Abstract
Escherichia coli SoxS activates transcription of the genes of the soxRS regulon, which provide the cell's defense against oxidative stress. In response to this stress, SoxS is synthesized de novo. Because the DNA binding site of SoxS is highly degenerate, SoxS efficiently activates transcription by the mechanism of prerecruitment. In prerecruitment, newly synthesized SoxS first forms binary complexes with RNA polymerase. These complexes then scan the chromosome for class I and class II SoxS-dependent promoters, using the specific DNA-recognition properties of SoxS and σ70 to distinguish SoxS-dependent promoters from the vast excess of sequence-equivalent soxboxes that do not reside in promoters. Previously, we determined that SoxS interacts with RNA polymerase in two ways, by making protein-protein interactions with the DNA-binding determinant of the α subunit and by interacting with σ70 region 4 (σ70 R4) both “on-DNA” and “off-DNA”. Here, we address the question of how SoxS and σ70 R4 co-exist at class II promoters, where the binding site for SoxS either partially or completely overlaps the -35 region of the promoter, which is usually bound by σ70 R4. To do so, we created a tri-alanine scanning library that covers all of σ70 R4. We determined that interactions between σ70 R4 and the DNA in the promoter's −35 region are required for activation of class I promoters, where the binding site lies upstream of the −35 hexamer, but they are not required at class II promoters. In contrast, specific three-amino acid stretches are required for activation of class I (lac) and class II (galP1) cyclic AMP receptor protein-dependent promoters. We conclude from these data that SoxS and σ70 R4 interact with each other in a novel way at class II SoxS-dependent promoters such that the two proteins do not accommodate one another in the −35 region but instead SoxS binding there occludes the binding of σ70 R4.
Keywords: SoxRS regulon, genetic epistasis, protein-DNA interactions, pre-recruitment
Introduction
In Escherichia coli, transcription is carried out by a single, DNA-dependent RNA polymerase holoenzyme (RNAP) which is comprised of a σ factor that specifies transcription initiation and a core set of five proteins (α2, β, β′, ω) that carries out transcription elongation. Although E. coli encodes seven σ factors, most transcription during exponential growth is dependent on the σ70 factor, whose binding to the core is a multistep and cooperative process1 2. σ70 cannot bind DNA by itself3.
Members of the σ70 family share four regions of amino acid sequence homology (regions 1–4), which are also conserved in structure and function4–10, Among the four σ70 domains, region 4 (σ70 R4), and in particular its 4.2 subdomain, play a predominant role in binding of the σ70 subunit to the −35 promoter element. Interactions between a hydrophobic pocket in σ70 R4 and a hydrophobic patch on the flap-tip helix of the β subunit are required to position region 4.2 properly11. These interactions also appear to stabilize the contacts between σ70 R4 and the Zn2+-binding domain (ZBD) of the β' subunit, which may further promote binding of subdomain 4.2 to the −35 element10–12. Many transcriptional activators are known to bind to, or close to, the −35 element and then recruit RNAP through interactions with σ70 R413. Conversely, some bacterial and bacteriophage-encoded proteins, like Rsd and AsiA, are known to repress RNAP activity by binding directly to σ70 R4 and other regions of σ70 involved in contacting the −35 element14–19. To date, however, no transcription factor has been reported that occludes the binding of RNAP to the −35 element during transcription activation.
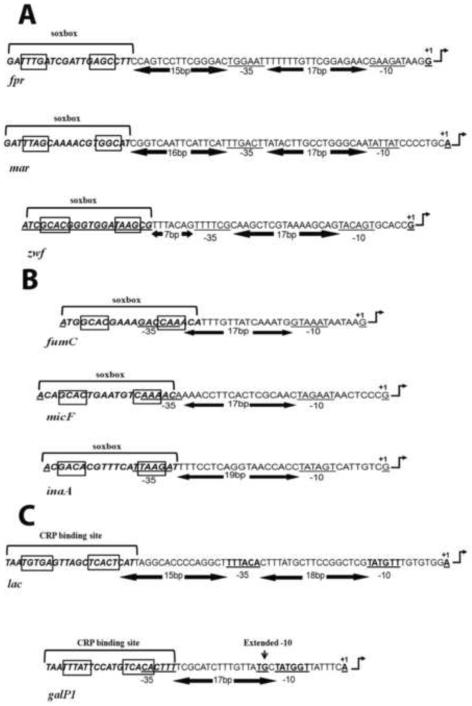
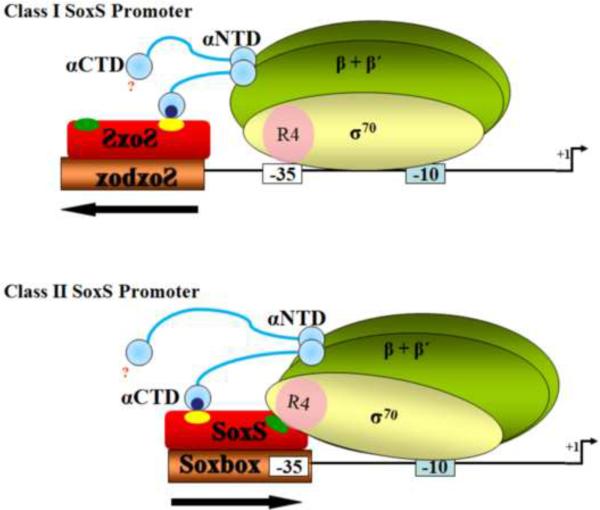
SoxS, a member of the AraC/XylS family of bacterial regulatory proteins20, is the direct transcription activator of the genes of the SoxRS regulon,21–25, whose products provide the cell's defense against the oxidative stress imposed by redox cycling compounds26 as well as other environmental threats such as antibiotics, heavy metals, organic solvents, bile salts and long chain fatty acids21; 24; 27–35. The promoters of the genes whose transcription is activated by SoxS fall into two classes22; 36; 37 (Figure 1): the SoxS binding site of class I promoters (e.g., fpr and mar) lies 15–16 bp or 26–27 bp upstream of the −35 promoter hexamer and in the “backward” orientation whereas in class II promoters (e.g., fumC and inaA) the binding site overlaps the −35 region of the promoter and lies in the “forward” direction36; 37.
Figure 1.
DNA sequences of the SoxS-dependent and CRP-dependent promoters used in this study and location of their activator binding sites and promoter elements. The sequence of the non-template strand of each promoter is shown. The −10 hexamer of each promoter is underlined and in bold. The −35 hexamers of the canonical SoxS-dependent class I promoters fpr and mar are underlined, as is the −35 hexamer of the CRP-dependent class I lac promoter. The −35 regions of the zwf promoter, the class II SoxS-dependent promoters fumC, micF and inaA and the class II CRP-dependent galP1 promoter are also underlined and in bold. The number of bp between the two promoter elements is given between opposing arrows, as is the number of bp between the – 35 element and the activator binding site of the fpr, mar, zwf and lac promoters. The binding sites for SoxS and CRP are enclosed within brackets. (A) The sequences of the canonical SoxS-dependent class I promoters fpr and mar and the non-canonical class I zwf promoter. The binding sites of SoxS of both class I and class II promoters contain two conserved recognition elements, RE1 and RE2, which are enclosed within boxes and an “invariant A” at position 1, which is not enclosed41. Note, then, that position 1 of the soxbox of canonical class I promoters is at the promoter-proximal end of the binding site, with RE1 lying four bp upstream. (B) The sequences of the canonical class II SoxS-dependent promoters fumC, micF and inaA. Here, the invariant A at position 1 of these class II promoters is at the promoter-distal end of the soxbox, with RE1 lying four bp downstream. (C) The sequences of a class I (lac) and a class II (galP1) CRP-dependent promoter. The two most conserved elements of the CRP binding sites are enclosed within boxes.
SoxS is endowed with several features that distinguish it from most other bacterial transcription activators. Thus, SoxS functions as a monomer38 and binds to a highly degenerate 20 bp DNA sequence36; 37; 39–42. Moreover, SoxS belongs to a non-canonical two-component system that functions in two stages43; 44. In this system, dimeric SoxR plays the role of a sensor-transmitter in that its 2Fe-2S centers are reduced during normal growth and become oxidized during oxidative stress45–48. This oxidation activates constitutively expressed SoxR49, which then activates the transcription of soxS. The ensuing transcription of soxS leads to the de novo synthesis of SoxS, which, as the response-regulator, then activates the transcription of the genes of the SoxRS regulon.
The mechanism of SoxS-dependent transcription activation of the regulon's genes is also unusual: it occurs by pre-recruitment50; 51, which is also called DNA-scanning52. In prerecruitment, newly synthesized SoxS molecules first form binary complexes with RNAP in solution and off DNA and then these complexes scan the chromosome for SoxS-dependent promoters that contain promoter elements recognized by σ70 and a properly positioned binding site for SoxS. The physiological advantage of this mechanism is that it enables newly synthesized molecules of SoxS to distinguish the degenerate soxboxes that reside in SoxS-dependent promoters from the excess of sequence-equivalent, non-functional soxboxes that do not lie in promoters50; 51.
Although the crystal structure of SoxS has not been determined, the protein has been characterized extensively at the molecular level. Thus, by subjecting SoxS to a comprehensive alanine scanning mutagenesis, the plate phenotypes of the mutants allowed the identification of two classes of mutants: some were defective in specific DNA binding while others were positive control (PC) mutants, which bound soxbox DNA normally but were defective in transcription activation of SoxS-dependent promoters53. Importantly, the two types of mutants could be located on a three-dimensional model of SoxS that was based on the co-crystal structure of MarA, a paralog of SoxS, bound to mar DNA54. The substitutions that confer defects in DNA binding were located within the recognition helices of the two helix-turn-helix (HTH) DNA binding motifs of SoxS53. Interestingly, the PC mutants located near but above the HTH motif in the N-terminal region of the protein are defective in SoxS-dependent activation of both class I and class II promoters whereas the PC mutants located near but above the HTH motif in the C-terminal region are only defective in SoxS-dependent activation of class II promoters53.
Moreover, a systematic mutagenesis of the soxbox of two class I promoters allowed the determination of the optimal binding site of SoxS, which contains: an A residue at position 1; recognition element 1 (RE1) with the sequence GCAC at positions 4–7; an A/T rich spacer at positions 8–14; recognition element 2 (RE2) with the sequence CAAA at positions 15–18; and two remaining bp with no information content but necessary for DNA binding in vitro36; 41.
As an “ambidextrous” transcription activator that activates transcription of both class I and class II promoters22, SoxS was expected to make protein-protein interactions with at least two surfaces of RNAP. Indeed, in vivo and in vitro evidence has been presented that SoxS contacts both the C-terminal domain (CTD) of the α subunit of RNAP and σ70 R4. Using a library of single alanine substitutions in the α-CTD from positions 255 to 329, we determined that alanine substitution of 10 amino acids reduce or enhance transcription activation of class I and/or class II promoters55 (K.L. Griffith, T.I. Wood and R.E.W., Jr., unpublished results). Moreover, the interactions found between SoxS and the α-CTD were shown to occur in solution and in the absence of specific DNA binding, interactions required for pre-recruitment50; 52. These interactions were the first examples of an activator binding to this portion of the α-CTD. .
In addition, we recently identified seven single alanine substitutions of the C-terminal tail of σ70 R4 that reduce SoxS-dependent transcription activation of either the class II promoters fumC and micF or the non-canonical zwf promoter. As expected, none of the σ70 R4 substitutions reduced activation of the canonical class I promoter, fpr, because its soxbox is too far (15 bp) from σ70 R4 to interact with it56. Furthermore, we determined that SoxS and σ70 R4 also interact in solution in the absence of specific DNA binding and that amino acids of the class I/II surface of SoxS are required for these “off-DNA” interactions56. In conclusion, these experiments provide evidence that the class I/II surface of SoxS makes protein-protein contacts with domains of two different subunits of RNAP, the α-CTD and σ70 R4, with some interactions likely to occur off-DNA and others on-DNA and/or to be dependent on the specific promoter being activated55; 56.
Since RE2 of the soxboxes of class II promoters (fumC, inaA, micF) either partially or completely overlaps the −35 hexamers, we wished to determine whether SoxS and σ70 can co-occupy this region of a promoter. To answer this question, we constructed a library of tri-alanine substitutions of σ70 R4 covering positions 531–590 and determined the effects of the substitutions on transcription at these class II SoxS-dependent promoters. The data obtained suggest that the binding of SoxS to a soxbox that overlaps the −35 hexamer occludes the binding of σ70 R4 to the −35 element.
We also conducted experiments to determine whether the position and orientation of SoxS on promoter DNA are similar to those of MarA and Rob. We found that the interactions between SoxS and promoter DNA is similar to the interactions observed in the crystal structure of MarA bound to mar DNA54. Thus, by determining the specific position of SoxS on soxbox DNA, these data provide further support for the ability of SoxS to interfere with the binding of σ70 R4 to the −35 hexamer, as inferred from the experiments with the tri-alanine substitutions of σ70 R4.
