Abstract
Identifying the dynamics of individual molecules along their reactive pathways remains a major goal of modern chemistry. For simple chemical reactions, the transition state position is thought to be highly localized. Conversely, in the case of more complex reactions involving proteins, the potential energy surfaces become rougher, resulting in heterogeneous reaction pathways with multiple transition state structures. Force-clamp spectroscopy experimentally probes the individual reaction pathways sampled by a single protein under the effect of a constant stretching force. Herein, we examine the distribution of conformations that populate the transition state of two different reactions; the unfolding of a single protein and the reduction of a single disulfide bond, both occurring within the same single protein. By applying the recently developed static disorder theory, we quantify the variance of the barrier heights, σ2, governing each distinct reaction. We demonstrate that the unfolding of the I27 protein follows a non-exponential kinetics, consistent with a high value of σ2 ~ 18 (pNnm)2. Interestingly, shortening of the protein upon introduction of a rigid disulfide bond significantly modulates the disorder degree, spanning from σ2 ~ 8 to ~21 (pNnm)2. These results are in sharp contrast with the exponential distribution of times measured for an SN2 chemical reaction, implying the absence of static disorder σ2 ~0 (pNnm)2. Our results demonstrate the high sensitivity of the force-clamp technique to capture the signatures of disorder in the individual pathways that define two distinct force-induced reactions, occurring within the core of a single protein.
Introduction
Elucidating the dynamics of reactant molecules along the particular pathway that connects them with the resulting products has become a cornerstone in modern chemistry, allowing detailed reconstruction of the potential energy surface of a chemical reaction. In general, for small systems composed of only a few molecules, the valley connecting reactants with products is narrow, the reaction pathways are well defined and the position of the transition state is highly localized 1,2. For example, the study of elementary chemical reactions such as the simple SN2 nucleophilic substitution, mainly using crossed molecular beams and ab-initio calculations, has provided detailed information regarding the position and height of the free-energy barriers determining the process 3–7. By contrast, in the case of more complex reactions involving bigger molecular systems such as proteins, the potential energy surfaces become rougher, which results in heterogeneous reaction pathways with multiple transition state structures1. The complexity of the resulting energy landscapes has been mainly revealed by the use of computational methods, which sample the individual (un)folding trajectories of a single protein molecule while under particular force-field conditions8–14. The high degree of static and dynamic heterogeneity between individual trajectories required statistical mechanical tools to interpret these data12,13. In sharp contrast with the in-silico observations, the macroscopic nature of traditional protein folding experiments conducted in the bulk averages out the pathways in the ensemble, thus resulting in a smooth free-energy landscape that mainly captures two stable thermodynamic states, namely the folded and unfolded conformations15–17. The conversion between these two states has been traditionally given the treatment of a first order chemical reaction, implying that the concentration of the reactant species –the protein native state– decreases exponentially with time15. This observation entails that each individual molecule has a well defined and unique native state, the unfolding path is the same and the saddle point defining the transition state is conserved.
The advent of single molecule techniques, primarily fluorescence spectroscopy, has allowed measuring the fluctuation dynamics of single enzyme molecules over time18. These experiments revealed a wide range of catalytic activity within individual enzymes present in the ensemble population (“static disorder”) and also large variations in the catalytic activity of an individual enzyme (“dynamic disorder”)18. Such a complex behavior, invisible to bulk experiments, manifests itself in the non-exponential kinetics measured for different enzymes19,20. Theoretical works describing these experiments have carefully identified the main features of the potential energy surfaces (often exhibiting multiple saddle points) that govern the large number of reactive paths measured in the experiment21–29. The combination of experimental and theoretical work is therefore required to ascribe the complexity in the kinetics of catalysis to the molecular mechanisms involved in the conformational changes of the single enzyme over time20,30. The complex kinetics experimentally measured for large biomolecules such as enzymes contrasts with the apparent simplicity of chemical reactions with small molecules, which can also be now investigated one molecule at a time31,32.
Thus, it becomes clear that the emergence of single molecule techniques is providing a new vista on chemical reactivity. Of particular relevance, single molecule force-clamp spectroscopy excels at probing the conformational dynamics of a single protein under force33. By applying a calibrated constant force to a single protein, the individual unfolding pathways of single proteins can be directly monitored34. In these experiments, the multidimensional energy surface can be projected onto a single dimension, the end-to-end length of the stretched protein, which defines the reaction coordinate with sub-Ångström resolution. Using a large pool of individual unfolding trajectories, force-clamp measurements revealed that the time-course of unfolding of the small protein ubiquitin clearly departed from the single exponential survival probability associated with a simple two-state unfolding scenario35–37. Follow-up experiments demonstrated an increased broadening in the distribution of unfolding pathways of ubiquitin as the constant pulling force was increased, scaling with F2. These latter results were interpreted in the framework of static disorder in the classical Arrhenius description 38. This theory provides the means to quantify the variance, σ2, in the amount of disorder in the barrier heights with respect to average value assuming a Gaussian distribution. The implications derived from the unfolding kinetics obtained from our single molecule experiments are deep, indicating that force-clamp spectroscopy can be now used to directly probe the broad conformational diversity defining the native state of proteins using the mechanical stability of each conformation as its structural fingerprint.
Contrary to these observations on the ubiquitin protein, mechanical unfolding of protein G resulted in a simple all-or-none two-state transition, consistent with an exponentially distribution of unfolding times at a given constant force39. These results suggest that the degree of disorder in the unfolding pathways measured for a single protein under force is likely to be highly protein specific, varying over a wide range. The challenge lies now on discovering the molecular origin of the non-exponential kinetics encountered in mechanical unfolding of proteins, and to distinguish it from a distinct molecular process that follows first-order kinetics occurring under the same experimental conditions, within the structure of a single protein.
