Abstract
Objectives
We previously reported vicriviroc (VCV) resistance in an HIV-infected subject and used deep sequencing and clonal analyses to track the evolution of V3 sequence forms over 28 weeks of therapy. Here, we test the contribution of gp120 mutations to CCR5 antagonist resistance and investigate why certain minority V3 variants emerged as the dominant species under drug pressure.
Methods
19 site-directed HIV-1 mutants were generated that contained gp120 VCV-resistance mutations. Viral sensitivities to VCV, maraviroc, TAK-779 and HGS004 were determined.
Results
Three patterns of susceptibilities were observed: sigmoid inhibition curves with IC50s similar to pre-treatment virus (07J-week 0 [W0]), single mutants with decreased IC50s compared to 07J-W0, and mutants that contained ≥5 of 7 VCV-resistance mutations with flattened inhibition curves and decreased or negative percent maximal inhibition. Substitutions such as S306P, which sensitized virus to CCR5 antagonists when present as single mutations, were not detected in the baseline virus population but were necessary for maximal resistance when incorporated into V3 backbones that included pre-existing VCV resistance mutations.
Conclusion
CCR5 antagonist resistance was reproduced only when a majority of V3 mutations were present. Minority V3 loop variants may serve as a scaffold upon which additional mutations lead to complete VCV resistance.
Keywords: HIV-1, CCR5 antagonists, antiviral therapy, HIV-1 resistance, maraviroc, vicriviroc, TAK-779
INTRODUCTION
Small molecule CCR5 antagonists, such as vicriviroc (VCV), maraviroc (MVC), and TAK779, are allosteric noncompetitive inhibitors that bind to CCR5 and prevent its interaction with the viral envelope glycoprotein gp120;1 MVC is FDA-approved for clinical use and VCV has been in later stages of development.2–7 Monoclonal antibodies (MAb) against CCR5 (e.g., HGS004 and PRO140) and derivatives of natural CCR5 ligands, such as PSC-RANTES, which down-regulates CCR5 expression on the cell surface, have also been developed but are not approved for clinical use.8–12 Resistance to small-molecule CCR5 antagonists has been described both in vitro and in vivo and occurs when the virus adapts to use inhibitor-bound CCR5 for viral entry.13–20 This resistance is phenotypically characterized by a flattening, or plateau, of the dose-response curve that reduces the achievable percent maximal inhibition (PMI), rather than increasing the 50% inhibitory concentration (IC50).16–17,21–24
To date, most data on CCR5 antagonist resistance have come from analyses of viruses selected during in vitro passage.15,17,19–20,23,25 There is a paucity of genotypic and phenotypic data from resistant clinical isolates. To date, an analysis of mutations leading to clinical VCV resistance has been reported for one subtype C and one subtype D isolate.16,26 Although in vitro selection studies of resistance mutations have provided valuable insight into clinical resistance for other antiretroviral drugs, in vivo extrapolation of these data can be limited. For examples, mutations have been identified from the in vitro passage of HIV-1 in the presence of protease inhibitors, non-nucleoside reverse transcriptase inhibitors and fusion inhibitors that are not major contributors to clinical resistance.27–32 CCR5 antagonist resistance-associated mutations are highly variable and context dependent, due in part to extensive sequence heterogeneity of HIV-1 env. The V3 loop of HIV-1 gp120, comprising positions 296–331 (numbered according to HXB2), is the principal determinant of co-receptor usage,33 and genotypic analyses of VCV-resistant viruses have focused primarily on this domain. In vitro studies have shown that mutations in the V3 loop of env appear to develop sequentially, ultimately leading to a complete loss of drug activity. The effect of V3 loop mutations on VCV or MVC susceptibility may depend, however, on mutations or polymorphisms elsewhere in the envelope glycoprotein, including the C4 domain of gp120 and the fusion peptide of gp41.13,15–16,25