Results
Effect of tri-alanine substitutions of σ70 R4 on transcription from SoxS-dependent promoters
Previous work has shown that SoxS and σ70 interact with one another and that amino acids in the distal portion of region 4.2 (residues 590–600) and the C-terminal tail (residues 591–612) are important for SoxS-dependent transcription activation of class II promoters fumC and micF and the non-canonical class I zwf promoter but not the class I fpr promoter56. Here, we investigated whether amino acids of σ70 that reside in the distal portion of region 3.2 (residues 531–540), in region 4.1 (residues 541–570) and in the N-terminal portion of region 4.2 (residues 571–590)4; 5; 10 play a role in transcription from SoxS-dependent promoters. To accomplish this goal, we used site-directed mutagenesis of plasmid pVR- σ57 to create a tri-alanine- scanning library of σ70 from positions 531–590 such that each mutant σ70 protein would have three successive amino acid residues substituted with alanine (e.g., P531A, L532A, and D533A). The effect of the tri-alanine substitutions on transcription from the various promoters was determined by assay of β-galactosidase activity. Transcription reduced to 80% or less of wild type transcription was considered to be a significant defect, as defined previously58; 59.
We first tested our library at three class II SoxS dependent promoters (fumC, inaA and micF), where the soxbox partially or completely overlaps the −35 hexamer (Fig. 1). Surprisingly, we found that none of the tri-alanine substitutions reduce SoxS-dependent transcription activation at any of these promoters (Table 1). Thus, we infer that the binding of SoxS to the soxbox of class II promoters prevents the binding of σ70 R4 to the −35 region of the promoter. The results of these experiments were startling since previous in vitro work with galP19T, a class II, cyclic AMP receptor protein (CRP)-dependent promoter, indicates that the binding of CRP has no effect on the position of σ70 R4 upon its subsequent binding to the −35 element of the promoter60.
Table 1.
The effects of tri-alanine substitutions of σ70 R4 on SoxS-dependent transcription activation of class I and class II promoters.
| Tri-Alanine Stretches of σ70 R4 | β-galactosidase activity (% of wild type) | |||||
|---|---|---|---|---|---|---|
| Class I Promoters | Class II Promoters | |||||
| fpr | mar | zwf | fumC | inaA | micF | |
| P531A-D533A | 127 ±5 | 127 ±9 | 93 ±4 | 123 ±13 | 102 ±5 | 102 ±4 |
| S534A-T536A | 126 ±8 | 97 ±9 | 109 ±6 | 102 ±8 | 110 ±3 | 98 ±11 |
| T537A-S539A | 88 ±6 | 88 ±5 | 98 ±8 | 156 ±4 | 86 ±12 | 93 ±6 |
| L540A-A542A | 116 ±9 | 112 ±8 | 97 ±5 | 104 ±8 | 116 ± 8 | 104 ±4 |
| A543A-H545A | 112 ±8 | 97 ±9 | 93 ±6 | 105 ±10 | 105 ±6 | 106 ±3 |
| D546A-L548A | 120 ±4 | 97 ±5 | 89 ±7 | 85 ±6 | 113 ±4 | 116 ±9 |
| A549A-L551A | 127 ±5 | 86 ±6 | 99 ±4 | 98 ±12 | 114 ±2 | 113 ±8 |
| T552A-R554A | 123 ±5 | 85 ±5 | 69 ±4 | 149 ±18 | 100 ±8 | 86 ±4 |
| E555A-K557A | 116 ± 3 | 77 ±5 | 83 ±5 | 93 ±12 | 83 ±2 | 89 ±8 |
| V558A-R560A | 109 ±3 | 83 ±8 | 84 ±10 | 101 ±11 | 92 ±4 | 111 ±10 |
| M561A-F563A | 78 ±7 | 84 ±7 | 115 ±5 | 154 ±8 | 105 ±5 | 116 ±8 |
| G564A-D566A | 87 ±8 | 91 ±9 | 86 ±2 | 158 ±7 | 120 ±12 | 108 ±6 |
| M567A-T569A | 101 ±8 | 83 ±6 | 70 ±1 | 134 ±15 | 113 ±7 | 112 ±7 |
| D570A-T572A | 89 ±9 | 95 ±4 | 83 ±2 | 121 ±16 | 130 ±13 | 88 ±7 |
| L573A-E575A | 70 ±4 | 79 ±7 | 101 ±2 | 159 ±14 | 90 ±3 | 91 ±3 |
| V576A-K578A | 78 ±6 | 83 ±7 | 69 ±1 | 95 ±8 | 151 ±14 | 131 ±4 |
| Q579A-D581A | 119 ±14 | 69 ±8 | 76 ±2 | 95 ±8 | 113 ±2 | 126 ±5 |
| V582A-R584A | 109 ±5 | 74 ±8 | 85 ±2 | 89 ±8 | 96 ±5 | 98 ±4 |
| E585A-I587A | 112 ± 2 | 85 ±5 | 85 ±1 | 88 ±3 | 101 ±3 | 121 ±5 |
| R588A-I590A | 70 ±5 | 88 ±9 | 91 ±3 | 104 ±7 | 138 ±8 | 127 ±12 |
The library of tri-alanine substitutions of σ70 R4 carried on plasmid pVR-σ70 was introduced into derivatives of E. coli strain N7840 (Δmor) [pBAD33-his6-SoxS] carrying transcriptional fusions of the above-named promoters to lacZ. SoxS expression was induced with 0.2% arabinose when the A600 reached 0.1–0.2; wild type and mutant σ70 proteins are constitutively expressed from a truncated galP1 promoter carried on plasmid pVR-σ70. The host chromosome contains rpoD and expresses wild type σ70 from it. Expression of the fusions was determined by assay of β-galactosidase activity as described in Materials and Methods. The data are expressed as the % of the wild type value and the standard deviations are given as the % of the mean. Wild type Miller units at SoxS dependent promoters were fpr (2110), mar (2980), zwf (2450), fumC (1196), inaA (1601) and micF (443). As described in the text, values for mutants that are ≤ 80% of the wild type activity are taken as a meaningful difference; these values are in bold and are highlighted in grey.
Next, we determined the effect of the library on SoxS-dependent transcription activation of class I promoters fpr and mar, and on zwf, the non-canonical class I promoter. We also determined the effect of the library on basal transcription from these promoters. Table 1 shows that eight tri-alanine substitutions reduce SoxS-dependent transcription of these class I promoters. Of these eight substitutions, two are in region 4.1 and six are in region 4.2. None of the mutants confer a strong phenotype in that transcription was reduced by less than twofold. These substitutions also reduce SoxS-independent transcription (data not shown). We rationalize these data by arguing that since the −35 elements of class I promoters typically reside at least 15–16 bp downstream of the SoxS binding site, they are almost certain to be fully available for contact with σ70 R4 under both SoxS-dependent and SoxS-independent conditions. Accordingly, tri-alanine substitutions that confer a defect in the binding of σ70 R4 to the respective −35 elements would be expected to reduce transcription from these promoters under the two conditions.
A few substitutions reduce transcription more under non-inducing than inducing conditions (data not shown). We do not know the basis for this effect. However, one explanation is that the 35° bend in the DNA induced by the binding of SoxS to the soxbox38 partially compensates for the defects in transcription initiation conferred by the tri-alanine substitutions of σ70 R4.
Given these data showing that members of the library can reduce transcription of class I promoters wherein the respective soxboxes do not overlap the −35 element, we expected that some library members would reduce basal, SoxS-independent transcription from the class II promoters. Table 2 shows that six substitutions significantly reduce basal transcription from the fumC promoter and three substitutions significantly reduce basal transcription from the inaA promoter. Curiously, no substitutions reduce transcription from the micF promoter (Table 2). We do not know why none of the substitutions reduces SoxS-independent transcription at the micF promoter. One possible explanation is that under non-inducing conditions, the contribution to the overall binding of σ70 to the micF promoter made by the interaction of σ70 R4 with the −35 element is small, because the affinity of σ70 R4 for this particular promoter element is low, If so, then loss of contact by a mutant of σ70 R4 would be inconsequential.
Table 2.
The effect of tri-alanine substitutions of σ70 R4 on basal transcription from the class II fumC, inaA and micF promoters.
| Tri-Alanine Stretches ofσ70R4 | β-galactosidase activity (% of wild type) | ||
|---|---|---|---|
| fumC | micF | inaA | |
| P531A-D533A | 134 ±6 | 132 ±18 | 124 ±4 |
| S534A-T536A | 98 ±15 | 88 ±10 | 118 ±5 |
| T537A-S539A | 81 ±13 | 84 ±11 | 137 ±8 |
| L540A-A542A | 88 ±22 | 125 ±9 | 116 ±4 |
| A543A-H545A | 116 ±18 | 105 ±14 | 98 ±4 |
| D46A-L548A | 56 ±5 | 100 ±7 | 117 ±6 |
| A549A-L551A | 174 ±18 | 121 ±10 | 92 ±4 |
| T552A-R554A | 177 ±13 | 107 ±4 | 97 ±6 |
| E555A-K557A | 10 ±12 | 122 ±10 | 110 ±3 |
| V558A-R560A | 151 ±8 | 108 ±8 | 89 ±9 |
| M561A-F563A | 59 ±5 | 130 ±6 | 120 ±6 |
| G564A-D566A | 82 ±6 | 130 ±4 | 124 ±3 |
| M567A-T569A | 72 ±10 | 137 ±5 | 73 ±8 |
| D570A-T572A | 215 ±14 | 183 ±13 | 126 ±5 |
| L573A-E575A | 90 ±17 | 163 ±8 | 104 ±5 |
| V576A-K578A | 62 ±3 | 209 ±24 | 86 ± 7 |
| Q579A-D581A | 290 ±14 | 208 ±8 | 100 ±3 |
| V582A-R584A | 64 ±7 | 135 ±4 | 108 ±6 |
| E585A-I587A | 54 ±5 | 156 ±7 | 75 ±4 |
| R588A-I590A | 219 ±18 | 169 ±7 | 78 ±3 |
The experiments were carried out with the same class II fusion strains and under the same conditions as those described in Table 1, except that no arabinose was added to the cultures such that SoxS expression was not induced. Expression of the fusions was determined by assay of β-galactosidase activity as described in Materials and Methods. The data are expressed as the % of the wild type value and the standard deviations are given as the % of the mean. Wild type Miller units at SoxS dependent promoters were fumC (287), inaA (77) and micF (123). As described in the text, values for mutants that are ≤ 80% of the wild type activity are taken as a meaningful difference; these values are in bold and are highlighted in grey.
Regardless of the problem with the micF promoter, these data show that members of the library can indeed interfere with functions essential to the basal, SoxS-independent transcription from the class II promoters fumC and inaA.
Protein-DNA Interactions between SoxS and the soxbox
To explore further the inference that SoxS competes with the binding of σ70 R4 to the −35 promoter element at class II SoxS-dependent promoters, we performed in vivo genetic epistasis tests61 between SoxS mutants defective in DNA binding53 and single base pair substitutions in the soxbox53. The goal of this experiment was not only to determine the orientation of SoxS when it is bound to the soxbox, but also to identify some specific contacts between amino acids of SoxS and base pairs within the soxbox so that we could accurately position SoxS on the DNA. The co-crystal structure of MarA bound to the marbox of the mar promoter54 served as a critical reference for SoxS, because MarA is a paralog of SoxS. Moreover, SoxS and MarA bind DNA as monomers38; 54, they bind to the same degenerate sites22; 36; 37; 39; 62 and they activate transcription of the same set of genes, although to different degrees28; 63–65. Thus, the amino acid residues of SoxS that make base-specific contacts with soxbox/marbox DNA are likely to be similar, if not identical, to the amino acids of MarA that make base-specific contacts with soxbox/marbox DNA54. Accordingly, for our genetic epistasis tests, we selected five alanine substitutions of SoxS (W36A, Q39A, R40A, T87A and R90A) which are homologous to five of the eight amino acids of MarA (W42, Q45, R46, T93 and R96) that are predicted by the MarA/mar co-crystal structure to contact specific bases of marbox DNA54.
For two reasons our epistasis tests were conducted with fpr, a canonical class I SoxS-dependent promoter, and the non-canonical zwf promoter. First, we did not use canonical class II promoters because RE2 of their soxboxes overlaps the −35 element22; as such, transcriptional defects arising by substitutions in that region would be difficult to interpret because they could be due to alterations in binding by SoxS or σ70 R4. However, the soxbox of the zwf promoter can serve as a surrogate of soxboxes within class II promoters because, like them, it is oriented in the forward direction. Second, we knew from our alanine scanning mutagenesis of SoxS that the above substitutions are defective in DNA binding and transcription activation in vivo at these two promoters53.