Besides studying protein (un)folding, the combination of protein engineering and force-clamp spectroscopy has allowed direct examination of the effect of a constant force on the potential energy surface of a chemical reaction, at the single bond level 40. Our results have demonstrated that the reduction of a single disulfide bond embedded within the core of a protein is activated by the pulling force in the presence of nucleophiles 41. These experiments have permitted reconstruction of the energy landscape of a chemical reaction under force, allowing capture of the most plausible conformation of the reaction transition state 41–43. Herein, we compare the effect of a mechanical constant force on the energy landscape of two different reactions; the unfolding of a single protein and the reduction of a single disulfide bond by a small nucleophile, both occurring within the same single protein. Our results demonstrate that, similar to ubiquitin, the transition state of unfolding of the 27th immunoglobulin-like domain of cardiac titin (I27), the mechanical properties of which have been amply studied 44,45, is not unique, but instead composed of a set of similar structures. Interestingly, shortening of the I27 protein by the presence of disulfide bonds has a strong effect on the measured heterogeneity in the unfolding pathways. Our results are quantitatively discussed within the framework of the static disorder theory. In sharp contrast with these results, the energy landscape of an SN2 chemical reaction, occurring within a much shorter length-scale, is smooth, implying a high degree of homogeneity within the individual reactive pathways connecting the reactants with the reaction products. Our results demonstrate the ability of the force-clamp technique, through the acquisition of a large statistical pool of data, to uncover fine details of the ensemble of transition state structures populating the saddle point of the potential energy surface governing protein (un)folding and chemical reactions, at the single molecule level.
Materials and methods
Protein engineering
PolyI27 (I278) was constructed by consecutive subcloning of the respective monomers, using the BamHI and BgIII restriction sites44. The disulfide bond mutants were constructed using a cysteine-free I27 protein in which native Cys47 and Cys63, which do not form a disulfide bond, were mutated to alanines. Using the QuickChange site-directed mutagenesis kit (Stratagene), we introduced additional mutations to form disulfide bonds at specific positions within the I27 protein sequence: Gly32 to Cys, Ala75 to Cys (I27G32C–A75C); Pro28 to Cys, Lys54 to Cys (I27P28C–K54C); Glu24 to Cys, Lys55 to Cys (I27E24C–K55C)46. As in the case of the wt-(I278) protein, we constructed an eight-domain N–C linked polyprotein for each I27 mutant. Such polyproteins were engineered by consecutive subcloning of the respective monomers using the BamH I, Kpn I and Bgl II restriction sites. The eight-domain polyproteins were cloned into the pQE80L (Qiagen) expression vector and transformed into the BLR DE3 Escherichia coli expression strain. Each polyprotein construct was finally purified by histidine metal-affinity chromatography with Talon resin (Clontech) and by gel filtration using Superdex 200 HR column (GE BioSciences).
Force Spectroscopy
Force-clamp AFM experiments were conducted at room temperature using a home-made set-up under force-clamp conditions described elsewhere34. The sample was prepared by depositing 1–10 ml of protein in PBS solution (at a concentration of 1–10 mg ml−1) onto a freshly evaporated gold coverslide. Each cantilever (Si3N4 Veeco MLCT-AUHW) was individually calibrated using the equipartition theorem, which gave a typical spring constant of 15 pN/nm. Single proteins were picked up from the surface by pushing the cantilever onto the surface with a contact force of 500–1,000 pN to promote the non-specific adhesion of the proteins on the cantilever surface. The piezoelectric actuator was then retracted to produce a set deflection (force), which was kept constant throughout the experiment using an external, active feedback mechanism while the extension was recorded. The force feedback was based on a proportional, integral and differential amplifier, the output of which was fed to the piezoelectric positioner. The feedback response is limited to ~2–5 ms. The high resolution piezoelectric actuator allowed our measurements of protein length a peak-to-peak resolution of ~0.5 nm. Data from the force traces were filtered using a pole Bessel filter at 1 kHz. The reaction rate constants that we measure (~ 1 s−1) are chosen such that they are not compromised by the feedback bandwidth or cantilever drift occurring over much longer timescales. In the chemical reduction experiments, a PBS solution containing 10 mM 1-mercapto-2-propanol was used, and the pH value was adjusted to 7.4.
Data analysis
All data were recorded and analysed using custom software written in Igor Pro 6.0 (Wavemetrics). The fingerprint of a single polyprotein in our unfolding experiments was considered to be at least six well-resolved steps of the corresponding height in each construct exhibiting long detachment times in order not to bias the unfolding probability37. In the case of the reduction experiments, no traces that included unfolding events during the second force pulse were included in the analysis. Only data with more than five reduction events in the second pulse were included in the analysis. The survival probability is defined as , where p(t)dt corresponds to the probability of unfolding during the period of time between t and t + dt. The survival probability at each force is obtained from the associated probability density histogram of unfolding times, p(t). The integral in the fitting function (Eq. 2) is evaluated by taking a sum of the integrand over the interval [− 6σ,6σ], with the interval width of dr = 0.001. All the fitting procedures are performed by the Levenberg-Marquardt least-squares algorithm, implemented in the Igor Pro 6 software package. The errors of k and σ2 are estimated by the bootstrap method 47.