We previously reported the emergence of CCR5 antagonist resistance in an HIV-1 subtype C isolate (07J-week 28 [W28]) from a patient with virologic failure on a VCV-containing antiretroviral regimen.16 Amino acid substitutions that included K305R, S306P, T307I, F318I, T320R, G321E, and H330Y accumulated sequentially on both sides of the V3 stem.16 Deep sequencing and clonal analysis demonstrated dramatic shifts in the population structure of the viral quasispecies after initiation of VCV. The V3 loop forms with VCV-associated resistance mutations that existed at baseline increased in frequency early in treatment and were subsequently replaced at later time points by V3 loop forms with greater VCV resistance.16,34 For example, the V3 sequences with K305R/T307I/F318I/T320R, K305R/T307I/F318I/T320R/H330Y and T307I/F318I/T320R/H330Y substitutions were present as minority variants at week 0 at a frequency of less than 1%. These sequences increased in frequency by week 12 to 25%, 2.6% and 15%, respectively, in the setting of drug pressure but by week 19 were replaced by emergence of a VCV-resistant virus that carried the V3 sequence with mutations K305R/T307I/F318I/T320R/G321E/H330Y.34 The resistant viruses, which demonstrated significant reductions in VCV PMIs, incorporated one or two additional V3 mutations into pre-existing, partially VCV-resistant, minority variant backbones.34
To elucidate the contribution of env mutations, singly and in combination, to clinical resistance and to investigate why certain variants present as minority or intermediate forms eventually emerged as the dominant species under drug pressure, we used a deep sequencing data set to guide the construction of a series of recombinant viruses representing majority, minority and intermediary V3 sequence forms that developed during VCV therapy. Viruses harboring these diverse loops in addition to a mutation outside V3 were examined for their sensitivity to CCR5 antagonists to determine whether the initial V3 loop sequence changes under drug pressure lead to changes in IC50s or decreased PMI and maximal resistance. Both gain and loss of function were studied by introducing V3 mutations from the resistant 07J-W28 into the VCV sensitive virus (07J-W0), and by reverting key V3 mutations in 07J-W28 env back to the 07J-W0, pre-treatment sequence. These site-directed mutant viruses were assessed for their sensitivity to VCV, MVC, TAK-779 and HGS004.
METHODS
Reagents
TAK-779 and MVC were obtained from the AIDS Research and Reference Program (NIH); HGS004 (Human Genome Sciences, Gaithersburg, MD) was provided by M. Subramanian; VCV (Schering-Plough Research Institute, Kennilworth, NJ) was provided by B.M. Baroudy.
Site-Directed Mutagenesis
Full-length envelopes from plasma virus obtained at the pre-treatment (week 0) and time of virologic failure (week 28) visits were cloned into PCR4-TOPO TA vector (Invitrogen, Carlsbad, CA). Site-directed mutagenesis using the Quickchange IIE protocol (Stratagene, La Jolla, CA) was performed to introduce specific changes within the 07J-W0 and 07J-W28 full-length envelopes. Bi-directional full-length env sequencing of recombinant clones was performed to verify the presence of the desired substitutions and the absence of adventitious mutations. The amino acid numbering used in this study was based on strict alignment with the HXB2 reference env sequence; for consistency, in our discussion of prior studies, amino acid positions have been renumbered to reflect the HXB2 alignment.
Recombinant Virus Production
Full-length env genes carrying V3 loop mutations described above were introduced into an NL4-3 backbone using a yeast-gap repair homologous recombination strategy as previously described with minor modifications.35 Briefly, the full-length HIV-1 clone NL4-3 from pNL4-3 was incorporated into the multiple cloning site (MCS) of pRS315 (New England Biolabs, Beverly, MA). Oligonucleotides 5’-CTATAGGGCGAATTGGAGCTCTACTTACACCAGGAAAGGCGCTACTTCTAGATGTACT-3’ and 5’-GGAACAAAAGCTGGGTACCGTGCAACCTCTACCTCCTGGGCGTACATCTAGAAGTAGC-3’ were used as templates in overlapping PCR reactions with primers 5’-CTATAGGGCGAATTGGAGCT –3’ and 5’-GGAACAAAAGCTGGGTACCG-3’. The resulting PCR product was inserted into XbaI-digested pRS315 using yeast gap-repair homologous