Presuming that SoxS would bind soxbox DNA like the binding of MarA to mar DNA in the co-crystal, we set up our genetic epistasis tests by combining each single alanine substitution of SoxS residing in the N-terminal HTH motif (W36A, Q39A and R40A) with a base pair substitution at each of the four positions in RE1 of the fpr and zwf promoters; similarly we combined each single alanine substitution of SoxS residing in the C-terminal HTH motif (T87A and R90A) with a base pair substitution at each of the four positions in RE2 of the two promoters. At each position in the REs, we chose a base pair substitution that reduces transcription activation of a given promoter by wild type SoxS to an intermediate level41, since small or large effects are problematic in genetic epistasis tests. We then carried out the epistasis tests by determining the effects of the various combinations of mutations on transcription in each strain.
The criterion for epistasis61 was that if the defect in transcription conferred by a double mutant is no greater than that conferred by either of the single mutations alone, the two mutations are considered to be epistatic. If two mutations are epistatic to one another, then their wild type counterparts contact each other. On the other hand, if the defect conferred by a double mutant is greater than the defect conferred by the two single mutations, the two mutations are considered to be non-epistatic61. If two mutations are non-epistatic, then their wild type counterparts do not contact one another.
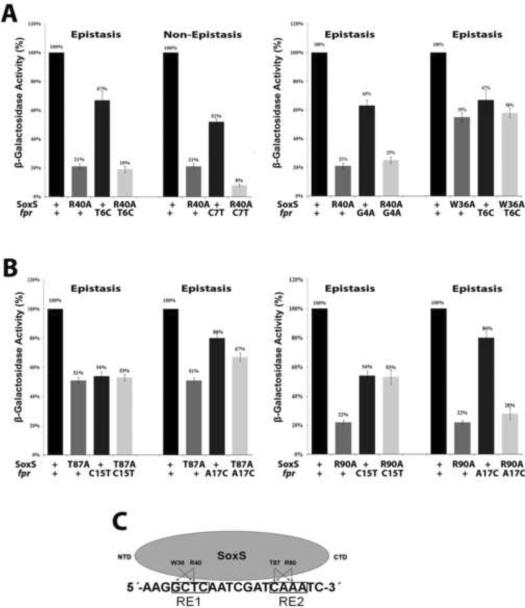
Figures 2 and 3 show the results of the epistasis tests with the fpr and zwf promoters, respectively. With the fpr promoter, W36A and R40A in the putative N-terminal HTH motif of SoxS are each epistatic to a base pair in RE1 that is predicted by the co-crystal structure to be contacted by W42 and R46, the homologous amino acids of MarA54. Similarly, T87A and R90A in the putative C-terminal HTH motif of SoxS are each epistatic to a base pair in RE2 that is predicted by the co-crystal structure to be contacted by T93 and R96, the homologous amino acids of MarA. In addition, R40A, T87A and R90A are each epistatic to a second base pair which is not predicted from the co-crystal structure to be contacted by the homologous amino acids of MarA. These additional contacts are not surprising because the sequences of the two binding sites are different and the epistasis tests were carried out in vivo whereas the co-crystal structure was determined with a purified complex. Regardless, these results show that the position and orientation of SoxS bound to soxbox DNA in vivo are the same as those observed in the structure of the MarA/mar co-crystal.
Figure 2.
Genetic epistasis experiments between single base pair substitutions of the soxbox of the fpr promoter and DNA binding mutants of SoxS. The sequence of the template strand of the fpr soxbox is shown in panel C; the invariant A of the soxbox is at the 5' end of the sequence. (A) (Left) Epistatic and non-epistatic interactions between the R40A substitution in the N-terminal HTH motif of SoxS and base pair substitutions T6C and C7T of RE 1, respectively. (Right) Epistatic interactions between the W36A and R40A substitutions in the N-terminal HTH motif of SoxS and substitutions T6C and G4A of RE 1, respectively. (B) (Left) Epistatic interactions between the T87A substitution in the C-terminal HTH motif of SoxS and base pair substitutions C15T and A17C of RE 2. (Right) Epistatic interactions between the R90A substitution in the C-terminal HTH motif of SoxS and substitutions C15T and A17C of RE 2. (C). A schematic representation of the interactions between amino acid residues of SoxS and nucleotide bases within the fpr soxbox, as determined by the genetic epistasis tests shown in (A) and (B). The epistasis data show that amino acids of the N- and C-terminal HTH motifs of SoxS contact one or two base pairs within RE 1 and RE 2, respectively, but whether the contacted base is on the template or non-template strand cannot be determined by these tests.
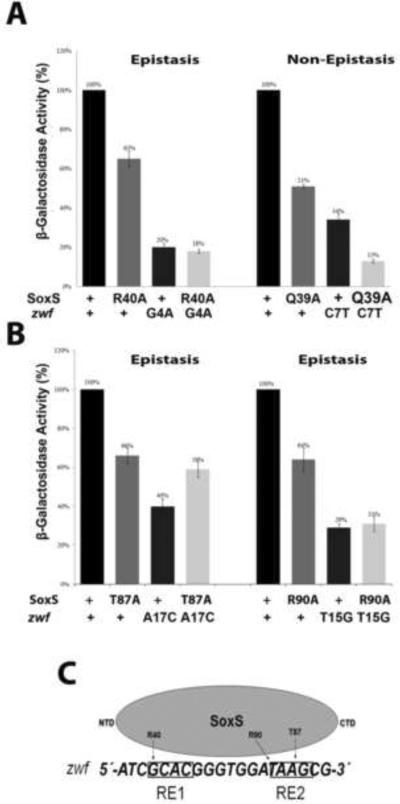
Figure 3.
Genetic epistasis experiments between single base pair substitutions of the soxbox of the zwf promoter and DNA binding mutants of SoxS. The sequence of the non-template strand of the zwf soxbox is shown in panel C; the invariant A of the soxbox is at the 5' end of the sequence. (A) (Left) Epistatic interaction between the R40A substitution in the N-terminal HTH motif of SoxS and base pair substitution G4A of RE 1. (Right) Non-epistatic interaction between the Q39A substitution in the N-terminal HTH motif of SoxS and substitution C7T of RE 1. (B) (Left) Epistatic interaction between the T87A substitution in the C-terminal HTH motif of SoxS and base pair substitution A17C of RE 1. (Right) Epistatic interaction between the R90A substitution of the C-terminal HTH motif of SoxS and substitution T15G of RE 2. (C) A schematic representation of the interactions between amino acid residues of SoxS and nucleotide bases within the zwf soxbox, as determined by the genetic epistasis tests shown in (A) and (B). The epistasis data show that amino acids of the N- and C-terminal HTH motifs of SoxS each contact one base pair within RE 1 and RE 2, respectively, but whether the contacted base is on the template or non-template strand cannot be determined by these tests.
Similar results were obtained with the zwf promoter (Fig. 3). Thus, R40A and R90A are epistatic to a base pair in RE1 and RE2, respectively; the same base pairs are predicted by the co-crystal structure to be contacted by R46 and R96, the homologous amino acids of MarA54. In addition, T87A is epistatic to a base pair in RE2 that is not predicted by the co-crystal structure to be contacted by T93 of MarA. Thus, the epistasis experiments conducted with the zwf promoter also place the N-terminal HTH motif on RE1 of a soxbox and the C-terminal HTH motif on RE2.
Effect of tri-alanine substitutions of σ70 R4 on CRP-dependent transcription activation of its class I and class II promoters
The surprising results of Table 1 led us to ask: “Is the absence of co-occupancy between SoxS and σ70 R4 at the −35 region of class II SoxS-dependent promoters a common property of bacterial transcription activators that activate class II promoters? As the system for answering this important question, we chose the best-studied transcription activator, CRP. Thus, we used the tri-alanine scanning library of σ70 R4 to determine whether the binding of CRP to the class II gal P1 promoter interferes with the binding of σ70 R4 to the −35 region. We also determined the effect of the tri-alanine substitutions of σ70 R4 on CRP-dependent transcription from the lac promoter and an on CRP-independent transcription from a lac promoter whose CRP binding site has been replaced by an UP element, which enhances transcription by interacting with the DNA-binding determinant of the α-CTD of RNAP66; 67.
Table 3 shows that five tri-alanine substitutions of σ70 R4, D570A-T572A, L573A-E575A, V576A-L578A, V582A-R584A and R588A-I590A, reduce CRP-dependent transcription from the lac promoter under inducing conditions. Previous work has shown that one or more amino acids within stretches V582-R584 and R588-I590 interact directly with the −35 promoter element68 and substitutions therein reduce lac transcription69 while single amino acid substitutions within the other three stretches either reduce or enhance transcription in a sequence-independent manner69. In addition, at the lac promoter and the related CC-61.5 promoter, amino acids within positions 573–604 of σ70 make protein-protein interactions with the 261 determinant of the α-CTD70. These contacts, together with the interactions between Activation Region 1 of CRP and the 287 determinant of the α-CTD, allow the α-CTD to serve as a bridge that connects RNAP bound at the lac promoter to CRP bound to a site centered at −61.570. Regardless of whether the five tri-alanine substitutions reduce CRP-dependent transcription from the lac promoter by interfering with the binding of σ70 R4 to promoter DNA or whether they function by disrupting the bridging interactions between the α-CTD and σ70 R4, the results obtained with them provide evidence that our data are consistent with previously published results69; 70. Importantly, four of the tri-alanine substitutions (L573A-E575A, V576A-L578A, V582A-R584A, R588A-I590A) that reduce CRP activation of the lac promoter also reduce SoxS-dependent transcription from at least one of the class I promoters examined (compare Table 1 and Table 3). As described below (Table 4), some of these alanine substitutions confer a growth-defective phenotype.
Table 3.
The effects of tri-alanine substitutions of σ70 R4 in strains containing transcriptional fusions of lacZ to a CRP-dependent class I promoter (strain W3110), a CRP-dependent class II promoter (strain DM00021) and a CRP− class I promoter where the CRP binding site has been replaced by an UP element (strain RLG4282).
| Tri-Alanine Stretches of σ70 R4 | β-galactosidase activity (% of wild tvpe) | ||
|---|---|---|---|
| lac (Class I) | UP+ CRP− lac | gal (Class II) | |
| P531A-D533A | 116 ±5 | 81 ±5 | 85 ±3 |
| S534A-T536A | 116 ±4 | 84 ±4 | 90 ±1 |
| T537A-S539A | 122 ±5 | 82 ±6 | 80 ±1 |
| L540A-A542A | 111 ±13 | 94 ±4 | 82 ±2 |
| A543A-H545A | 124 ±4 | 94 ±2 | 84 ±3 |
| D546A-L548A | 95 ± 9 | 86 ±1 | 80 ±3 |
| A549A-L551A | 92 ± 10 | 94 ±3 | 77 ±3 |
| T552A-R554A | 98 ± 8 | 85 ±1 | 77 ±1 |
| E555A-K557A | 95 ±11 | 67 ±3 | 97 ±3 |
| V558A-R560A | 107 ±2 | 80 ±2 | 87 ±7 |
| M561A-F563A | 103 ±7 | 68 ±3 | 85 ±3 |
| G564A-D566A | 103 ±10 | 78 ±3 | 86 ±5 |
| M567A-T569A | 117 ±5 | 82 ±5 | 71 ±4 |
| D570A-T572A | 67 ±8 | 90 ±5 | 78 ±3 |
| L573A-E575A | 72 ±7 | 88 ±10 | 80 ±4 |
| V576A-K578A | 55 ±6 | 57 ±9 | 85 ±5 |
| Q579A-D581A | 83 ±11 | 67 ±3 | 106 ±5 |
| V582A-R584A | 63 ±3 | 77 ±7 | 78 ±6 |
| E585A-I587A | 94 ±14 | 83 ±3 | 93 ±5 |
| R588A-I590A | 68 ±4 | 58 ±4 | 104 ±2 |
The experiments were carried out as described in the legend to Table 1, except that the cultures of strains W3110 and DM00021 were treated with 1 mM IPTG to induce transcription from the lac promoters and cultures of strain RLG4282 were treated with 0.2% D-galactose to induce transcription from the galP1 promoter. Wild type Miller units were lac (5740), UP+lac (928) and gal (460).
Table 4.
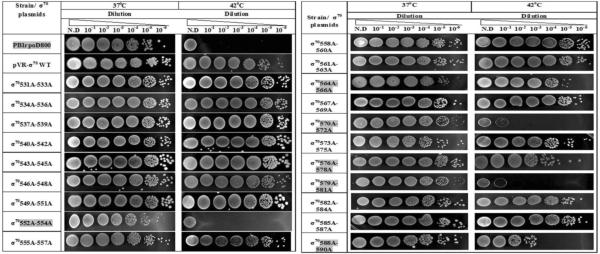
Effect of overexpression of the tri-alanine substitutions of σ70 R4 on growth at 42°C in strain PB1rpoD800.
|
Several mutants conferred temperature-lethal phenotypes (T552A-R554A, D570-T572-Q579-D581) while others conferred partial defects (G564A-T566A, V576A-K578A, D588A-I590A). The mutants conferring growth defects are highlighted in grey. N.D., no dilution.