Results
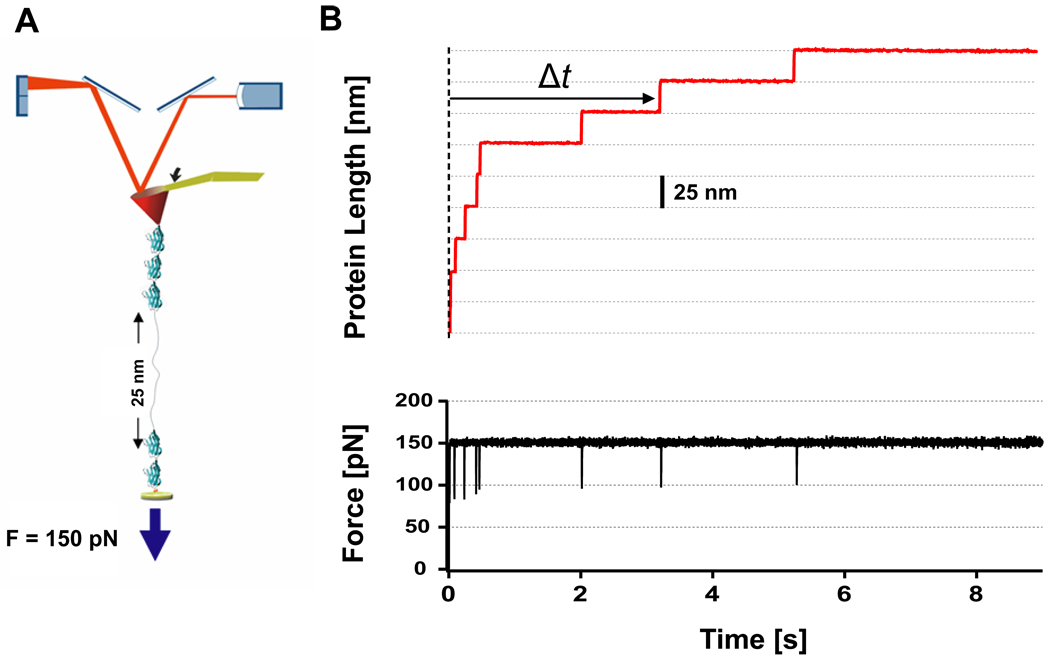
In our force-clamp essay, we make use of polyproteins composed of 8 identical domains of the I27 protein (Fig. 1A). Under a constant force of 150 pN, each individual protein module in the polyprotein chain unfolds yielding a step increase in length of ~25 nm (Fig. 1B and Figure 2A, red trace)37. The resulting unfolding trajectories resemble a staircase, where the unfolding of each particular domain occurs stochastically at a time Δt from the moment that the force is applied to the protein (vertical dotted line in Fig. 1B). By capturing 319 individual unfolding trajectories such as those shown in Fig.1B and Fig. 2A, we collected 2,296 independent unfolding events, the dwell time distribution of which is shown in Fig. 2B. The average unfolding rate calculated as k =1/<t>, where <t>, corresponds to the average time for unfolding, is k = 0.83 s−1. However, a single exponential distribution with a rate constant of 0.83 s−1 (black line in Fig. 2B) fails to capture the full time distribution of events. Similar conclusions can be drawn from the cumulative distribution of unfolding times shown in Fig. SI1A; the single exponential fit to the experimental data (dotted black line) does not capture the distribution of unfolding times. Notably, the experimental distribution of unfolding times shows a high population of events that occur faster than that predicted by a single exponential distribution as shown in the normalized residuals plot in Fig. SI1A (inset) and also in Fig. 2B.
Figure 1. Unfolding the wild-type I278 polyprotein in a force-clamp.
A) Schematics of the experimental set-up. B) Unfolding the I27 polyprotein under a constant force of 150 pN results in a staircase-like elongation, where the unfolding of each monomer in the chain occurs stochastically at a time Δt after the application of force eliciting steps of ~25 nm in length.
Figure 2. Probing the kinetics of protein unfolding and disulfide reduction within the same single molecule.
A) Unfolding the (I27G32C-A75C)8 polyprotein under a constant force of 180 pN (blue trace) results in a stepwise elongation of 11 nm (inset), corresponding to the extension of the protein up to the position of the mechanically rigid disulfide bond. Exposing the disulfide bond to a solution containing 10 mM 1-mercapto-2-propanol at 300 pN triggers the reduction of the disulfide bond, which is marked by a ~14 nm step-wise elongation (orange trace) corresponding to the extension of the amino acids trapped behind the disulfide bond. The total protein elongation of the (I27G32C-A75C)8 construct after the unfolding and reduction reactions matches the length of the unfolded wild-type I27 protein (red trace). B) A histogram corresponding to 2296 unfolding events of the wt-I27 protein measures the probability of unfolding at a constant pulling force of 150 pN. At short dwell times the distribution deviates significantly from a single exponential (black line). C) A histogram of 1269 unfolding events of the (I27G32C-A75C)8 protein at a constant pulling force of 130 pN cannot be reproduced by a single exponential distribution with an associated characteristic time k =1/<t> (black line). D) Time-course distribution corresponding to 897 individual disulfide reduction events occurring under a constant force of 300 pN. The distribution can be precisely captured by a single exponential fit (black line).
Unlike classical unfolding experiments using chemicals or large temperature jumps, the mechanical unfolding of proteins is a highly localized process mainly involving the rupture of a few key hydrogen bonds within the structure of the protein native state, which constitute the crucial structural motif that provides the protein with mechanical stability48,49. In the case of the I27 protein, such mechanical clamp is placed between the A´G strands, encompassing amino acids 9–15 and 83–8749. While mutations in the amino acids constituting the protein transition state are known to significantly alter the mechanical stability of the protein48, the nature of the amino acids composing the peripheral regions beyond such a mechanical clamp and the total length of the protein are generally believed not to considerably affect its mechanical resistance. However, it is plausible that changes in the protein length involve little rearrangements in the conformation of the protein’s native state, which would remain invisible to ensemble experiments, especially when the 3D crystal structure is not available. In order to test this hypothesis, we engineered a double mutation (Gly32Cys and Ala75Cys) into the core of the I27 protein, which spontaneously forms a disulfide bond that is buried and inaccessible to the external solvent environment 46. We constructed and expressed a polyprotein that consisted of eight identical repeats of this mutated domain, (I27G32C-A75C)8. Pulling this construct under a constant force of 130 pN results in a staircase in which the length of each step measures ~ 11 nm (Fig. SI3B). Such protein extension corresponds to the unfolding of the protein up to the position of the disulfide bond, which acts as an internal covalent mechanical clamp that prevents the protein from completely unraveling once the hydrogen bond motif has been disrupted upon the application of force. We gathered 180 individual full unfolding trajectories containing 1,269 unfolding events, which yielded an average unfolding rate of k = 0.99 s−1. As before, a single exponential distribution with a rate constant of 0.99 s−1 (black line in Fig. 2C) does not fully capture the time distribution of unfolding events (Fig. SI1B).