recombination. Transformed yeast were selected in leucine dropout media. This newly engineered pRS315 was digested with XbaI and combined with AatII-digested pNL4-3 to transform and select yeast in leucine dropout media. Plasmids with NL4-3 within the MCS of pRS315 (pRS315-NL4-3) were confirmed by restriction digestion. A yeast auxotrophic selection marker (HIS3) was amplified from pRS313 (New England Biolabs) using primers 5’-GACGTCATTGGATGGCCTGCTGTAAGGTCGCGCGTTTCGGTGATGACG-3’ and 5’-TCAGCAGTTCTTGAAGTACTCCGCTTTAAATAATCGGTGTCAC-3’. This HIS3 PCR product was substituted for the NL4-3 nef gene using BspE1 partially digested pRS315-NL4-3 and yeast gap-repair homologous recombination. Yeast with the resulting plasmid (pRS315-NL4-3-Δnef-HIS) were selected on leucine and histidine dropout media. The yeast auxotrophic marker URA3 was amplified from pRS316 (New England Biolabs) using primers 5’-AGAAAGAGCAGAAGACAGTGGCAATGAAGATTGTACTGAGAGTGCAC-3’ and 5-TTTTGACCACTTGCCACCCATGGTATTTCACACCGCAGGG-3’. The pNL4-3 full-length envelope in pRS315-NL4-3-Δnef-HIS was replaced with the URA3 PCR product using a StuI digested pRS315-NL4-3-Δnef-HIS and yeast gap repair. Yeast with the recombined plasmid (pRS315-NL4-3-Δnef-HIS-ΔEnv-URA) were selected on leucine, uracil and histidine drop out media. Subsequently, different HIV-1 envelopes of interest were introduced in the place of URA within pRS315-NL4-3-Δnef-HIS-ΔEnv-URA. Briefly, pRS315-NL4-3-Δnef-HIS-ΔEnv-URA was digested with StuI and combined with subject-derived env amplicons. Yeast with the recombined plasmid (pRS315-NL4-3-Δnef-HIS+ENV) were selected in leucine, histidine dropout media supplemented with fluoroorotic acid (FOA). All yeast transformations were done by a standard lithium acetate procedure. All plasmids rescued from yeast colonies were used to transform STBL4 E. coli (Invitrogen) to generate larger plasmid quantities.
Virus stocks were generated and titered as previously described with minor modifications 16,35. Briefly, virus stocks were generated by transfecting 293T human embryonic kidney fibroblasts with full-length recombinant NL4-3 plasmids containing the desired env mutations using the FuGene 6 protocol (Roche Molecular Biochemicals, Indianapolis, IN). Supernatants were collected 48 hours later. Relative viral infectivity was determined by endpoint dilution on TZM-bl cells obtained from the NIH AIDS Research and Reference Program.36–37 The Galacto-Light Plus chemiluminescent reporter gene assay system (Applied Biosystems, Foster City, CA) was used to detect β-galactosidase activity in infected TZM-bl cells after 48 hours of incubation in Dulbecco's Modified Eagle’s Medium supplemented with 10% fetal bovine serum and penicillin/streptomycin (DMEM complete). Relative light units (RLU) were measured for serial dilutions of virus, and a linear regression was performed to obtain RLU per unit volume of the viral stock.
Drug Susceptibility Assays
Susceptibilities of recombinant viruses to entry inhibitors were determined using a TZM-bl cell-based assay. Two-fold serial drug dilutions and approximately 1.0 × 104 TZM-bl cells and 20µM DEAE-dextran in 100 µL of DMEM complete media were added to each well of a 96-well microtiter plate and incubated for one hour. The cells were then inoculated with an amount of virus sufficient to produce 1.5 × 105 RLU on a PerkinElmer (Waltham, Massachusetts) 1450 Betacounter luminescent reader and incubated for 48 hours before β-galactosidase activity was measured as above. The percent inhibition was calculated as the ratio of RLU in drug-containing wells compared to the drug-free control wells. Each recombinant virus was assayed in triplicate, and each assay was performed at least two times. Inter-assay variability was determined by best-fit values from reproducible dose-response curves.
Statistical Analyses
Graphical analysis of drug susceptibility experiments was performed with GraphPad Prism 5 (Graphpad software, La Jolla, CA). To allow inter-assay comparison of each recombinant virus, mean IC50 and 95% confidence intervals (CI) for each sigmoid dose response regression curve were calculated from best-fit values from the regression models.