As a control, we wanted to determine whether the tri-alanine substitutions of σ70 R4 that reduce CRP-dependent transcription from the lac promoter also reduce CRP-independent transcription from this promoter. However, the level of CRP-independent lac transcription in a strain carrying a crp deletion and growing in the presence of IPTG71, is only about one percent of the amount of induced lac transcription in a wild type strain; as such, it would be difficult to determine accurately the effect of mutants of σ70 R4 on lac transcription in such a strain. Accordingly, we used strain RLG4282, wherein DNA upstream of position −37 of the lac promoter, which includes the CRP binding site, has been deleted and replaced by the UP element of the rrnBP1 promoter72; introduction of the UP element enhances CRP-independent transcription by ~30-fold72. Following the introduction of the library into strain RLG4282, we found that eight of the tri-alanine substitutions of σ70 R4 reduce transcription from the UP+/CRP− lac promoter to 80% or less of the amount obtained with the strain carrying the plasmid encoding wild type σ70 R4 (Table 3). Of these, five also reduce SoxS-dependent transcription activation of at least one class I promoter (see Table 1).
Interestingly, three of these substitutions, E555A-K557A, M561A-F563A and G564A-F566A, have no effect on CRP-dependent transcription from the wild type lac promoter (compare columns 1 and 2 of Table 3). Moreover, all three substitutions reside in region 4.1 of σ70, the region that interacts with the hydrophobic patch of the flap-tip helix of the β subunit and thereby positions region 4.2 so that it can bind a −35 promoter element11. Thus, in some way, the ability of CRP to bind to its site in the lac promoter masks the effect of these substitutions on lac transcription. One possibility is that at the wild type lac promoter where Activation Region 1 in the promoter-proximal subunit of DNA-bound CRP interacts with the 287 determinant of the α-CTD73, the concurrent interaction between the 261 determinant of the α-CTD and amino acids of σ70 R470 stabilizes the binding of region 4.2 to the −35 promoter element.
None of the 20 tri-alanine substitutions of σ70 R4 reduces SoxS-dependent transcription activation of the class II promoters fumC, inaA and micF (Table 1), as if the binding of the activators to its respective binding sites occludes the binding of σ70 R4. Thus, we very much wanted to determine whether CRP would have a similar effect on the class II CRP-dependent galP1 promoter, where the CRP binding site partially overlaps the −35 promoter hexamer (Fig. 1). To do this, we introduced the library into strain DM00021, which carries a galP1-lacZ fusion, and determined the effects of the substitutions on expression from the CRP-dependent promoter. Significantly, we found that the data obtained with CRP are completely different from those obtained with SoxS in that five tri-alanine substitutions of σ70 R4 (A549A-L551A, T552A-R554A, M567A-T569A, D570A-T572A, V582A-R584A) reduce transcription of the class II galP1 promoter under CRP-dependent conditions (Table 3) whereas none of the library's tri-alanine substitutions reduces SoxS-dependent transcription at the class II promoters (Table 1). Thus, the data in Table 3 indicate that CRP and σ70 R4 can co-bind the −35 region of the class II galP1 promoter. Indeed, in vitro Fe-BABE footprinting studies by Bown et al. 60 showed that CRP and σ70 R4 can co-exist at the −35 region of the gal P1 promoter.
Promoters that contain an extended −10 element, TGn, can initiate transcription without the specific binding of σ70 R4 to the −35 region74–78. Since the galP1 promoter contains an extended −10 element and since none of the bases in the −35 region match those of the consensus −35 hexamer (see Fig. 1), we do not know how the five tri-alanine substitutions of σ70 R4 reduce CRP-dependent transcription from the galP1 promoter. This leads us to point out that just because promoters with an extended −10 element do not require interactions between the −35 region and σ70 R4, does not mean that interactions between base pairs within the −35 region and σ70 R4 cannot occur and thereby enhance overall transcription78; 79. We also note that amino acids R554 and R584 are known to interact directly with DNA within the −35 region68. Thus, the absence of an interaction between the −35 region and one or more amino acids within these two tri-alanine stretches could be responsible for the defect.
Effect of the tri-alanine substitutions of σ70 R4 on growth
We determined the effect of the tri-alanine substitutions of σ70 R4 on growth by introducing the library into strain PB1rpoD800, which carries rpoD800, a temperature-sensitive allele of the gene encoding σ70 80. For this complementation analysis, we carried out two assays at the non-permissive temperature of 42°C with the set of partial diploids: growth in liquid medium; and a qualitative determination of the colony-forming units per ml when the cells were grown at 37°C, a permissive temperature80, and plated on LB agar at 42°C and 37°C.
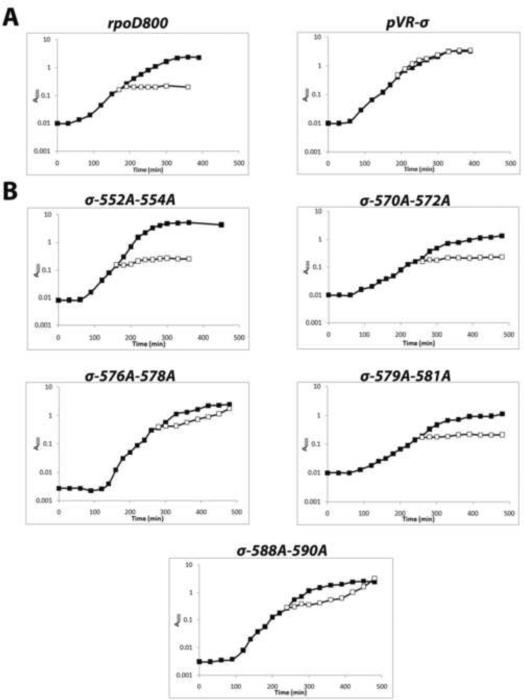
As expected, parental strain PB1rpoD800 grew normally in liquid LB medium at 37°C and growth ceased within 30 min after shifting the culture to 42°C (Fig. 4); the growth lethal phenotype conferred by the rpoD800 mutation was also observed when the parental strain was plated on LB agar at 42°C (Table 4). Also as expected, plasmid pVR-σ57 carrying the wild type rpoD gene complemented the rpoD800 mutation during growth of the partial diploid at 42°C in liquid medium (Fig. 4) and on LB agar (Table 4).
Figure 4.
Identification of members of the library of tri-alanine substitutions of σ70 R4 carried on plasmid pVR-σ that fail to complement the temperature-sensitive rpoD800 mutation in strain PB1rpoD800 during growth at 42°C, the non-permissive temperature. (A) (Left) Demonstration of the growth-lethal effect of the rpoD800 mutation of strain PB1rpoD800 upon a shift from growth at the permissive temperature of 37°C (clear square) to 42°C (black square). (Right) Demonstration of the ability of plasmid pVR-σ carrying the wild type rpoD allele to complement the growth-lethal effect of the chromosomal rpoD800 mutation during growth at 42°C. (B)Growth curves demonstrating the failure of five members of the library of tri-alanine substitutions of σ70 R4 to complement the growth-lethal effect of the rpoD800 mutation during growth at 42°C. Note that the growth curves of the partial diploids carrying the other 15 members of the library looked like that of PB1rpoD800[pVR-σ], i.e., full complementation (data not shown).
With the set of PB1rpoD800 strains carrying the library of tri-alanine substitutions, we carried out the test of growth in liquid medium at the non-permissive temperature and the test of the ability to form colonies on plates a 42°C. The two tests produced the same results with each partially diploid strain. Thus, plasmids carrying mutants T552A-R554A, D570A-T572A and Q579A-D581A failed to complement the rpoD800 allele in that the growth of the respective liquid cultures ceased within 30 min when the cultures were shifted from 37°C to 42°C (Fig. 4) and the lethal phenotype of rpoD800 was also observed when the respective diploid strains were plated at 42°C (Table 4). In addition, mutants V576A-K578A and R588A-I590A partially complemented the rpoD800 mutation in the two tests. Thus, each of these five tri-alanine substitutions alter or completely disrupt an essential function of σ70 and therefore cell growth. Indeed, amino acids R554 and R588 are known to directly contact DNA in the −35 promoter hexamer68 and hence transcription of many housekeeping genes would be significantly reduced. Also, mutation G577S is defective in transcription from the lac and phage P22 ant promoters69; accordingly, transcription from many essential genes is likely to be affected by the G577A substitution. The nature of the defects in the function of σ70 conferred by the other two tri-alanine substitutions, D570A-T572A and Q579A-D581A, is unknown, but they may alter the proper conformation of the σ70 factor.
We note that four of the tri-alanine substitutions, T552A-R554A, V576A-K578A, Q579A-D581A and R588A-I590A, also reduce transcription at the class I SoxS-dependent promoters in the presence of the wild type rpoD gene (see Table 1). Importantly, although substitutions T552A-R554A and D570A-T572A reduce transcription from the class II, CRP-dependent galP1 promoter (Table 3), none of the five tri-alanine substitutions reduce transcription of the class II SoxS-dependent promoters, fumC, inaA and micF (Table 1). Thus, these observations provide additional evidence that the binding of σ70 R4 to the −35 region of these three class II promoters is inessential for SoxS-dependent activation of their transcription, while binding to the −35 element of class I promoters is required for activation.
Location of the amino acid substitutions of σ70 R4 that reduce transcription on structural models of holo-RNAP and of σA bound to promoter DNA
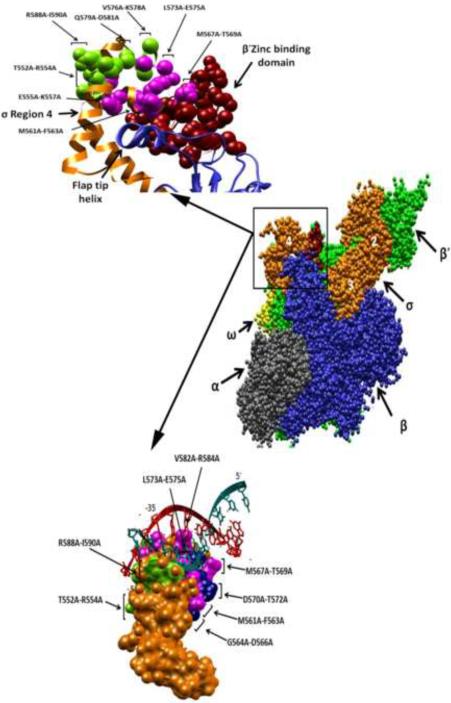
We sought a means of understanding better the basis of the effects of the tri-alanine substitutions of σ70 R4 on growth and on SoxS-dependent transcription of class I promoters. Accordingly, we located the amino acids that confer these defects in growth and transcription on the crystal structures of holo-RNAP of Thermus thermophilus10 and on the binary complex of Thermus aquaticus σA bound to DNA containing a consensus −35 element68 (Fig. 5).
Figure 5.
Location of the equivalent positions of amino acids substituted with alanine in the trialanine scanning library of σ70 R4 on the crystal structures of holo-RNAP from Thermus thermophilus10 and on the crystal structure of Thermus aquaticus σA bound to DNA containing a consensus −35 element68. Only substitutions that reduce SoxS-dependent transcription activation of class I promoters or confer a defect in growth in liquid medium or on plates are identified. A space-filling model of the T. thermophilus RNAP holoenzyme is shown in the center, with the different subunits labeled and color-coded: σA subunit, orange; β subunit, blue; β' subunit, green; α subunit, grey; and ω subunit, yellow. The region displaying the interaction between σA R4 and the β flap-tip helix is enlarged in the upper left corner. The colors of the subunits are same as those in the model of holo-RNAP. Shown in magenta are the amino acid residues of σA that are equivalent to those within four (E555-K557, M561-F563, M567-T569 and L573-E575) of the five tri-alanine stretches of σ70 R4 that reduce SoxS-dependent transcription activation of class I promoters but have no effect on growth; the amino acids equivalent to those in the other stretch (V582-R584) do not appear in the crystal structure. The amino acids of σA that are equivalent to those within the four stretches of tri-alanine substitutions σ70 R4 that reduce SoxS-dependent transcription activation and also confer a defect in growth (T552-R554, V576-K578, Q579-D581 and R588-I590) are shown in green. Not appearing in this crystal structure are the amino acids of σA that are equivalent to those within the two tri-alanine stretches that confer a defect in growth (G564-D566, D570-T572) but not in SoxS-dependent transcription of class I promoters. The lower left corner displays the model of the crystal structure of σA R4 bound to DNA containing a −35 promoter hexamer. The amino acids residues of σA that are equivalent to those within four (M561-F563, M567-T569, L573-E575, V582-R584) of the five stretches of trialanine substitutions of σ70 R4 that reduce SoxS-dependent transcription activation of class I promoters but have no effect on growth are shown in magenta; the amino acids equivalent to those within the other stretch (E555-K557) do not appear in the crystal structure. The amino acids σA that are equivalent to two (T552-R554 and R588-I590) of the four stretches of trialanine substitutions that reduce transcription activation and also confer a growth defect are shown in green; the amino acids of σA that are equivalent to those within the other two tri-alanine stretches do not appear in this crystal structure. The amino acids of σA that are equivalent to those within the two tri-alanine stretches that confer a defect in growth (G564-D566, D570-T572) but not in SoxS-dependent transcription of class I promoters are shown in blue. The modeling was carried out with the Chimera molecular modeling software89, as described in Materials and Methods.