The results presented here demonstrate that the unfolding of both the wt-I27 protein (Fig. 2B) and the shorter disulfide bond truncated form (Fig. 2C) exhibit unfolding pathways that are heterogeneous, evocative of a native state composed of an ensemble of similar conformations with slightly different energies35. Therefore, the unfolding process exhibits a higher degree of complexity than expected for a simple first-order chemical reaction with a single reactant species exhibiting a smooth energy surface. Besides allowing investigation of the individual unfolding pathways (Fig. 2C), the (I27G32C-A75C)8 polyprotein provides the perfect platform to experimentally investigate, at the single bond level, the individual reaction pathways of the simple SN2 chemical reaction that involves the reduction of the disulfide bond that shortcuts the I27 protein in the presence of a nucleophile. Following the experimental approach of our previous studies40,41, we used a double pulse protocol to study the force-dependent kinetics of disulfide bond reduction (Fig. 2A). The first force pulse (180 pN) rapidly unfolded the modules in the polyprotein, resulting in the ~11 nm steps (blue trace). Unfolding the protein exposes the buried disulfide bond to the solution, which allows the chemical attack provided that a nucleophile agent is present in the solution. A second force pulse at a higher force (300 pN) is applied to the protein shortly after all the modules unfolded. In the presence of 10 mM of 1-mercapto-2-propanol in the solution, the reduction of each disulfide bond is marked by a ~14 nm elongation step (Fig. 2A, orange trace), that corresponds to the total length of the residues trapped behind the disulfide bond. The total length of the unfolded protein after complete disulfide bond reduction exactly matches the length of the I27 wild-type protein (red trace). Figure 2D shows the time distribution corresponding to 897 individual disulfide reduction events stemming from 144 individual reactive trajectories. In sharp contrast with the time distribution corresponding to the unfolding process (Fig. 2C), the reduction of a disulfide bond follows a perfect exponential time-course distribution, as revealed by the single exponential fit (black line) to the data, yielding a reduction rate, k = 1.03 s−1, which perfectly corresponds to the reaction rate calculated as k =1/<t> = 1.01 s−1. This result suggests that the reduction of a disulfide bond by an S-based nucleophile in solution follows a rather precise pathway with a conserved transition state structure. We then examined whether the specific location of the disulfide bond within the I27 structure had a role in the disulfide bond reactivity. We repeated the experiments described in Fig. 2A,C–D with the (I27E24C-K55C)8 construct, where the disulfide bond shortcuts the protein between positions 24 and 55. In this case, unfolding the protein up to the disulfide bond elicits steps of ~14.5 nm. The reduction of the disulfide bond, studied during the second force pulse at 300 pN, is identified by a step increase in length of ~10.4 nm, which corresponds again to the length of the amino acids trapped behind the disulfide bond (Figure SI2). As before, the time-course distribution of the unfolding events in this case follows a perfect single exponential, demonstrating that the apparent simplicity observed for the disulfide bond reduction is independent of the chemical environment of the reactive site.
In our experiments, we chose the particular conditions of force and nucleophile concentration such that the three different processes studied here –unfolding of the wild-type protein, unfolding of the truncated protein and reduction of the disulfide bond–occur at the same rate (~ 1 s−1, see Fig. 2B, 2C, 2D and Fig. SI1). However, the time-course distribution of the individual events in the case of protein unfolding or disulfide reduction is significantly different. The subtleties emerge only when the individual pathways are investigated under constant force conditions at the single molecule level. In order to quantify the diversity in the number of pathways and transition state structures visited during the reaction, we recently developed a generalized Arrhenius equation50, based on Zwanzig’s static disorder theory51,52, to account for the non exponential survival probability that we observe in our experiments38(Fig. 2B and 2C). From the molecular perspective, proteins are known to exhibit conformational diversity in the native state ensemble53. In this scenario, static disorder entails that the interconversion rates between the different structural conformers is slow compared to the reaction rate. Such structural complexity is likely to result in a diverse set of unfolding pathways, which would lead to fluctuations in the height of the free-energy barriers and also in the conformation of the reaction transition state.
Adapting the Arrhenius/Bell formalism 50,54 to include the disorder in the barrier height, σΔG, and in the transition state conformation, σΔx, the mean rate of a reaction is given by:
| [1] |
where r is the amount of disorder in the barrier heights with respect to the average value, ΔGavg − FΔxavg. In this equation, A stands for the preexponential factor, kB is the Boltzmann constant, T is the absolute temperature, ΔG is the height of the free-energy barrier of reaction in the absence of force, and Δx represents the actual distance from the native, initial conformation to the transition state conformation along the reaction coordinate. In our force-clamp experiments, we measure the ensemble-averaged survival probability (Fig. 2BCD). In the presence of static disorder, the ensemble-averaged survival probability 〈S(t,F)〉 is given by the superposition of the survival probability for each reaction pathway, weighted by the probability of the corresponding pathway, giving rise to:
| [2] |
where
| [3] |
In the absence of disorder (σ →0; f (r)→δ (r)), the survival probability becomes a single exponential, in good agreement with the classical Arrhenius conception. Conversely, in the presence of disorder (σ > 0), the survival probability becomes non-exponential. Using this refined statistical approach 38, we tested this theory on our experimental data on protein unfolding and chemical reaction. The histograms of measured unfolding dwell times in each case (Fig. 2BC) correspond to the probability of unfolding, p(t). From these histograms, we simply calculate the ensemble-averaged survival probability as . Figure 3A and 3B show the plot of the ln[− ln 〈S(t)〉] versus ln t obtained for the wild-type and the (I27G32C-A75C)8 polyproteins, respectively. The fit of the static disorder model (Eq. 2) reproduces quite well the experimental data in both cases, yielding values for k and σ2 that are listed in Table 1. The errors in the fit parameters were estimated using the bootstrap method47. For comparison, the single exponential fit is included in the plots as a continuous black line. The obtained value for σ2 combines the contribution to the disorder arising from the fluctuations in both the height of the energy barrier and the distance to the transition state. While in the experiments described here we cannot decouple both effects, σ2 is a quantitative reporter of the degree of heterogeneity in the transition state sampled in each particular force-activated reaction. The obtained high values of σ2 for the wt-I27 protein (17.5 ± 1.7) and for the truncated I27G32C-A75C form (7.7 ± 1.3) quantitatively confirm the high degree of heterogeneity measured in the protein unfolding pathways under force. In apparent contrast with these results, fitting the static disorder model (Eq.2) to the experimental time distribution corresponding to the disulfide bond reduction under force (Fig. 2C) results in a value of σ2 = 3.41·10−5 ± 0.2 (Table 1), thus implying the absence of disorder and the precise agreement with a single exponential time distribution consistent with simple first order kinetics (Fig 3C).
Figure 3. Surviving probability for the unfolding and reduction reactions is well described by static disorder theory.