RESULTS
V3 Loop Mutants
We generated a series of V3 loop mutants that either introduced VCV-resistance mutations into the VCV-sensitive (07J-W0) env or reverted V3 loop mutations from the VCV-resistant env (07J-W28) back to their original pre-treatment amino acids. Mutations were added progressively to each side of the 07J-W0 V3 loop, until the 07J-W28 V3 loop carrying the K305R/S306P/T307I/F318I/T320R/G321E/H330Y mutations was recreated in the 07J-W0 backbone. Individual mutations and combinations of mutations were selected for susceptibility testing based on their detection in samples studied by deep sequencing and/or clonal analysis at weeks 0, 12, 16 and 19 as described.16,34 Two additional mutants were constructed that incorporated an S365V substitution from VCV-resistant virus that lay outside the V3 loop. This mutation, which was absent from pre-treatment env clones, was present in 11 of 20 full-length env clones of the week 28 virus and was lost within 20 weeks of VCV discontinuation [Genbank accession numbers EU664612 to EU664675].16
Effect of V3 Loop Mutations on CCR5 Antagonist Susceptibility
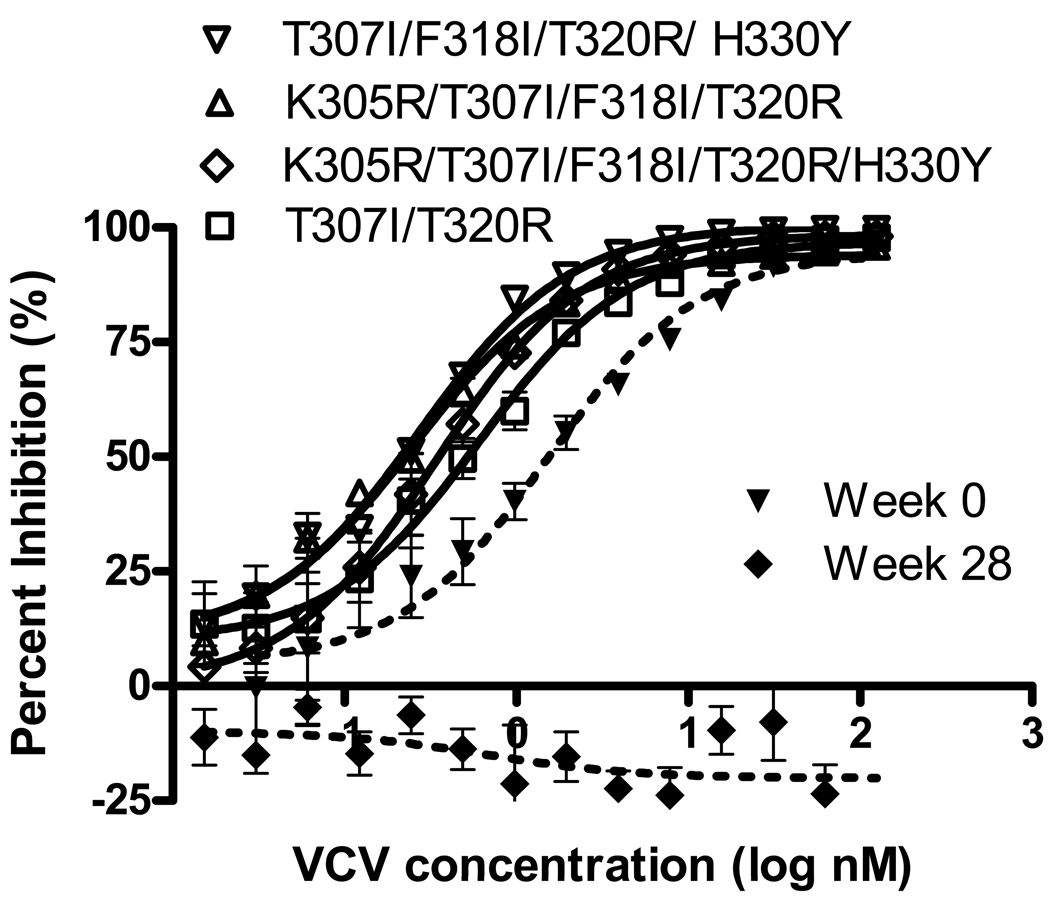
Recombinant viruses carrying 1 to 4 mutations in V3 were completely inhibited by each of the small molecule CCR5 antagonists tested. The inhibition curves for viruses with V3 mutations representing minority variants identified by deep-sequencing and clonal analysis of the week 0 sample are shown in Figure 1. Table 1 shows the IC50s of CCR5 antagonists for recombinant viruses carrying V3 mutations in the 07J-W0 env backbone. The T307I and T307I/T318R mutants did not demonstrate a significant change in IC50 for any of the small-molecule inhibitors tested (T307I was not tested in presence of VCV), whereas the T307I/F318I/T320R/H330Y, K305R/T307I/F318I/T320R and K305R/T307I/F318I/T320R/H330Y mutants demonstrated reduced IC50s to VCV and TAK-779.). The T307I/F318I/T320R/H330Y, and K305R/T307I/F318I/T320R/H330Y mutants demonstrated reduced IC50s to MVC as well.
