/S0006-3495(93)81335-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters G. H., Olsen O. H., Svendsen A., and Wade R. C., Biophys. J. 71, 119 (1996). 10.1016/S0006-3495(96)79207-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C. E., Shen T., Trylska J., Tozzini V., and McCammon J. A., Biophys. J. 90, 3880 (2006). 10.1529/biophysj.105.074575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorfe A. A., Chang C. E., Ivanov I., and McCammon J. A., Biophys. J. 94, 1144 (2008). 10.1529/biophysj.107.117879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swift R. V. and McCammon J. A., J. Am. Chem. Soc. 131, 5126 (2009). 10.1021/ja808064g [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H. X., Wlodek S. T., and McCammon J. A., Proc. Natl. Acad. Sci. U.S.A 95, 9280 (1998). 10.1073/pnas.95.16.9280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorfe A. A., Lu B., Yu Z., and McCammon J. A., Biophys. J. 97, 897 (2009). 10.1016/j.bpj.2009.05.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade R. C., Luty B. A., Demchuk E., Madura J. D., Davis M. E., Briggs J. M., and McCammon J. A., Nat. Struct. Biol. 1, 65 (1994). 10.1038/nsb0194-65 [DOI] [PubMed] [Google Scholar]
- Kim J. H. and Lee S., J. Chem. Phys. 131, 014503 (2009). 10.1063/1.3158469 [DOI] [PubMed] [Google Scholar]
- Zhou H.-X., Biophys. J. 98, L15 (2010). 10.1016/j.bpj.2009.11.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smoluchowski M. V., Z. Phys. Chem. 92, 129 (1917). [Google Scholar]
- Zhou H.-X., J. Phys. Chem. 94, 8794 (1990). 10.1021/j100388a010 [DOI] [Google Scholar]
- Kramers H. A., Physica 7, 284 (1940). 10.1016/S0031-8914(40)90098-2 [DOI] [Google Scholar]
- Zhou H. X., Biophys. J. 73, 2441 (1997). 10.1016/S0006-3495(97)78272-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoup D., Lipari G., and Szabo A., Biophys. J. 36, 697 (1981). 10.1016/S0006-3495(81)84759-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H.-X., J. Chem. Phys. 105, 7235 (1996). 10.1063/1.472530 [DOI] [Google Scholar]
- Ermak D. L. and McCammon J. A., J. Chem. Phys. 69, 1352 (1978). 10.1063/1.436761 [DOI] [Google Scholar]
- Eletsky A., Kienhofer A., Hilvert D., and Pervushin K., Biochemistry 44, 6788 (2005). 10.1021/bi0474259 [DOI] [PubMed] [Google Scholar]
- Sullivan S. M. and Holyoak T., Proc. Natl. Acad .Sci. U.S.A. 105, 13829 (2008). 10.1073/pnas.0805364105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H.-X. and Szabo A., J. Phys. Chem. 100, 2597 (1996). 10.1021/jp952376i [DOI] [Google Scholar]
- Hammes G. G., Chang Y. C., and Oas T. G., Proc. Natl. Acad. Sci. U.S.A. 106, 13737 (2009). 10.1073/pnas.0907195106 [DOI] [PMC free article] [PubMed] [Google Scholar]