Plot of ln[− ln 〈S(t)〉] versus ln t corresponding to the unfolding of the wt-I27 protein at 150 pN (A), the unfolding of the truncated (I27G32C-A75C)8 protein at 130 pN (B), and the reduction of the disulfide bond in the (I27G32C-A75C)8 protein at 300 pN (C). The solid color lines represent the fits of the static disorder survival probability (Eq. 2) to the data in each case with the reaction rate k and the variance of the barrier heights σ2 as fit parameters. The obtained values in each case are listed in Table 1. For comparison, the fits to a single exponential are included in the figures (black traces). While the single exponential fit captures well the time-course distribution for the disulfide bond reduction (C), it clearly fails to reproduce the data corresponding to the unfolding reaction (A, B).
Table 1. Kinetic parameters for protein unfolding and disulfide bond reaction from the static disorder model fit.
The reaction rate (unfolding o chemical reduction) k, and the variance of the barrier heights, σ2, were obtained from the static disorder model fit to the data. The errors were estimated using the bootstrap method.
| k [s−1] | σ2 [(pNnm)2] | |
|---|---|---|
| I27wt unfolding | 1.1 ± 0.1 | 17.5 ± 1.7 |
| I27G32C-A75C unfolding | 1.2 ± 0.1 | 7.7 ± 1.3 |
| I27 G32C-A75C reduction | 1.0 ± 0.1 | 3.4×10−4 ± 0.2 |
| I27E24C-K55C unfolding | 1.9 ± 0.1 | 21.3 ± 2.4 |
| I27P28C-K54C unfolding | 3.5 ± 0.2 | 15.7 ± 2.1 |
| I27E24C-K55C reduction | 1.0 ± 0.1 | 9.8 ×10−4 ± 0.7 |
While the difference in the measured values for disorder, σ2, between protein unfolding and chemical reduction reactions is evident, differences within the disorder values obtained for the unfolding process of both I27 protein forms are also noticeable. Since the σ2 value obtained for the short I27G32C-A75C form is smaller than that corresponding to the wt-I27, it is conceivable that the fluctuations in the protein unfolding pathways directly correlate with the protein length. In order to test this hypothesis, we investigated the distribution of unfolding pathways of a set of different I27 protein mutants where the position of the disulfide bond is varied within the structure, namely in positions 32–75, 24–55 and 28–54. Thus, the number of amino acids being extended upon unfolding of the protein (the unsequestered amino acids) is different in each case, namely 46 amino acids for the (I27G32C-A75C) construct, 58 amino acids for the (I27E24C-K55C) protein and 63 amino acids in the case of the (I27P28C-K54C) protein. The wild-type I27 protein is composed of 89 residues. Examples of experimental trajectories obtained for each I27 protein mutant are shown in Fig. SI3.The distribution of unfolding times corresponding to the unfolding of the (I27G32C-A75C)8, (I27E24C-K55C)8 and (I27P28C-K54C)8 polyproteins at 130 pN are shown in Figures 4A–C. In none of the cases, a single exponential time distribution with an associated time constant k =1/<t> (black line) fully reproduces the experimental distribution of unfolding times. Figures 4D–F show the fit of the static disorder theory for each protein mutant. The values of the resulting fitting parameters are included in Table 1. While the shortest protein, (I27G32C-A75C)8 exhibits the lower value of dispersion σ2 = 7.7 ± 1.3, the highest departure from the single exponential time distribution, σ2 = 21.3 ± 2.4 (Fig. 4D–F) does not correspond to the longest wild-type form, but instead to the (I27E24C-K55C)8 protein, which extends only 58 amino acids. Hence, we do not observe correlation between the heterogeneity in the unfolding pathways and the protein length, suggesting that, rather than the protein length, the heterogeneity of unfolding pathways seems to be mainly dictated by the precise conformation of the protein native state, which is subtly modified by the presence of the shortcutting disulfide bonds present in the protein mutants.
Figure 4. The measured disorder in the unfolding pathways of I27 does not correlate with the protein length.
Time-course distribution corresponding to the unfolding of the (A) (I27G32C-A75C)8 construct (n = 1269), (B) the (I27E24C-K55C)8 construct (n = 1528) and (C) the (I27P28C-K54C)8 polyprotein (n = 1099). In none of the cases, a single exponential time distribution with an associated unfolding rate k =1/<t> fully reproduces the experimental distribution of unfolding times. (D–E) Associated plots of ln[− ln 〈S(t)〉] versus ln t corresponding to the unfolding of the (I27G32C-A75C)8, (I27E24C-K55C)8 and (I27E24C-K55C)8 constructs, respectively. The fits of the static disorder survival probability (colored lines) and of the single exponential (black line) clearly show the adequacy of the static disorder theory to fit the experimental results. The obtained values for the fitting parameters k and σ2 are listed in Table 1. Notably, the values of σ2 do not correlate with the number of amino acids being extended after mechanical unfolding of the protein.
Discussion
Here we investigated the effect of applying a constant stretching force on two complete different reactions; the unfolding of a single protein and the reduction of a disulfide bond, both occurring within the structure of the same single molecule. Notably, both reactions give rise to distinct results, highlighting in each case the distribution of sampled transition state structures. It has been long recognized, mostly using NMR relaxation techniques53,55, that proteins are dynamic systems56, and that the fluctuations of both the backbone and side-chains and their interplay with the water molecules that solvate the protein shell give rise to an ensemble of nearly isoenergetic conformations that define the protein’s native state. However, direct structural characterization defining each distinct protein conformation, together with their timescale of interconversion, has been mainly restricted to the information provided by all-atom molecular dynamics simulations57. Since the “liquid-like” side-chain fluctuations are measured to occur within a 10 ns timescale57, it is extremely unlikely that they can be captured as ‘frozen’ conformations in our experiments. By contrast, it is plausible that the mechanically distinct sub-states that we capture stem from the interplay of the slower motions of the “solid” backbone with the surrounding water molecules, which altogether define each independent protein conformation58. Indeed, in the case of mechanical unfolding of the I27protein, the transition state is composed of a set of key hydrogen bonds that form the mechanical clamp (Figure 5A). Crucially, the solvent molecules are intricately related to the transition state conformation59, and it is likely that the heterogeneity of pathways that we measure have their origin in an ensemble of transition state structures with similar energy that differ in the number and dynamics of water molecules governing the forced unfolding reaction38.
Figure 5. Schematics of the transition state structures of the two distinct reactions probed in our experiments.
