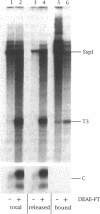
Abstract
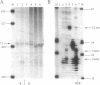
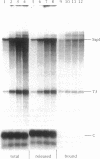
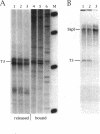
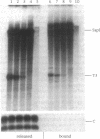
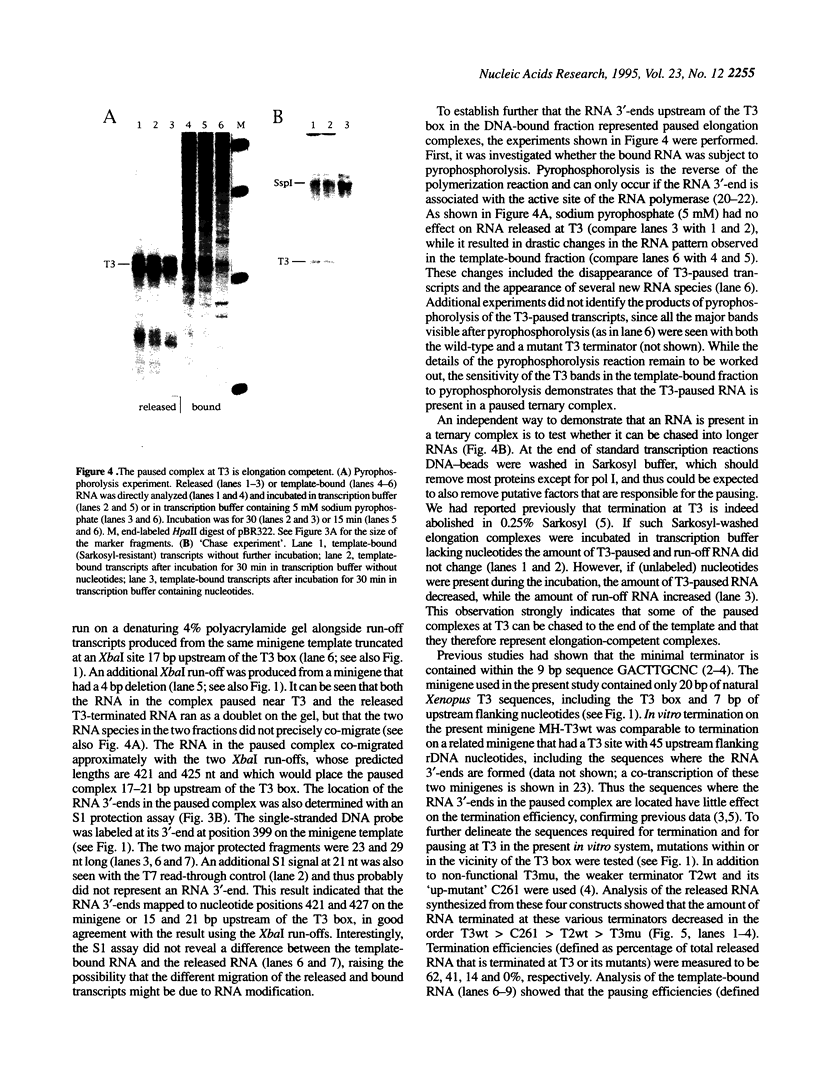
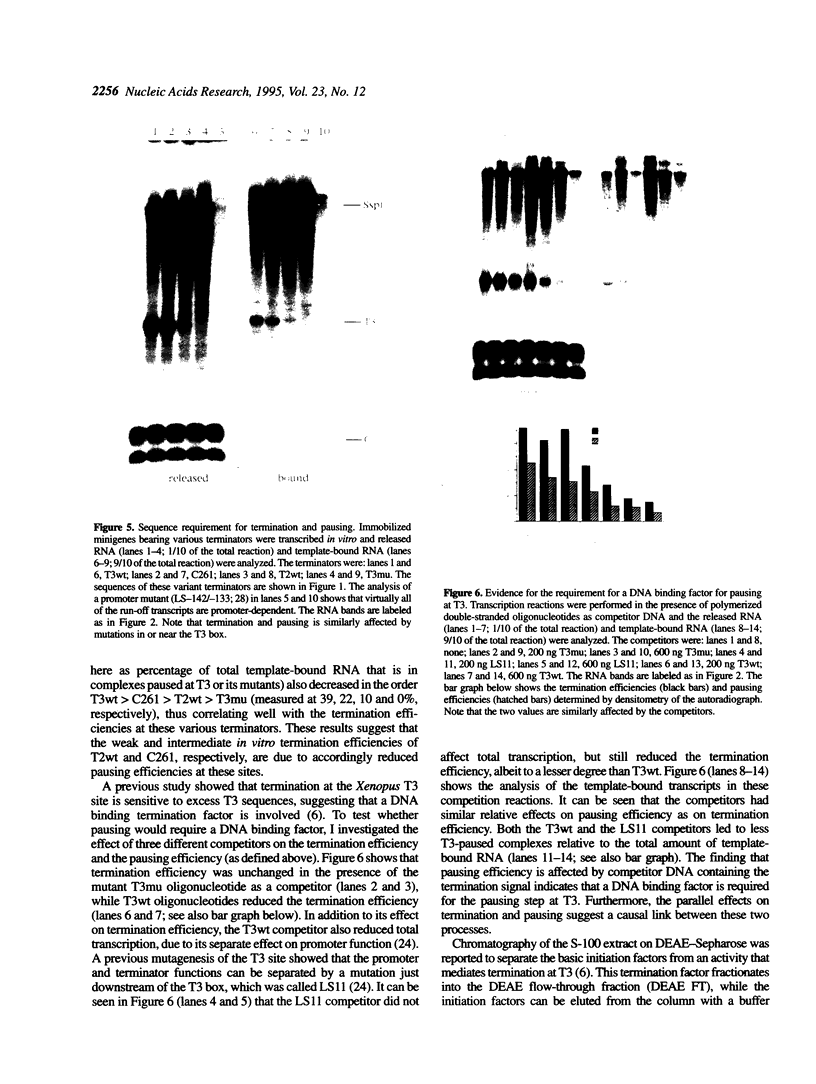
In Xenopus, termination by RNA polymerase I (pol I) is mediated by the 9 bp sequence GACTTGCNC and RNA 3'-ends are formed -15-20 nt upstream of this terminator element. Here I show that this 9 bp element, also called the 'T3 box', is a pause signal for the elongating transcription complex. The two major transcripts in the paused complex have 3'-ends mapping to 15 and 21 nt upstream of the T3 box, remain bound to the template in 0.25% Sarkosyl, are subject to pyrophosphorolysis and can be chased into longer transcripts. Mutations that reduce overall termination also affect pausing, indicating that pausing is a limiting step in the termination process. Oligonucleotide competition experiments, furthermore, suggest that pausing requires a DNA binding factor. The data support a model in which the first step leading to transcription termination by pol I in Xenopus is pausing of the elongation complex upstream of the T3 box.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bartsch I., Schoneberg C., Grummt I. Purification and characterization of TTFI, a factor that mediates termination of mouse ribosomal DNA transcription. Mol Cell Biol. 1988 Sep;8(9):3891–3897. doi: 10.1128/mcb.8.9.3891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell F. E., Jr, Setzer D. R. Transcription termination by RNA polymerase III: uncoupling of polymerase release from termination signal recognition. Mol Cell Biol. 1992 May;12(5):2260–2272. doi: 10.1128/mcb.12.5.2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Winter R. F., Moss T. The ribosomal spacer in Xenopus laevis is transcribed as part of the primary ribosomal RNA. Nucleic Acids Res. 1986 Aug 11;14(15):6041–6051. doi: 10.1093/nar/14.15.6041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggermont J., Proudfoot N. J. Poly(A) signals and transcriptional pause sites combine to prevent interference between RNA polymerase II promoters. EMBO J. 1993 Jun;12(6):2539–2548. doi: 10.1002/j.1460-2075.1993.tb05909.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evers R., Smid A., Rudloff U., Lottspeich F., Grummt I. Different domains of the murine RNA polymerase I-specific termination factor mTTF-I serve distinct functions in transcription termination. EMBO J. 1995 Mar 15;14(6):1248–1256. doi: 10.1002/j.1460-2075.1995.tb07108.