Genetic, biochemical and structural studies have identified many amino acids that play a variety of functional roles in transcription initiation, either directly or indirectly. With respect to the work presented here, we note that amino acids R541, T544 and L607 of σ70 R4 interact with the β-flap of RNAP81; 82, a critical interaction that helps position σ70 R4 on the core RNAP so that it can make specific contacts with the −35 promoter element11; 83. Moreover, an interaction between the ZBD of the β' subunit of RNAP and σ70 R4 enhances its binding to promoter DNA, in part by stabilizing the β-flap interaction with σ70 R411. Additional work conducted by Geszvain et al.11 provided evidence that amino acid R554 of σ70 R4 interacts with the ZBD of the β subunit. Lastly, the co-crystal structure of σ70 R4 bound to a consensus −35 element indicates that amino acid residues R554, R562, L573, E574, T583, R584, E585, R586, R588 and K593 of σ70 R4 make direct or water-mediated contacts with the bases, the phosphate backbone and deoxyribose68.
Most of the amino acids residues mentioned above that contact the β flap-tip helix, the ZBD of the β' subunit or the −35 hexamer have been substituted with alanine in our library of trialanine substitutions and many members have been shown in this work to alter transcription initiation, or growth, or both. For example, in region 4.1 of σ70, substitution T552A-R554A reduces transcription at the mar promoter (Table 1) and also confers a temperature-lethal phenotype (Table 4). In addition, amino acid R554 appears to contact the ZBD of the β′ subunit11 as well as the phosphate backbone just upstream of the −35 element68; the absence of any of these functions could be the basis for the defect in mar transcription. Similarly, substitution E555A-K557A reduces SoxS-dependent activation from the zwf promoter. This defect could be because these residues can no longer make productive interactions with the core RNAP2 or with the ZBD on the β' subunit11. Substitution, G564A-D566A, has a mild growth defect in the plate assay (Table 4), but no effects on SoxS-dependent transcription were observed. Although amino acid I565 is known to contact the RNAP core2, the effect of substituting alanine for isoleucine at this position is rather small, as was the case for several other substitutions examined by Gezvain et al.11. However, substitution G564A may be responsible for the growth defect because of the change in local protein structure effected by replacing glycine, which disrupts α-helices, with alanine. Interestingly, previous work did not reveal a role in either DNA binding or in interaction with core-RNAP for the amino acids M567-T569 of σ70 R4. However, we found that alanine substitutions of these amino acids reduce transcription activation of mar (Table 1), basal transcription of fumC and inaA (Table 2) and CRP-dependent transcription from galP1. In agreement, the crystal structure of holo-RNAP shows that these amino acids are close to the ZBD of the β' subunit10, which likely accounts for their effects on transcription.
Tri-alanine substitutions in region 4.2 that caused a defect in transcription activation, growth, or both included D570A-T572A, L573A-E575, V576-L578A, Q579A-D581A V582-R584A and R588A-I590A. Previous studies implicate substitution D570A-T572A in reducing transcription at the lac promoter69. These amino acids are close to the core of RNAP and to promoter DNA and thus are capable of making direct protein-DNA or protein-protein interactions. Alternatively, they may stabilize the functional conformation of the surrounding amino acids so that they can make those contacts. Furthermore, Q579A-D581A conferred a temperature-lethal phenotype, presumably due to the loss of a stabilizing effect of D581 on the interaction of σ70 R4 with the β-flap and the −35 hexamer81; 82. Two other substitutions, V576AK578A and R588A-I590A, confer a mild growth defect and reduce transcription activation by SoxS. The V576-K578 patch is buried between amino acids that either contact DNA or are involved in contacting the β-flap. Thus, as with G564A in the G564A-D566A patch, the G577A substitution within the V576A-K578A patch may also disrupt the positioning of the surrounding amino acids and thereby inhibit their ability to make protein-DNA or protein-protein interactions. Also, these amino acids may directly contact DNA at SoxS-dependent promoters. Finally, studies have suggested that amino acids L573, E574 and E575 contact DNA68; 69; 84. In our experiments, alanine substitutions of these amino acid residues reduced transcription at class I SoxS-dependent promoters (Table 1) but not basal transcription of class II promoters (Table 2). According to the crystal structure determined by Campbell et al.68 and the genetic studies of Siegele et al.69, amino acid R584A should directly contact the C/G bp in the −35 hexamer of the lac promoter. At the zwf promoter, a C/G bp is located at the same position and, consistent with the crystal structure, substitutions, V582A-R584A reduce transcription from it, suggesting that the patch interacts with the −35 element.
Discussion
The construction described herein of a library of tri-alanine substitutions of σ70 from positions 531–590 enabled us to identify yet another novel property of SoxS: when SoxS binds to the soxbox of at least three class II promoters, which partially or completely overlaps the -35 region of the promoter (and resides in the forward orientation), σ70 R4 does not bind to this region of the promoter. Thus, neither substitutions of amino acids of σ70 R4 that are known to directly contact base pairs in the −35 region68, nor substitutions of amino acids that are known to be involved in the proper positioning of σ70 R4 on core RNAP10; 11 have any effect on SoxS-dependent transcription activation of the three studied class II promoters (Table 1). However, growth experiments showed that at least five amino acids within five different stretches subjected to tri-alanine scanning mutagenesis are essential for growth (Table 4).
We also determined the effect of the substitutions on basal, SoxS-independent transcription at the three class II promoters. Importantly, several substitutions reduce basal transcription at the fumC and inaA promoters under non-inducing conditions (Table 2). Two conclusions can be drawn from these data. First, the substitutions of σ70 R4 function as expected in that substitutions of amino acids of σ70 R4 known to be important to binding to the −35 hexamer68 reduce transcription when the −35 element is not bound by SoxS, i.e., under SoxS-independent conditions. Second, since the substitutions of σ70 R4 function as expected, then the failure of any of the substitutions to reduce SoxS-dependent transcriptions strongly indicates that the binding of SoxS to the class II soxboxes occludes the binding of σ70 R4 to the −35 hexamer.
The genetic epistasis tests conducted in vivo allowed us to determine whether the binding of SoxS to the soxbox of a class II promoter can actually occlude the binding of σ70 R4 to base pairs of the −35 region that would otherwise be bound by σ70 R4 under non-inducing conditions. With these experiments, we determined that amino acids W36 and R40 of SoxS within its N-terminal HTH motif contact base pairs in RE1 of the fpr soxbox and that amino acids T87 and R90 of the C-terminal HTH motif contact base pairs in RE2. Our epistasis tests with the zwf soxbox produced the same results. Thus, since the DNA sequences of canonical class II promoters show that RE 2 partially or completely overlaps the −35 hexamer and that RE 1 lies upstream, these data fix the orientation of SoxS on soxbox DNA of class II promoters. Moreover, the orientation deduced from these experiments agrees with the orientation of MarA bound to the marbox of the class I mar promoter in the crystal structure of the complex54, i.e., RE 1 is bound by the N-terminal HTH motif and RE 2 is bound by the C-terminal motif. Thus, these experiments support the hypothesis that the binding of SoxS to the soxbox of class II promoters prevents the co-binding of σ70 R4 to the −35 region.
To determine if SoxS also prevents the binding of σ70 R4 to class I promoters, we tested the canonical class I promoters, fpr and mar, and the non-canonical zwf promoter. We show that members of the library affecting either the binding of σ70 R4 to the −5 region and/or the positioning of σ70 R4 on core RNAP significantly reduce transcription from these three promoters. Thus, the failure of any of the 20 tri-alanine substitutions of σ70 R4 to reduce transcription from the canonical class II promoters strongly indicates that the binding of SoxS to the respective −35 regions blocks the binding of σ70 R4 to the region. Moreover, the data also suggest that the normal function of σ70 R4, i.e., binding to the −35 region of promoters and facilitating and stabilizing the binding of σ70 R4 to core RNAP, is not required for SoxS-dependent transcription activation of these class II promoters.
Is the interference with the binding of σ70 R4 to class II promoters by the binding of SoxS to the soxboxes of such promoters a general property of activators of class II promoters? To address this question, we tested the effect of the substitutions on CRP-dependent activation of the class II galP1 promoter fused to lacZ. Importantly, and in contrast with the effect of SoxS binding to class II SoxS-dependent promoters on the binding of σ70 R4, six members of the library of tri-alanine substitutions significantly reduce transcription from the class II galP1 promoter (Table 3). This genetic result indicating that both CRP and σ70 R4 can co-bind to the −35 region of the galP1 promoter is consistent with the FeBABE in vitro cleavage experiments of Bown et al. 60 who showed that by an unknown mechanism CRP and σ70 R4 can accommodate one another in simultaneously binding to the −35 region of the promoter. Thus, the contrasting results between the effects of the binding of SoxS and CRP to their respective sites within the −35 regions of class II promoters on the binding there of σ70 R4 demonstrates another unusual characteristic of SoxS in transcription activation.
Is there precedence for an activator binding preferentially at or near the −35 promoter element and excluding σ70 R4 from the promoter? Even though there are some examples of proteins that inhibit the binding of σ70 R4 to its −35 element, SoxS is to our knowledge the first example of an activator that blocks the binding of σ70 R4 to the −35 hexamer by binding to its target binding site in class II promoters. An example of a system with some resemblance to the effect of SoxS on the function of σ70 R4 at class II promoters is transcription from the middle promoters of bacteriophage T4. In the T4 system, the binding of phage-encoded AsiA to σ70 R4 diverts the binding of host RNAP from host promoters to MotA-dependent “middle genes” of the phage18; 19; 85; 86. Thus, like the binding of SoxS to soxbox DNA prevents σ70 R4 from exerting its normal DNA binding function, the direct binding of AsiA to σ70 R4 has the same effect. The difference between the two systems is that SoxS appears to exert its inhibitory function by physically blocking the binding of σ70 R4 to a specific set of promoters whereas AsiA functions by binding to and remodeling σ70 R4 such that it cannot bind efficiently to promoters lacking DNA-bound MotA.
Another system that somewhat resembles that of SoxS involves the effect of transcription activator Spo0A on the precise positioning of region 4 of the sigma A (σA R4) subunit of Bacillus subtilis RNAP on the −35 region of the spoIIG promoter. With RNAP containing FeBABE covalently bound to the amino acid of σA that is homologous to R588 of σ70 R4, Kumar et al. 87 found that in the absence of Spo0A, σA-containing holo-RNAP forms a closed complex that leads to the cleavage of DNA at a consensus −35 sequence of the spoIIG promoter whose 3' end is 22 bp upstream of the −10 hexamer. However, when cleavage was carried out after the introduction of Spo0A to the in vitro reaction mixture, a primary cleavage site was 4 bp downstream of the site obtained in the absence of the activator. With σA R4 now residing only 18 bp upstream of the −10 element, σA is better able to interact with the −10 hexamer and thereby form a stable closed complex that can subsequently lead to open complex formation87. Thus, in this system, the binding of the activator repositions σA so that transcription initiation can proceed. In contrast, the binding of SoxS to a soxbox at class II promoters prevents the binding of σ70 R4 to the DNA, while protein-protein interactions between amino acids of the two positive control surfaces of SoxS and the distal end of region 4.2 of σ70 anchor the C-terminal region of σ70 in a position that allows region 2.4 to bind the −10 hexamer and form the open complex that leads to transcription initiation (Fig. 6).
Figure 6.