(A) Unfolding of the I27 protein involves the breakage of a set of key hydrogen bonds (white, inset). Upon disruption, the protein structure (blue region) is extended up to the disulfide bond, which acts as a mechanical covalent clamp. B) The reduction of the exposed disulfide bond encompasses the back-side attack of the 1-mercapto-2-propanol nucleophile and the sub-Ångström elongation of the S-S bond under force. Upon reduction, the trapped amino acids (orange region) are elongated under the presence of the pulling force. The diversity of conformations defining the transition state for mechanical protein unfolding contrasts with the structural simplicity measured for the SN2 chemical reaction under force.
Our results demonstrate that shortening the I27 protein by the presence of a disulfide bond, although placed in a region away from the key A´G patch, greatly affects the distribution of unfolding pathways. Interestingly, the breadth of the unfolding time distribution does not correlate with the protein length. These results suggest that slight changes in the protein tertiary conformation have major implications on the distribution of the transition state conformations. We speculate that the change in the protein backbone imposed by the rigid disulfide bond restricts the orientation and dynamics of the water molecules defining the unfolding transition state. It is therefore tempting to hypothesize that the structure and diversity in the transition state structures sampled in each protein might correspond to a natural mechanism that regulates protein fragility from a functional viewpoint, which would be of particular importance in cardiac I27 protein. In the case of protein unfolding, the heterogeneity in the unfolding pathways is interpreted here using the static disorder theory in terms of the multiple conformations that populate the transition state structures38. An alternative explanation for the measured distribution of unfolding pathways would emerge from the conformational diversity of the native state of the protein, which would lead to an ensemble of structures with slightly different energy barriers35. It is plausible that the interplay of both scenarios, i.e., the diversity in both the transition state structures and also in the native state conformation account for the diversity of unfolding pathways that we measure in our experiments. The heterogeneity measured for protein unfolding contrasts with the simplicity and univocal conformation of the transition state corresponding to the reduction of the disulfide bond that shortcuts the I27 protein. In this case, the transition state geometry of the SN2 chemical reaction involving the 1-mercapto-2-propanol back-side attack (Figure 5B), and occurring on the Ångström lengthscale, is highly conserved. Indeed, the high degree of directionality imposed by the back-side attack that describes the SN2 chemical reaction restricts the conformational space available for the successful collision to take place. Such stereospecificity is likely to account for the homogeneity in the transition state structures that we measure in our experiments. Alternatively, it is also possible that the reaction transition state involves a set of similar structures rather than a unique one. However, due to the fleeting nature of the reaction transition state conformation (occurring on the femtosecond timescale60) the rate of interconversion between the different structures would be much faster than the rates measured in our experiments. In such a case, the dynamic disorder in the sampled transition state structures would explain the exponential distribution of times that we measure. The apparent simplicity of the chemical reaction under force using a small nucleophile such as 1-mercapto-2-propanol contrasts with the complex kinetics observed for the same disulfide bond reduction when an enzyme such as thioredoxin is used43,61,62. In any case, our experiments probing chemical reactivity under force using small nucleophiles confirm the adequacy of the simple first-order kinetic law to fully capture the kinetics of the statistically broad set of reactive trajectories, at the single bond level.
The apparent simplicity of the chemical reactions under force contrasts with the complexity in the unfolding mechanism, both reactions being measured within the same molecule and under identical experimental conditions. This experiment serves as a control that rules out the possibility that the origin of the measured non-exponential kinetics for protein unfolding results from the limitations of our experimental set-up. These potential sources of experimental error 63,64 include the instrumental fluctuations in the constant force applied to the proteins, the finite time response of the force feedback, the averaging of data obtained during independent experimental days using different cantilever probes, or the thermal fluctuations of the polyproteins.
The heterogeneity in the distribution of unfolding pathways of the native I27 protein closely resembles that found for ubiquitin (σ2 = 18.5 ± 1.5) at the same pulling force38. The challenge is now to identify the interplay between structural motifs within the protein (such as the number and position of hydrogen bonds) and the medium conditions (such as solvent or temperature), which could altogether affect the distribution of unfolding pathways that can be captured at the single molecule level. Most importantly, according to our results, it is apparent that protein unfolding is a much complex reaction, for which its kinetic treatment as a simple first order chemical reaction is an oversimplification that does not capture the essential physical properties underlying the process. Regarding the chemical reactions, it would be interesting to indentify signatures of complexity at the single bond length-scale using small nucleophiles by studying reactions that could proceed along two concomitant and parallel reaction mechanisms, exhibiting distinct transition state conformations, as it would be the case of a reducing agent that could either proceed through a nucleophilic attack or following an addition/elimination mechanism 65,66.
Our results demonstrate the capability of force-clamp spectroscopy to capture a large set of individual reactive trajectories that allow direct identification of the degree of heterogeneity in the transition state structures sampled in each particular reaction. The high statistical throughput that can be now achieved with the technique is therefore perfectly suited to indentify fine details in the potential energy landscape, which can only be unveiled at the single molecule level. Crucially, here we demonstrated that the variance of the barrier heights, σ2, is a very sensitive parameter that can vary over a wide range within the structure of a single protein. Indeed, the heterogeneity in the unfolding transition state ensemble is very sensitive to mutations that introduce disulfide bonds in different locations throughout the protein. By contrast, disulfide bond reduction by a small nucleophile such as 1-mercapto-2-propanol shows no disorder, implying a single well defined transition state structure that is insensitive to its location within the same protein. The long-term goal is to build a comprehensive formalism to reconstruct the full energy landscape of a folding protein, and to correlate it with the protein’s architecture. The diversity of conformations that we found in the most prominent energy minimum of the landscape, corresponding to the native state, might have severe implications on the functionality of proteins with mechanical function, of common occurrence in nature.
Supplementary Material
Acknowledgments
This work was supported by the Fundación IberCaja (S.G-M.) and by National Institute of Health Grants HL66030 and HL61228 (to J.M.F).