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firek S., Read C., Smith D. R., Moss T. The Xenopus laevis ribosomal gene terminator contains sequences that both enhance and repress ribosomal transcription. Mol Cell Biol. 1989 Sep;9(9):3777–3784. doi: 10.1128/mcb.9.9.3777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W., Powell W., Mote J., Jr, Reines D. Nascent RNA cleavage by arrested RNA polymerase II does not require upstream translocation of the elongation complex on DNA. J Biol Chem. 1993 Dec 5;268(34):25604–25616. [PMC free article] [PubMed] [Google Scholar]
- Izban M. G., Luse D. S. Transcription on nucleosomal templates by RNA polymerase II in vitro: inhibition of elongation with enhancement of sequence-specific pausing. Genes Dev. 1991 Apr;5(4):683–696. doi: 10.1101/gad.5.4.683. [DOI] [PubMed] [Google Scholar]
- Kuhn A., Grummt I. 3'-end formation of mouse pre-rRNA involves both transcription termination and a specific processing reaction. Genes Dev. 1989 Feb;3(2):224–231. doi: 10.1101/gad.3.2.224. [DOI] [PubMed] [Google Scholar]
- Labhart P. Identification of two steps during Xenopus ribosomal gene transcription that are sensitive to protein phosphorylation. Mol Cell Biol. 1994 Mar;14(3):2011–2020. doi: 10.1128/mcb.14.3.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labhart P. Negative and positive effects of CpG-methylation on Xenopus ribosomal gene transcription in vitro. FEBS Lett. 1994 Dec 19;356(2-3):302–306. doi: 10.1016/0014-5793(94)01291-1. [DOI] [PubMed] [Google Scholar]
- Labhart P., Reeder R. H. A 12-base-pair sequence is an essential element of the ribosomal gene terminator in Xenopus laevis. Mol Cell Biol. 1987 May;7(5):1900–1905. doi: 10.1128/mcb.7.5.1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labhart P., Reeder R. H. A point mutation uncouples RNA 3'-end formation and termination during ribosomal gene transcription in Xenopus laevis. Genes Dev. 1990 Feb;4(2):269–276. doi: 10.1101/gad.4.2.269. [DOI] [PubMed] [Google Scholar]
- Labhart P., Reeder R. H. Characterization of three sites of RNA 3' end formation in the Xenopus ribosomal gene spacer. Cell. 1986 May 9;45(3):431–443. doi: 10.1016/0092-8674(86)90329-6. [DOI] [PubMed] [Google Scholar]
- Labhart P., Reeder R. H. Functional difference between the sites of ribosomal 40S precursor 3' end formation in Xenopus laevis and Xenopus borealis. Nucleic Acids Res. 1990 Sep 11;18(17):5271–5277. doi: 10.1093/nar/18.17.5271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang W. H., Morrow B. E., Ju Q., Warner J. R., Reeder R. H. A model for transcription termination by RNA polymerase I. Cell. 1994 Nov 4;79(3):527–534. doi: 10.1016/0092-8674(94)90261-5. [DOI] [PubMed] [Google Scholar]