Cartoon representation of protein-protein interactions at class I and class II SoxS-dependent promoters. The N-terminal and C-terminal domains of the two α subunits, which are connected by a flexible linker, and the β, β' and σ70 subunits of holo-RNAP are labeled and colored as are the −10 and −35 promoter hexamers; the relative position of the start site of transcription is numbered and identified by an arrow. Region 4 and the C-terminal tail of the σ70 subunit are denoted by a pink circle lying within the σ70 subunit and labeled “R4”. SoxS is colored red and the yellow and green circles lying on it represent the class I/II and class II positive control surfaces, respectively53. The soxbox is colored bronze. The backward orientation of SoxS bound to the soxbox and the soxbox itself at canonical class I promoters is denoted by writing “SoxS” from right to left. The protein-protein interactions between the class I/II surface of SoxS and the DNA-binding (265) determinant of the α-CTD55 are denoted by the intersection of the yellow and dark blue circles, respectively. The location of the second α-CTD is unknown and thus is denoted by a question mark. The ability of σ70 R4 to bind to the −35 promoter element of class I promoters and the absence of effects of tri-alanine substitutions on SoxS-dependent transcription activation at these promoters (Table 1) is denoted by the intersection of σ70 R4 (a pink circle) and the promoter hexamer (a colorless rectangle). At canonical class II promoters, the binding of SoxS to the soxbox prevents amino acid residues within σ70 R4 from binding to the −35 region (Table 1). However, amino acid residues within the distal end of R4 and the C-terminal end of σ70 make specific contacts with amino acids within the class I/II surface of SoxS56. For clarity, the interaction between the class II surface of SoxS (colored green) and the amino acids σ70 R4 with which it interacts are not positioned accurately.
In conclusion, we can now ask whether the above described interaction between SoxS and the C-terminal region of σ70 form a binary complex that lands on class II promoters during prerecruitment. Alternatively, a different binary complex might find a class II promoter and then the complex would rearrange to the above-described complex between a positive control surface(s) of SoxS and the distal end of region 4.2 of σ70. FeBABE cleavage experiments would help determine whether the binding of SoxS to the soxbox of a class II promoter prevents the binding of σ70 R4 to the −35 region, while in vitro transcription experiments22 using SoxS positive control mutants53 and cross-linking experiments may help determine whether either of the above two models is correct.
Methods and Materials
Bacterial strains, plasmids, and tri-alanine scanning mutagenesis
Table 5 lists the Escherichia coli strains used in this study and Table S1 lists the plasmids carrying members of the library of tri-alanine substitutions of σ70 R4 as well as other plasmids used in this study. The tri-alanine substitutions of σ70 R4 from positions 531–590 were introduced into the rpoD gene of plasmid pVR-σ57 by QuikChange site-directed mutagenesis (Stratagene) using primer pairs whose sequences are given in Table S2. The DNA sequences of the mutants were confirmed by the UMBC core sequencing facility using sense and antisense sequencing primers 5`-GTTGGCAAGCTTTTA-3' and 5`-GCTTTTAATCGTCCA-3', respectively. Each member of the library of tri-alanine substitutions of σ70 R4 was transformed by electroporation into derivatives of strain N7840 [pBAD33-his6-SoxS] carrying transcriptional fusions of lacZ to the fpr, mar, zwf, fumC, micF, and inaA promoters. The chromosome of N7840 carries the wild type allele of rpoD and thus constitutively expresses wild type σ70. Members of the library of tri-alanine substitutions are expressed constitutively from a truncated galP1 promoter carried on pVR- σ. The library was also introduced into strains W3110, DM0021 and RLG4282.
Table 5.
The strains used in this study.
| Strains | Relevant Genotype | Source or Reference |
|---|---|---|
| DH5α | endA1 hsdR17 supE44 thi-1 recA1 gyrA relAl Δ(argF-lac)U169 deoR(Φ80 lacZΔM15) | Lab stock |
| N7840 – fumC* | λ fumC ∷ lacZ Δmar Δ(argF-lac)U169 | R. G. Martin 36 |
| N7840 – micF* | λ micF ∷ lacZ Δmar Δ(argF-lac)U169 | R.G. Martin36 |
| N7840 – inaA* | λ inaA ∷ lacZ Δmar Δ(argF-lac)U169 | R.G. Martin36 |
| N7840 – fpr* | λ fpr ∷ lacZ Δmar Δ(argF-lac)U169 | R.G. Martin36 |
| N7840 – zwf* | λ zwf ∷ lacZ Δmar Δ(argF-lac)U169 | R.G. Martin36 |
| N7840 – marA* | λ mar ∷ lacZ Δmar Δ(argF-lac)U169 | R.G. Martin36 |
| RLG4282 (lac UP+) | NK5031/λ rrnB P1(−88 to −38,Δ72)-lac(−37 to +52)-lacZ | R.L. Gourse72 |
| W3110 | λ− IN (rrnD-rrnE)1 rph-1 | Lab stock |
| DM0021 | galP1 galE∷lacZΔ(galT)galKgalM | Lewis et al.90 |
| PB1rpoD800 | galK− rpoD800 zgh/Tn10 | Liebke et al.80 |
| GC4468 | Δ lacU169 rpsL | B. Demple91 |
The asterisks denote the name of the promoter to which lacZ is transcriptionally fused.
Assay of β-galactosidase activity in strains carrying a promoter-lacZ transcriptional fusion, a member of the pVR-σ70 library of tri-alanine substitutions and in some cases a plasmid expressing his6-SoxS
Cultures of the set of derivatives of strain N7840 [pBAD33-his6-SoxS] each carrying a member of the library of tri-alanine substitutions and one of the six SoxS-dependent promoters fused to lacZ were grown overnight in LB medium containing chloramphenicol (25 μ-g/ml) and ampicillin (100 mg/ml) at 37°C, diluted 1:100 in the same medium and grown at 37°C to a density at A600 of 0.1–0.2, at which point 0.2% arabinose was added to induce SoxS expression from the PBAD promoter of the plasmid. After growth under inducing conditions for 1 hr, expression of the transcriptional fusions in each strain was determined by duplicate assays of the β-galactosidase activity within duplicate samples taken from duplicate cultures using the high-throughput method of Griffith and Wolf88. Three independent experiments were carried out. The averages were determined and the standard errors of the mean were calculated. As described in the text, average values of β-galactosidase activity produced from a strain expressing a mutant of σ70 R4 that were ≤ 80% of the wild type activity were taken as being significantly different from the strain expressing wild type σ70; these values are shown in bold and are highlighted in the respective tables. In addition, the experiments determining the effect of the library on expression from the lac promoter (strain W3110), the galP1 promoter (strain DM0021) and the UP+ CRP-lac promoter (RLG 4282) were carried out as above, except that the cultures of W3110 and DM0021were induced with 1 mM IPTG or 0.2% D-galactose, respectively, when the A600 of the cultures reached ~0.2; cultures of strain RLG4282 were untreated.
Genetic Epistasis Tests
The epistasis experiments to define specific in vivo interactions between amino acids of SoxS and base pairs within the fpr and zwf soxboxes were conducted in strain GC4468 (Δ lac) carrying two compatible plasmids. One was pBAD33-his6-SoxS encoding wild type his6-SoxS or alanine substitutions of it, with expression of his6-SoxS being under the control of the arabinose-inducible PBAD promoter55; 88. The second plasmid was either pfpr-6 carrying the fpr promoter fused to lacZ with wild type22 or mutant41 soxboxes or pZ5 carrying the zwf promoter fused to lacZ with wild type40 or mutant41 soxboxes. The pBAD33-his6-SoxS plasmids carrying the single alanine substitutions of SoxS were prepared by digesting pBAD18-his6-SoxS carrying the respective substitutions53 with HindIII and XbaI, purifying the resulting SoxS-containing fragments and cloning them into pBAD33 digested with the same two enzymes. The plasmids are listed in Table S1.
The pairs of compatible plasmids were introduced into strain N7840 by electroporation and grown at 37°C in LB broth containing ampicillin (100 μg/ml) and chloramphenicol (25 μg/ml) to A600 of ~0.2, at which point 0.2% arabinose was added to induce SoxS synthesis. After incubation for 1 hr, samples were taken and assayed for β-galactosidase activity as described above.
The following plasmid combinations were tested for epistasis: (i) plasmid pZ5 carrying a zwf-lac fusion with the wild type, G4A and C7T alleles of the zwf soxbox together with plasmid pBAD33-his6-SoxS carrying the wild type, W36A, Q39A and R40A alleles of SoxS; (ii) plasmid pZ5 carrying a zwf-lac fusion with the wild type, C15T, A16C, A17C, A18T and A18C alleles of the zwf soxbox together with plasmid pBAD33-his6-SoxS carrying the wild type, T87A and R90A alleles of SoxS; plasmid pfpr6 carrying an fpr-lac fusion carrying the wild type, G4A, T6C and C7T alleles of the fpr soxbox together with plasmid pBAD33-his6-SoxS carrying the wild type, W36A, Q39A and R40A alleles of SoxS; and (iv) plasmid pfpr-6 carrying the wild type, C15T, A17C, A18C and A18T and A18G alleles of SoxS together with plasmid pBAD33-his6-SoxS carrying the wild type, T87A and R90A alleles. The cells were grown, SoxS synthesis was induced and samples were taken and assayed for β-galactosidase activity as described above.
Assays of overexpression toxicity on plates and measurement of growth in liquid medium
To determine whether any of the tri-alanine substitutions in the library of substitutions of σ70 R4 confer a general defect on growth, we transformed the plasmids into strain PB1rpoD80080 (Table 5) whose rpoD800 allele produces an in-frame deletion within σ70 that renders the cell temperature-sensitive for growth at 42°C; we then carried out two growth tests on each partially diploid strain. For the assay of toxicity upon overexpression of the members of the library of trialanine substitutions of σ70 R4, overnight cultures of each partially diploid strain grown in LB medium containing ampicillin (100 μg/ml) at the permissive temperature of 37°C were subjected to six serial tenfold dilutions. Then, using a multi-channel pipettor, 10μl of each dilution of a given culture was spotted onto two LB agar plates containing ampicillin (100μg/ml). One plate was incubated overnight at 37°C and the other at 42°C. This procedure was carried out for the strains carrying each member of the library. The results were recorded with a digital camera. For determining the effect of the substitutions on growth in liquid medium, the overnight cultures were diluted 1:100 into the same medium and incubated at 37°C. Initially, the A600 value for each culture was determined every 30 min but when the A600 reached a value of 0.1–0.2, the cultures were divided into two portions. One culture was incubated at 37°C and the other at 42°C, with measurements of the A600 values being taken every 20 min. The data were collected and the growth curves for each substitution at the permissive and non-permissive temperatures were plotted semi-logarithmically on the same graph.
Supplementary Material
Acknowledgements
We are grateful to Bryce Nickels, Sankar Adhya and Rick Gourse for providing strains used in this study and Tim Ford for taking the images in Table 4. We would also like to thank Steve Busby and Bryce Nickels for their useful comments and suggestions on this research. Funds for this research came from Public Health Service grant GM27113 from the National Institutes of Health and from the Designated Research Initiative Fund of University of Maryland Baltimore County, both of which were awarded to R.E.W. Funds supporting Neus Sanchez-Alberola were provided by the Fundació Cellex.