Footnotes
Supporting Information Available. Additional figures illustrating the different behaviour of force on the energy landscape of protein unfolding and chemical reactions are available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Dobson CM, Sali A, Karplus M. Angewandte Chemie-International Edition. 1998;37:868–893. doi: 10.1002/(SICI)1521-3773(19980420)37:7<868::AID-ANIE868>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 2.R.D. L. Molecular Reaction Dynamis. Cambridge University Press; 2005. [Google Scholar]
- 3.Brauman JI. Science. 2008;319:168. doi: 10.1126/science.1152387. [DOI] [PubMed] [Google Scholar]
- 4.Chabinyc ML, Craig SL, Regan CK, Brauman JI. Science. 1998;279:1882–1886. doi: 10.1126/science.279.5358.1882. [DOI] [PubMed] [Google Scholar]
- 5.Sun L, Song K, Hase WL. Science. 2002;296:875–878. doi: 10.1126/science.1068053. [DOI] [PubMed] [Google Scholar]
- 6.Bach RD, Dmitrenko O, Thorpe C. J Org Chem. 2008;73:12–21. doi: 10.1021/jo702051f. [DOI] [PubMed] [Google Scholar]
- 7.Hase WL. Science. 1994;266:998–1002. doi: 10.1126/science.266.5187.998. [DOI] [PubMed] [Google Scholar]
- 8.Dill KA, Chan HS. Nat Struct Biol. 1997;4:10–19. doi: 10.1038/nsb0197-10. [DOI] [PubMed] [Google Scholar]
- 9.Thirumalai D. Journal De Physique I. 1995;5:1457–1467. [Google Scholar]
- 10.Wolynes PG, Onuchic JN, Thirumalai D. Science. 1995;267:1619–1620. doi: 10.1126/science.7886447. [DOI] [PubMed] [Google Scholar]
- 11.Camacho CJ, Thirumalai D. Proc Natl Acad Sci U S A. 1995;92:1277–1281. doi: 10.1073/pnas.92.5.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bryngelson JD, Wolynes PG. Biopolymers. 1990;30:177–188. [Google Scholar]
- 13.Bryngelson JD, Wolynes PG. Proc Natl Acad Sci U S A. 1987;84:7524–7528. doi: 10.1073/pnas.84.21.7524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bryngelson JD, Onuchic JN, Socci ND, Wolynes PG. Proteins. 1995;21:167–195. doi: 10.1002/prot.340210302. [DOI] [PubMed] [Google Scholar]
- 15.Fersht A. Structure and Mechanism in Protein Science. New York: 1999. [Google Scholar]
- 16.Mayne L, Englander SW. Protein Sci. 2000;9:1873–1877. doi: 10.1110/ps.9.10.1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zwanzig R. Proc Natl Acad Sci U S A. 1997;94:148–150. doi: 10.1073/pnas.94.1.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roeffaers MB, De Cremer G, Uji-i H, Muls B, Sels BF, Jacobs PA, De Schryver FC, De Vos DE, Hofkens J. Proc Natl Acad Sci U S A. 2007;104:12603–12609. doi: 10.1073/pnas.0610755104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smiley RD, Hammes GG. Chem Rev. 2006;106:3080–3094. doi: 10.1021/cr0502955. [DOI] [PubMed] [Google Scholar]
- 20.Ferrer S, Tunon I, Marti S, Moliner V, Garcia-Viloca M, Gonzalez-Lafont A, Lluch JM. J Am Chem Soc. 2006;128:16851–16863. doi: 10.1021/ja0653977. [DOI] [PubMed] [Google Scholar]
- 21.Basner JE, Schwartz SD. J Am Chem Soc. 2005;127:13822–13831. doi: 10.1021/ja043320h. [DOI] [PubMed] [Google Scholar]
- 22.Pu J, Gao J, Truhlar DG. Chem Rev. 2006;106:3140–3169. doi: 10.1021/cr050308e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu HP, Xun L, Xie XS. Science. 1998;282:1877–1882. doi: 10.1126/science.282.5395.1877. [DOI] [PubMed] [Google Scholar]
- 24.Xue Q, Yeung ES. Nature. 1995;373:681–683. doi: 10.1038/373681a0. [DOI] [PubMed] [Google Scholar]
- 25.Yang H, Luo G, Karnchanaphanurach P, Louie TM, Rech I, Cova S, Xun L, Xie XS. Science. 2003;302:262–266. doi: 10.1126/science.1086911. [DOI] [PubMed] [Google Scholar]
- 26.van Oijen AM, Blainey PC, Crampton DJ, Richardson CC, Ellenberger T, Xie XS. Science. 2003;301:1235–1238. doi: 10.1126/science.1084387. [DOI] [PubMed] [Google Scholar]
- 27.English BP, Min W, van Oijen AM, Lee KT, Luo G, Sun H, Cherayil BJ, Kou SC, Xie XS. Nat Chem Biol. 2006;2:87–94. doi: 10.1038/nchembio759. [DOI] [PubMed] [Google Scholar]
- 28.Velonia K, Flomenbom O, Loos D, Masuo S, Cotlet M, Engelborghs Y, Hofkens J, Rowan AE, Klafter J, Nolte RJ, de Schryver FC. Angew Chem Int Ed Engl. 2005;44:560–564. doi: 10.1002/anie.200460625. [DOI] [PubMed] [Google Scholar]
- 29.Flomenbom O, Velonia K, Loos D, Masuo S, Cotlet M, Engelborghs Y, Hofkens J, Rowan AE, Nolte RJ, Van der Auweraer M, de Schryver FC, Klafter J. Proc Natl Acad Sci U S A. 2005;102:2368–2372. doi: 10.1073/pnas.0409039102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garcia-Viloca M, Gao J, Karplus M, Truhlar DG. Science. 2004;303:186–195. doi: 10.1126/science.1088172. [DOI] [PubMed] [Google Scholar]
- 31.Lu S, Li WW, Rotem D, Mikhailova E, Bayley H. Nature Chemistry. 2010;2:921–928. doi: 10.1038/nchem.821. [DOI] [PubMed] [Google Scholar]
- 32.Garcia-Manyes S. Nature Chemistry. 2010;2:905–906. doi: 10.1038/nchem.882. [DOI] [PubMed] [Google Scholar]
- 33.Fernandez JM, Li H. Science. 2004;303:1674–1678. doi: 10.1126/science.1092497. [DOI] [PubMed] [Google Scholar]
- 34.Schlierf M, Li H, Fernandez JM. Proc Natl Acad Sci U S A. 2004;101:7299–7304. doi: 10.1073/pnas.0400033101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brujic J, Hermans RI, Walther KA, Fernandez JM. Nature Physics. 2006;2:282–286. [Google Scholar]