- Lang W. H., Reeder R. H. The REB1 site is an essential component of a terminator for RNA polymerase I in Saccharomyces cerevisiae. Mol Cell Biol. 1993 Jan;13(1):649–658. doi: 10.1128/mcb.13.1.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McStay B., Reeder R. H. A DNA-binding protein is required for termination of transcription by RNA polymerase I in Xenopus laevis. Mol Cell Biol. 1990 Jun;10(6):2793–2800. doi: 10.1128/mcb.10.6.2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McStay B., Reeder R. H. An RNA polymerase I termination site can stimulate the adjacent ribosomal gene promoter by two distinct mechanisms in Xenopus laevis. Genes Dev. 1990 Jul;4(7):1240–1251. doi: 10.1101/gad.4.7.1240. [DOI] [PubMed] [Google Scholar]
- Nudler E., Goldfarb A., Kashlev M. Discontinuous mechanism of transcription elongation. Science. 1994 Aug 5;265(5173):793–796. doi: 10.1126/science.8047884. [DOI] [PubMed] [Google Scholar]
- Proudfoot N. J. How RNA polymerase II terminates transcription in higher eukaryotes. Trends Biochem Sci. 1989 Mar;14(3):105–110. doi: 10.1016/0968-0004(89)90132-1. [DOI] [PubMed] [Google Scholar]
- Reeder R. H., Labhart P., McStay B. Processing and termination of RNA polymerase I transcripts. Bioessays. 1987 Mar;6(3):108–112. doi: 10.1002/bies.950060304. [DOI] [PubMed] [Google Scholar]
- Reeder R. H., Pennock D., McStay B., Roan J., Tolentino E., Walker P. Linker scanner mutagenesis of the Xenopus laevis ribosomal gene promoter. Nucleic Acids Res. 1987 Sep 25;15(18):7429–7441. doi: 10.1093/nar/15.18.7429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reines D., Ghanouni P., Li Q. Q., Mote J., Jr The RNA polymerase II elongation complex. Factor-dependent transcription elongation involves nascent RNA cleavage. J Biol Chem. 1992 Aug 5;267(22):15516–15522. [PMC free article] [PubMed] [Google Scholar]
- Rudd M. D., Izban M. G., Luse D. S. The active site of RNA polymerase II participates in transcript cleavage within arrested ternary complexes. Proc Natl Acad Sci U S A. 1994 Aug 16;91(17):8057–8061. doi: 10.1073/pnas.91.17.8057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnapp A., Grummt I. Transcription complex formation at the mouse rDNA promoter involves the stepwise association of four transcription factors and RNA polymerase I. J Biol Chem. 1991 Dec 25;266(36):24588–24595. [PubMed] [Google Scholar]
- Wang D., Hawley D. K. Identification of a 3'-->5' exonuclease activity associated with human RNA polymerase II. Proc Natl Acad Sci U S A. 1993 Feb 1;90(3):843–847. doi: 10.1073/pnas.90.3.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Hippel P. H., Yager T. D. The elongation-termination decision in transcription. Science. 1992 Feb 14;255(5046):809–812. doi: 10.1126/science.1536005. [DOI] [PubMed] [Google Scholar]