Abbreviations used
- RNAP
RNA polymerase
- σ70 R4
region 4 of σ70
- CTD
C-terminal domain
- NTD
N-terminal domain
- HTH
helix-turn-helix
- RE
Recognition Element
- CRP
cyclic AMP receptor protein
- ZBD
zinc binding domain
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gruber TM, Markov D, Sharp MM, Young BA, Lu CZ, Zhong HJ, Artsimovitch I, Geszvain KM, Arthur TM, Burgess RR, Landick R, Severinov K, Gross CA. Binding of the initiation factor sigma(70) to core RNA polymerase is a multistep process. Mol Cell. 2001;8:21–31. doi: 10.1016/s1097-2765(01)00292-1. [DOI] [PubMed] [Google Scholar]
- 2.Sharp MM, Chan CL, Lu CZ, Marr MT, Nechaev S, Merritt EW, Severinov K, Roberts JW, Gross CA. The interface of sigma with core RNA polymerase is extensive, conserved, and functionally specialized. Genes Dev. 1999;13:3015–26. doi: 10.1101/gad.13.22.3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dombroski AJ, Walter WA, Record MT, Jr., Siegele DA, Gross CA. Polypeptides containing highly conserved regions of transcription initiation factor σ70 exhibit specificity of binding to promoter DNA. Cell. 1992;70:501–512. doi: 10.1016/0092-8674(92)90174-b. [DOI] [PubMed] [Google Scholar]
- 4.Gruber TM, Gross CA. Multiple sigma subunits and the partitioning of bacterial transcription space. Annu Rev Microbiol. 2003;57:441–66. doi: 10.1146/annurev.micro.57.030502.090913. [DOI] [PubMed] [Google Scholar]
- 5.Lonetto M, Gribskov M, Gross CA. The sigma 70 family: sequence conservation and evolutionary relationships. J Bacteriol. 1992;174:3843–9. doi: 10.1128/jb.174.12.3843-3849.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borukhov S, Nudler E. RNA polymerase holoenzyme: structure, function and biological implications. Curr Opin Microbiol. 2003;6:93–100. doi: 10.1016/s1369-5274(03)00036-5. [DOI] [PubMed] [Google Scholar]
- 7.Gross CA, Chan C, Dombroski A, Gruber T, Sharp M, Tupy J, Young B. The functional and regulatory roles of sigma factors in transcription. Cold Spring Harb Symp Quant Biol. 1998;63:141–55. doi: 10.1101/sqb.1998.63.141. [DOI] [PubMed] [Google Scholar]
- 8.Murakami KS, Darst SA. Bacterial RNA Polymerases: the whole story. Curr. Op. Struct. Biol. 2003;13:31–39. doi: 10.1016/s0959-440x(02)00005-2. 2003. 13: p. 31–39. [DOI] [PubMed] [Google Scholar]
- 9.Murakami KS, Masuda S, Campbell EA, Muzzin O, Darst SA. Structural basis of transcription initiation: an RNA polymerase holoenzyme-DNA complex. Science. 2002;296:1285–1290. doi: 10.1126/science.1069595. [DOI] [PubMed] [Google Scholar]
- 10.Vassylyev DG, Sekine S, Laptenko O, Lee J, Vassylyeva MN, Borukhov S, Yokoyama S. Crystal structure of a bacterial RNA polymerase holoenzyme at 2.6 A resolution. Nature. 2002;417:712–9. doi: 10.1038/nature752. [DOI] [PubMed] [Google Scholar]
- 11.Geszvain K, Gruber TM, Mooney RA, Gross CA, Landick R. A hydrophobic patch on the flap-tip helix of E.coli RNA polymerase mediates sigma(70) region 4 function. J Mol Biol. 2004;343:569–87. doi: 10.1016/j.jmb.2004.08.063. [DOI] [PubMed] [Google Scholar]
- 12.Murakami KS, Masuda S, Darst SA. Structural basis of transcription initiation: RNA polymerase holoenzyme at 4 A resolution. Science. 2002;296:1280–1284. doi: 10.1126/science.1069594. [DOI] [PubMed] [Google Scholar]
- 13.Dove SL, Darst SA, Hochschild A. Region 4 of s as a target for transcription regulation. Mol. Microbiol. 2003;48:863–874. doi: 10.1046/j.1365-2958.2003.03467.x. [DOI] [PubMed] [Google Scholar]
- 14.Campbell EA, Westblade LF, Darst SA. Regulation of bacterial RNA polymerase s factor activity: a structural perspective. Curr. Opin. Microbiol. 2008;11:121–127. doi: 10.1016/j.mib.2008.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Patikoglou GA, Westblade LF, Campbell EA, Lamour V, Lane WJ, Darst SA. Crystal structure of the Escherichia coli regulator of sigma70, Rsd, in complex with sigma70 domain 4. J. Mol. Biol. 2007;372:649–659. doi: 10.1016/j.jmb.2007.06.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baxter K, Lee J, Minakhin L, Severinov K, Hinton DM. Mutational analysis of s70 region 4 needed for appropriation by the bacteriophage T4 transcription factors AsiA and MotA. J. Mol. Biol. 2006;363:931–944. doi: 10.1016/j.jmb.2006.08.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Minakhin L, Bhagat S, Brunning A, Campbell EA, Darst SA, Ebright RH, Severinov K. Bacterial RNA polymerase subunit omega and eukaryotic RNA polymerase subunit RPB6 are sequence, structural, and functional homologs and promote RNA polymerase assembly. Proc. Natl. Acad. Sci. USA. 2001;98:892–897. doi: 10.1073/pnas.98.3.892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lambert LJ, Wei Y, Schirf V, Demeler B, Werner MH. T4 AsiA blocks DNA recognition by remodeling sigma70 region 4. EMBO J. 2004;23:2952–62. doi: 10.1038/sj.emboj.7600312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hinton DM, Pande S, Wais N, Johnson XB, Vuthoori M, Makela A, Hook-Barnard I. Transcriptional takeover by s appropriation: remodelling of the s70 subunit of Escherichia coli RNA polymerase by the bacteriophage T4 activator MotA and co-activator AsiA. Microbiol. 2005;151:1729–1740. doi: 10.1099/mic.0.27972-0. [DOI] [PubMed] [Google Scholar]
- 20.Gallegos M-T, Schleif R, Bairoch A, Hofmann K, Ramos JL. AraC/XylS family of transcriptional regulators. Microbiol. Molec. Biol. Rev. 1997;61:393–410. doi: 10.1128/mmbr.61.4.393-410.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Amábile-Cuevas CF, Demple B. Molecular characterization of the soxRS genes of Escherichia coli: two genes control a superoxide stress regulon. Nucleic Acids Res. 1991;19:4479–4484. doi: 10.1093/nar/19.16.4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jair K-W, Fawcett WP, Fujita N, Ishihama A, Wolf RE., Jr. Ambidextrous transcriptional activation by SoxS: requirement for the C-terminal domain of the RNA polymerase alpha subunit in a subset of Escherichia coli superoxideinducible genes. Mol. Microbiol. 1996;19:307–317. doi: 10.1046/j.1365-2958.1996.368893.x. [DOI] [PubMed] [Google Scholar]
- 23.Tsaneva IR, Weiss B. soxR, a locus governing a superoxide response regulon in Escherichia coli K-12. J. Bacteriol. 1990;172:4197–4205. doi: 10.1128/jb.172.8.4197-4205.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu J, Weiss B. Two divergently transcribed genes, soxR and soxS, control a superoxide response regulon of Escherichia coli. J. Bacteriol. 1991;173:2864–2871. doi: 10.1128/jb.173.9.2864-2871.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blanchard JL, Wholey WY, Conlon EM, Pomposiello PJ. Rapid changes in gene expression dynamics in response to superoxide reveal SoxRS-dependent and independent transcriptional networks. PLoS ONE. 2007;2:e1186. doi: 10.1371/journal.pone.0001186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gu M, Imlay JA. The SoxRS response of Escherichia coli is directly activated by redox-cycling drugs rather than by superoxide. Mol. Microbiol. 2010 doi: 10.1111/j.1365-2958.2010.07520.x. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ariza RR, Li Z, Ringstad N, Demple B. Activation of multiple antibiotic resistance and binding of stress-inducible promoters by Escherichia coli Rob protein. J. Bacteriol. 1995;177:1655–1661. doi: 10.1128/jb.177.7.1655-1661.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barbosa TM, Levy SB. Differential expression of over 60 chromosomal genes in Escherichia coli by constitutive expression of MarA. J. Bacteriol. 2000;182:3467–3474. doi: 10.1128/jb.182.12.3467-3474.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cohen SP, Hächler H, Levy SB. Genetic and functional analysis of the multiple antibiotic resistance (mar) locus in Escherichia coli. J. Bacteriol. 1993;175:1484–1492. doi: 10.1128/jb.175.5.1484-1492.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Asako H, Nakajima H, Kobayashi K, Kobayashi M, Aono R. Organic solvent tolerance and antibiotic resistance increased by overexpression of marA in Escherichia coli. Appl. Environ. Microbiol. 1997;63:1428–1433. doi: 10.1128/aem.63.4.1428-1433.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakajima H, Kobayashi K, Kobayashi M, Asako H, Aono R. Overexpression of the robA gene increases organic solvent tolerance and multiple antibiotic and heavy metal ion resistance in Escherichia coli. Appl. Environ. Microbiol. 1995;61:2302–2307. doi: 10.1128/aem.61.6.2302-2307.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.White DG, Goldman JD, Demple B, Levy SB. Role of the acrAB locus in organic solvent tolerance mediated by expression of marA, soxS, or robA in Escherichia coli. J. Bacteriol. 1997;179:6122–6126. doi: 10.1128/jb.179.19.6122-6126.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rosenberg EY, Bertenthal D, Nilles ML, Bertrand KP, Nikaido H. Bile salts and fatty acids induce the expression of Escherichia coli AcrAB multidrug efflux pump through their interaction with Rob regulatory protein. Mol. Microbiol. 2003;48:1609–1619. doi: 10.1046/j.1365-2958.2003.03531.x. [DOI] [PubMed] [Google Scholar]
- 34.Nikaido H. Multidrug efflux pumps of gram-negative bacteria. J. Bacteriol. 1996;178:5853–5859. doi: 10.1128/jb.178.20.5853-5859.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee E-H, Collatz E, Podglajen I, Gutman L. A rob-like gene of Enterobacter cloacae affecting porin synthesis and susceptibility to multiple antibiotics. Antimicrob. Agents Chemother. 1998;40:6299–6313. doi: 10.1128/aac.40.9.2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martin RG, Gillette WK, Rhee S, Rosner JL. Structural requirements for marbox function in transcriptional activation of mar/sox/rob regulon promoters in Escherichia coli: sequence, orientation and spatial relationship to the core promoter. Mol. Microbiol. 1999;34:431–441. doi: 10.1046/j.1365-2958.1999.01599.x. [DOI] [PubMed] [Google Scholar]
- 37.Wood TI, Griffith KL, Fawcett WP, Jair K-W, Schneider TD, Wolf RE., Jr. Interdependence of the position and orientation of SoxS binding sites in the transcriptional activation of the class I subset of Escherichia coli superoxide-inducible promoters. Mol. Microbiol. 1999;34:414–430. doi: 10.1046/j.1365-2958.1999.01598.x. [DOI] [PubMed] [Google Scholar]
- 38.Jair K-W, Yu X, Skarstad K, Thöny B, Fujita N, Ishihama A, Wolf RE., Jr. Transcriptional activation of promoters of the superoxide and multiple antibiotic resistance regulons by Rob, a binding protein of the Escherichia coli origin of chromosomal replication. J. Bacteriol. 1996;178:2507–2513. doi: 10.1128/jb.178.9.2507-2513.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fawcett WP, Wolf RE., Jr. Purification of a MalE-SoxS fusion protein and identification of the control sites of Escherichia coli superoxide-inducible genes. Mol. Microbiol. 1994;14:669–679. doi: 10.1111/j.1365-2958.1994.tb01305.x. [DOI] [PubMed] [Google Scholar]
- 40.Fawcett WP, Wolf RE., Jr. Genetic definition of the Escherichia coli zwf “soxbox,” the DNA binding site for SoxS-mediated induction of glucose 6-phosphate dehydrogenase in response to superoxide. J. Bacteriol. 1995;177:1742–1750. doi: 10.1128/jb.177.7.1742-1750.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Griffith KL, Wolf RE., Jr. Systematic mutagenesis of the DNA binding sites for SoxS in the Escherichia coli zwf and fpr promoters: identifying nucleotides required for DNA binding and transcription activation. Mol. Microbiol. 2001;40:1141–1154. doi: 10.1046/j.1365-2958.2001.02456.x. Erratum Mol. Microbiol. 42:571. [DOI] [PubMed] [Google Scholar]
- 42.Li Z, Demple B. Sequence specificity for DNA binding by Escherichia coli SoxS and Rob proteins. Mol. Microbiol. 1996;20:937–945. doi: 10.1111/j.1365-2958.1996.tb02535.x. [DOI] [PubMed] [Google Scholar]
- 43.Nunoshiba T, Hidalgo E, Amábile Cuevas CF, Demple B. Two-stage control of an oxidative stress regulon: the Escherichia coli SoxR protein triggers redox-inducible expression of the soxS regulatory gene. J. Bacteriol. 1992;174:6054–6060. doi: 10.1128/jb.174.19.6054-6060.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu J, Weiss B. Two-stage induction of the soxRS (superoxide response) regulon of Escherichia coli. J. Bacteriol. 1992;174:3915–3920. doi: 10.1128/jb.174.12.3915-3920.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ding H, Demple B. In vivo kinetics of a redox-regulated transcriptional switch. Proc. Natl. Acad. Sci. USA. 1997;94:8445–8449. doi: 10.1073/pnas.94.16.8445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ding H, Hidalgo E, Demple B. The redox state of the [2Fe-2S] clusters in SoxR protein regulates its activity as a transcription factor. J. Biol. Chem. 1996;271:33173–33175. doi: 10.1074/jbc.271.52.33173. [DOI] [PubMed] [Google Scholar]