- 36.Brujic J, Hermans RI, Garcia-Manyes S, Walther KA, Fernandez JM. Biophys J. 2007;92:2896–2903. doi: 10.1529/biophysj.106.099481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Garcia-Manyes S, Brujic J, Badilla CL, Fernandez JM. Biophys J. 2007;93:2436–2446. doi: 10.1529/biophysj.107.104422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kuo TL, Garcia-Manyes S, Li J, Barel I, Lu H, Berne BJ, Urbakh M, Klafter J, Fernandez JM. Proc Natl Acad Sci U S A. 2010;107:11336–11340. doi: 10.1073/pnas.1006517107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cao Y, Kuske R, Li H. Biophys J. 2008;95:782–788. doi: 10.1529/biophysj.107.128298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wiita AP, Ainavarapu SR, Huang HH, Fernandez JM. Proc Natl Acad Sci U S A. 2006;103:7222–7227. doi: 10.1073/pnas.0511035103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ainavarapu SRK, Wiita AP, Dougan L, Uggerud E, Fernandez JM. Journal of the American Chemical Society. 2008;130:6479–6487. doi: 10.1021/ja800180u. [DOI] [PubMed] [Google Scholar]
- 42.Garcia-Manyes S, Liang J, Szoszkiewicz R, Kuo TL, Fernandez JM. Nature Chemistry. 2009;1:236–242. doi: 10.1038/nchem.207. [DOI] [PubMed] [Google Scholar]
- 43.Szoszkiewicz R, Ainavarapu SR, Wiita AP, Perez-Jimenez R, Sanchez-Ruiz JM, Fernandez JM. Langmuir. 2008;24:1356–1364. doi: 10.1021/la702368b. [DOI] [PubMed] [Google Scholar]
- 44.Carrion-Vazquez M, Oberhauser AF, Fowler SB, Marszalek PE, Broedel SE, Clarke J, Fernandez JM. Proc Natl Acad Sci U S A. 1999;96:3694–3699. doi: 10.1073/pnas.96.7.3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li H, Linke WA, Oberhauser AF, Carrion-Vazquez M, Kerkvliet JG, Lu H, Marszalek PE, Fernandez JM. Nature. 2002;418:998–1002. doi: 10.1038/nature00938. [DOI] [PubMed] [Google Scholar]
- 46.Ainavarapu SR, Brujic J, Huang HH, Wiita AP, Lu H, Li L, Walther KA, Carrion-Vazquez M, Li H, Fernandez JM. Biophys J. 2007;92:225–233. doi: 10.1529/biophysj.106.091561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Efron B. The Jacknife, the Bootstrap, and Other Resampling Plans. Philedelphia: 1982. [Google Scholar]
- 48.Li H, Carrion-Vazquez M, Oberhauser AF, Marszalek PE, Fernandez JM. Nat Struct Biol. 2000;7:1117–1120. doi: 10.1038/81964. [DOI] [PubMed] [Google Scholar]
- 49.Lu H, Schulten K. Biophys J. 2000;79:51–65. doi: 10.1016/S0006-3495(00)76273-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Arrhenius S. Z Phys Chem. 1889;4:226–248. [Google Scholar]
- 51.Zwanzig R. Abstracts of Papers of the American Chemical Society. 1989;198 154-PHYS. [Google Scholar]
- 52.Zwanzig R. Journal of Chemical Physics. 1992;97:3587–3589. [Google Scholar]
- 53.Lindorff-Larsen K, Best RB, Depristo MA, Dobson CM, Vendruscolo M. Nature. 2005;433:128–132. doi: 10.1038/nature03199. [DOI] [PubMed] [Google Scholar]
- 54.Bell GI. Science. 1978;200:618–627. doi: 10.1126/science.347575. [DOI] [PubMed] [Google Scholar]
- 55.Wuthrich K, Wagner G. FEBS Lett. 1975;50:265–268. doi: 10.1016/0014-5793(75)80504-7. [DOI] [PubMed] [Google Scholar]
- 56.Frauenfelder H, Sligar SG, Wolynes PG. Science. 1991;254:1598–1603. doi: 10.1126/science.1749933. [DOI] [PubMed] [Google Scholar]
- 57.Shaw DE, Maragakis P, Lindorff-Larsen K, Piana S, Dror RO, Eastwood MP, Bank JA, Jumper JM, Salmon JK, Shan Y, Wriggers W. Science. 2010;330:341–346. doi: 10.1126/science.1187409. [DOI] [PubMed] [Google Scholar]
- 58.Zhou Y, Vitkup D, Karplus M. J Mol Biol. 1999;285:1371–1375. doi: 10.1006/jmbi.1998.2374. [DOI] [PubMed] [Google Scholar]
- 59.Dougan L, Feng G, Lu H, Fernandez JM. Proc Natl Acad Sci U S A. 2008;105:3185–3190. doi: 10.1073/pnas.0706075105. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 60.Zewail AH. Angew Chem Int Ed Engl. 2000;39:2586–2631. doi: 10.1002/1521-3773(20000804)39:15<2586::aid-anie2586>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 61.Wiita AP, Perez-Jimenez R, Walther KA, Grater F, Berne BJ, Holmgren A, Sanchez-Ruiz JM, Fernandez JM. Nature. 2007;450:124–127. doi: 10.1038/nature06231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Perez-Jimenez R, Li J, Kosuri P, Sanchez-Romero I, Wiita AP, Rodriguez-Larrea D, Chueca A, Holmgren A, Miranda-Vizuete A, Becker K, Cho SH, Beckwith J, Gelhaye E, Jacquot JP, Gaucher EA, Sanchez-Ruiz JM, Berne BJ, Fernandez JM. Nat Struct Mol Biol. 2009;16:890–896. doi: 10.1038/nsmb.1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Borgia A, Williams PM, Clarke J. Annu Rev Biochem. 2008;77:101–125. doi: 10.1146/annurev.biochem.77.060706.093102. [DOI] [PubMed] [Google Scholar]
- 64.Huang Z, Boulatov R. Pure and Applied Chemistry. 2010;82:931–951. [Google Scholar]
- 65.Bickelhaupt FM, Baerends EJ, Nibbering NMM. Chemistry-a European Journal. 1996;2:196–207. [Google Scholar]
- 66.Ensing B, Klein ML. Proc Natl Acad Sci U S A. 2005;102:6755–6759. doi: 10.1073/pnas.0408094102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.