- 47.Gaudu P, Moon N, Weiss B. Regulation of the soxRS oxidative stress regulon. Reversible oxidation of the Fe-S centers of SoxR in vivo. J. Biol. Chem. 1997;272:5082–5086. doi: 10.1074/jbc.272.8.5082. [DOI] [PubMed] [Google Scholar]
- 48.Gaudu P, Weiss B. SoxR, a [2Fe-2S] transcription factor, is active only in its oxidized form. Proc. Natl. Acad. Sci. USA. 1996;93:10094–10098. doi: 10.1073/pnas.93.19.10094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hidalgo E, Demple B. An iron-sulfur center essential for transcriptional activation by the redox-sensing SoxR protein. EMBO J. 1994;13:138–146. doi: 10.1002/j.1460-2075.1994.tb06243.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Griffith KL, Shah IM, Myers TE, O'Neill MC, Wolf RE., Jr. Evidence for “pre-recruitment” as a new mechanism for transcription activation in Escherichia coli: the large excess of SoxS binding sites per cell relative to the number of SoxS molecules per cell. Biochem. Biophys. Res. Commun. 2002;291:979–986. doi: 10.1006/bbrc.2002.6559. Erratum. Biochem. Biophys. Res. Commun. 294:1191. [DOI] [PubMed] [Google Scholar]
- 51.Griffith KL, Wolf RE., Jr. Genetic evidence for pre-recruitment as the mechanism of transcription activation by SoxS of Escherichia coli: the dominance of DNA binding mutations of SoxS. J. Mol. Biol. 2004;344:1–10. doi: 10.1016/j.jmb.2004.09.007. [DOI] [PubMed] [Google Scholar]
- 52.Martin RG, Gillette WK, Martin NI, Rosner JL. Complex formation between activator and RNA polymerase as the basis for transcription activation by MarA and SoxS in Escherichia coli. Mol. Microbiol. 2002;43:355–370. doi: 10.1046/j.1365-2958.2002.02748.x. [DOI] [PubMed] [Google Scholar]
- 53.Griffith KL, Wolf RE., Jr. A comprehensive alanine scanning mutagenesis of the Escherichia coli transcriptional activator SoxS: identifying amino acids important for DNA binding and transcription activation. J. Mol. Biol. 2002;322:237–257. doi: 10.1016/s0022-2836(02)00782-9. [DOI] [PubMed] [Google Scholar]
- 54.Rhee S, Martin RG, Rosner JL, Davies DR. A novel DNA-binding motif in MarA: the first structure for an AraC family transcriptional activator. Proc. Natl. Acad. Sci. USA. 1998;95:10413–10418. doi: 10.1073/pnas.95.18.10413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shah IM, Wolf RE., Jr. Novel protein-protein interaction between Escherichia coli SoxS and the DNA binding determinant of the RNA polymerase a subunit: SoxS functions as a co-sigma factor and redeploys RNA polymerase from UP-element-containing promoters to SoxS-dependent promoters during oxidative stress. J. Mol. Biol. 2004;343:513–532. doi: 10.1016/j.jmb.2004.08.057. [DOI] [PubMed] [Google Scholar]
- 56.Zafar A, Shah IM, Wolf RE., Jr. Protein-protein interactions between s70 region 4 of RNA polymerase and Escherichia coli SoxS, a transcription activator that functions by the pre-recruitment mechanism: evidence for “Off-DNA” and “On-DNA” interactions. J. Mol. Biol. 2010;410:13–32. doi: 10.1016/j.jmb.2010.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rhodius VA, Busby SJ. Interactions between activating region 3 of the Escherichia coli cyclic AMP receptor protein and region 4 of the RNA polymerase sigma70 subunit: application of suppression genetics. J. Mol. Biol. 2000;299:311–24. doi: 10.1006/jmbi.2000.3737. [DOI] [PubMed] [Google Scholar]
- 58.Bhende PM, Egan SM. Genetic evidence that transcription activation by RhaS involves specific amino acid contacts with sigma 70. J. Bacteriol. 2000;182:4959–4969. doi: 10.1128/jb.182.17.4959-4969.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Grainger DC, Belyaeva TA, Lee DJ, Hyde EI, Busby SJW. Transcription activation at the Escherichia coli melAB promoter: interactions of MelR with the C-terminal domain of the RNA polymerase a subunit. Mol. Microbiol. 2004;51:1311–1320. doi: 10.1111/j.1365-2958.2003.03930.x. [DOI] [PubMed] [Google Scholar]
- 60.Bown JA, Kolb A, Meares CF, Ishihama A, Minchin SD, Busby SJ. Positioning of region 4 of the Escherichia coli RNA polymerase sigma(70) subunit by a transcription activator. J Bacteriol. 2000;182:2982–4. doi: 10.1128/jb.182.10.2982-2984.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bhende PM, Egan SM. Amino acid-DNA contacts by RhaS: an AraC family transcription activator. J Bacteriol. 1999;181:5185–92. doi: 10.1128/jb.181.17.5185-5192.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jair K-W, Martin RG, Rosner JL, Fujita N, Ishihama A, Wolf RE., Jr. Purification and regulatory properties of MarA protein, a transcriptional activator of Escherichia coli multiple antibiotic and superoxide resistance promoters. J. Bacteriol. 1995;177:7100–7104. doi: 10.1128/jb.177.24.7100-7104.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Martin RG, Gillette WK, Rosner JL. Promoter discrimination by the related transcriptional activators MarA and SoxS: differential regulation by differential binding. Mol. Microbiol. 2000;35:623–634. doi: 10.1046/j.1365-2958.2000.01732.x. [DOI] [PubMed] [Google Scholar]
- 64.Martin RG, Rosner JL. Genomics of the marA/soxS/rob regulon of Escherichia coli: identification of directly activated promoters by application of molecular genetics and informatics to microarray data. Mol. Microbiol. 2002;44:1611–1624. doi: 10.1046/j.1365-2958.2002.02985.x. [DOI] [PubMed] [Google Scholar]
- 65.Pomposiello PJ, Bennik MH, Demple B. Genome-wide transcriptional profiling of the Escherichia coli responses to superoxide stress and sodium salicylate. J. Bacteriol. 2001;183:3890–3902. doi: 10.1128/JB.183.13.3890-3902.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ross W, Gosink KK, Salomon J, Igarashi K, Zou C, Ishihama A, Severinov K, Gourse RL. A third recognition element in bacterial promoters: DNA binding by the alpha subunit of RNA polymerase. Science. 1993;262:1407–13. doi: 10.1126/science.8248780. [DOI] [PubMed] [Google Scholar]
- 67.Blatter EE, Ross W, Tang H, Gourse RL, Ebright RH. Domain organization of RNA polymerase alpha subunit: C-terminal 85 amino acids constitute a domain capable of dimerization and DNA binding. Cell. 1994;78:889–96. doi: 10.1016/s0092-8674(94)90682-3. [DOI] [PubMed] [Google Scholar]
- 68.Campbell EA, Muzzin O, Chlenov M, Sun JL, Olson CA, Weinman O, Trester-Zedlitz ML, Darst SA. Structure of the bacterial RNA polymerase promoter specificity s subunit. Mol. Cell. 2002;9:527–539. doi: 10.1016/s1097-2765(02)00470-7. [DOI] [PubMed] [Google Scholar]
- 69.Siegele DA, Hu JC, Walter WA, Gross CA. Altered promoter recognition by mutant forms of the sigma 70 subunit of Escherichia coli RNA polymerase. Journal of Molecular Biology. 1989;260:591–603. doi: 10.1016/0022-2836(89)90568-8. [DOI] [PubMed] [Google Scholar]
- 70.Chen H, Tang H, Ebright RH. Functional interaction between RNA polymerase a subunit C-terminal domain and s70 in UP-element and activator-dependent transcription. Mol. Cell. 2003;11:1621–1633. doi: 10.1016/s1097-2765(03)00201-6. [DOI] [PubMed] [Google Scholar]
- 71.Schwartz D, Beckwith JR. Mutants missing a factor necessary for the expression of catabolite-sensitive operons in E. coli. In: Zipser D, Beckwith JR, editors. The Lac Operon. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, N.Y.: 1970. pp. 417–422. [Google Scholar]
- 72.Ross W, Aiyar S, Salomon J, Gourse R. Escherichia coli promoters with UP elements of different strengths: Modular structure of bacterial promoters. J. Bacteriol. 1998;180:5375–5383. doi: 10.1128/jb.180.20.5375-5383.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Savery NJ, Lloyd GS, Busby SJW, Thomas MS, Ebright RH, Gourse RL. Determinants of the C-terminal domain of the Escherichia coli RNA polymerase a subunit important for transcription at class I cyclic AMP receptor protein-dependent promoters. J. Bacteriol. 2002;184:2273–2280. doi: 10.1128/JB.184.8.2273-2280.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kumar A, Malloch RA, Fujita N, Smillie DA, Ishihama A, Hayward RS. The minus 35-recognition region of Escherichia coli sigma 70 is inessential for initiation of transcription at an “extended minus 10” promoter. Journal of Molecular Biology. 1993;232:406–418. doi: 10.1006/jmbi.1993.1400. [DOI] [PubMed] [Google Scholar]
- 75.Burns HD, Belyaeva TA, Busby SJ, Minchin SD. Temperature-dependence of open-complex formation at two Escherichia coli promoters with extended −10 sequences. Biochem. J. 1996;317:305–311. doi: 10.1042/bj3170305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Keilty S, Rosenberg M. Constitutive function of a positively regulated promoter reveals new sequences essential for activity. J Biol Chem. 1987;262:6389–95. [PubMed] [Google Scholar]
- 77.Ponnambalam S, Webster C, Bingham A, Busby S. Transcription initiation at the Escherichia coli galactose operon promoters in the absence of the normal −35 region sequences. J Biol Chem. 1986;261:16043–8. [PubMed] [Google Scholar]
- 78.Hook-Barnard IG, Hinton DM. Transcription initiation by mix and match elements:flexibility for polymerase binding to bacterial oromoters. Gene. Regul. Syst. Bio. 2007;1:275–193. [PMC free article] [PubMed] [Google Scholar]
- 79.Mitchell JE, Zheng D, Busby SJW, Michin SD. Identification and analysis of “extended −10” promoters in Escherichia coli. Nucleic Acid Research. 2003;31:4689–4695. doi: 10.1093/nar/gkg694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Liebke H, Gross C, Walter W, Burgess R. A new mutation rpoD800, affecting the sigma subunit of E. coli RNA polymerase is allelic to two other sigma mutants. Mol Gen Genet. 1980;177:277–82. doi: 10.1007/BF00267439. [DOI] [PubMed] [Google Scholar]
- 81.Nickels BE, Garrity SJ, Mekler V, Minakhin L, Severinov K, Ebright RH, Hochschild A. The interaction between sigma70 and the beta-flap of Escherichia coli RNA polymerase inhibits extension of nascent RNA during early elongation. Proc Natl Acad Sci U S A. 2005;102:4488–93. doi: 10.1073/pnas.0409850102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nickels BE, Dove SL, Murakami KS, Darst SA, Hochschild A. Protein-protein and protein-DNA interactions of sigma 70 region 4 involved in transcription activation by lambda cI. J Mol Biol. 2002;324:17–34. doi: 10.1016/s0022-2836(02)01043-4. [DOI] [PubMed] [Google Scholar]
- 83.Kuznedelov K, Minakhin L, Niedziela-Majka A, Dove SL, Rogulja D, Nickels BE, Hochchild A, Heyduk T, Severinov K. A role for interaction of the RNA polymerase flap domain with the s subunit in promoter recognition. Science. 2002;295:855–857. doi: 10.1126/science.1066303. [DOI] [PubMed] [Google Scholar]
- 84.Kuldell N, Hochschild A. Amino acid substitutions in the −35 recognition motif of sigma 70 that result in defects in phage lambda repressor-stimulated transcription. J. Bacteriol. 1994;176:2991–2998. doi: 10.1128/jb.176.10.2991-2998.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hinton DM. Molecular gymnastics: distortion of an RNA polymerase s factor. Trends Microbiol. 2005;13:140–143. doi: 10.1016/j.tim.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 86.Gregory BD, Nickels BE, Garrity SJ, Severinova E, Minakhin L, Urbauer RJ, Urbauer JL, Heyduk T, Severinov K, Hochschild A. A regulator that inhibits transcription by targeting an intersubunit interaction of the RNA polymerase holoenzyme. Proc Natl Acad Sci U S A. 2004;101:4554–9. doi: 10.1073/pnas.0400923101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kumar A, Moran CP., Jr. Promoter activation by repositioning of RNA polymerase. J Bacteriol. 2008;190:3110–7. doi: 10.1128/JB.00096-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Griffith KL, Wolf RE., Jr. Measuring b-galactosidase activity in bacteria: cell growth, permeabilization, and enzyme assays in 96-well arrays. Biochem. Biophys. Res. Commun. 2002;290:397–402. doi: 10.1006/bbrc.2001.6152. [DOI] [PubMed] [Google Scholar]
- 89.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–12. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 90.Lewis DE, Geanacopoulos M, Adhya S. Role of HU and DNA supercoiling in transcription repression: specialized nucleoprotein repression complex at gal promoters in Escherichia coli. Mol Microbiol. 1999;31:451–61. doi: 10.1046/j.1365-2958.1999.01186.x. [DOI] [PubMed] [Google Scholar]
- 91.Greenberg JT, Monach P, Chou JT, Demple B. Positive control of a global antioxidant defense regulon activated by superoxide-generating agents in Escherichia coli. Proc. Natl. Acad. Sci. USA. 1990;87:6181–6185. doi: 10.1073/pnas.87.16.6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.