FIG 1.
Inhibition curves for recombinant viruses carrying V3 loop mutations identified in minority variants present prior to VCV treatment inserted into the 07J-W0 envelope. Data shown are the means and standard errors of duplicate assays. VCV susceptibility of recombinant viruses expressing full-length env from 07J-W0 (week 0) and 07J-W28 (week 28) is shown for reference.
TABLE 1.
Susceptibility of mutant viruses to small-molecule CCR5 antagonists*
| V3 loop sequences of recombinant viruses | TAK-779 | MVC | VCV | |||
|---|---|---|---|---|---|---|
| IC50† (95% CI) | FC | IC50 (95% CI) | FC | IC50 (95% CI) | FC | |
| CTRPGNNTRKSTRIGPGQTFFATGDIIGDIRQAHC‡ | 22.2 (15.5, 31.9) | 1.00 | 0.65 (0.48, 0.88) | 1.00 | 1.53 (0.89, 2.62) | 1.00 |
| K305R | 19.1 (11.8, 31.1) | 0.86 | 0.99 (0.8, 1.2) | 1.52 | NT | - |
| S306P | 1.0 (0.7, 1.5) | 0.04 | 0.12 (0.09, 0.16) | 0.18 | 0.20 (0.16, 0.24) | 0.13 |
| T307I§ | 29.3 (15.9, 54.1) | 1.32 | 0.64 (0.38, 1.08) | 0.98 | NT | - |
| F318I | 1.4 (0.8, 2.3) | 0.06 | 0.38 (0.30, 0.50) | 0.58 | 0.29 (0.22, 0.38) | 0.20 |
| G321E | 3.6 (2.8, 5.0) | 0.16 | 0.53 (0.4, 0.71) | 0.82 | 0.40 (0.29, 0.54) | 0.26 |
| K205R/T307I | 11.2 (7.7, 16.1) | 0.50 | 1.08 (0.88, 1.33) | 1.66 | NT | - |
| T307I/T320R§ | 17.5 (12.8, 23.9) | 0.79 | 0.97 (0.66, 1.4) | 1.49 | 0.61 (0.39, 0.94) | 0.40 |
| T307I/F318I/T320R | 4.9 (3.4, 7.2) | 0.22 | 0.42 (0.32, 0.56) | 0.65 | 0.24 (0.18, 0.32) | 0.16 |
| T307I/F318I/T320R/H330Y§ | 5.8 (4.6, 7.2) | 0.26 | 0.24 (0.18, 0.31) | 0.37 | 0.27 (0.22, .034) | 0.18 |
| K305R/T307I/F318I/T320R§ | 6.3 (4.9, 8.2) | 0.28 | 0.33 (0.22, 0.5) | 0.51 | 0.25 (0.20, 0.31) | 0.16 |
| K305R/T307I/F318I/T320R/H330Y§ | 5.6 (4.7, 6.7) | 0.25 | 0.29 (0.23, 0.37) | 0.45 | 0.34 (0.27, 0.43) | 0.22 |
| T307I/F318I/T320R/G321E | 8.8 (6.7, 11.6) | 0.40 | 0.49 (0.37, 0.66) | 0.75 | 0.48 (0, 0.63) | 0.31 |
Note: IC50= 50% inhibitory concentration, CI = Confidence intervals, FC = Fold change in IC50compared to 07J-W0, NT = not tested. Data shown are means of at least two determinations.
Includes only those mutants without significant reductions in percent maximal inhibition (i.e., confidence intervals for PMI overlap 100%)
IC50 measured in nM of drug
Full-length 07J-W0 envelope; amino acid V3 sequence shown (starting at HXB2 position 296)
V3 sequences detected as minority variants in the week 0 virus population [Tsibris et al. 2009 34]
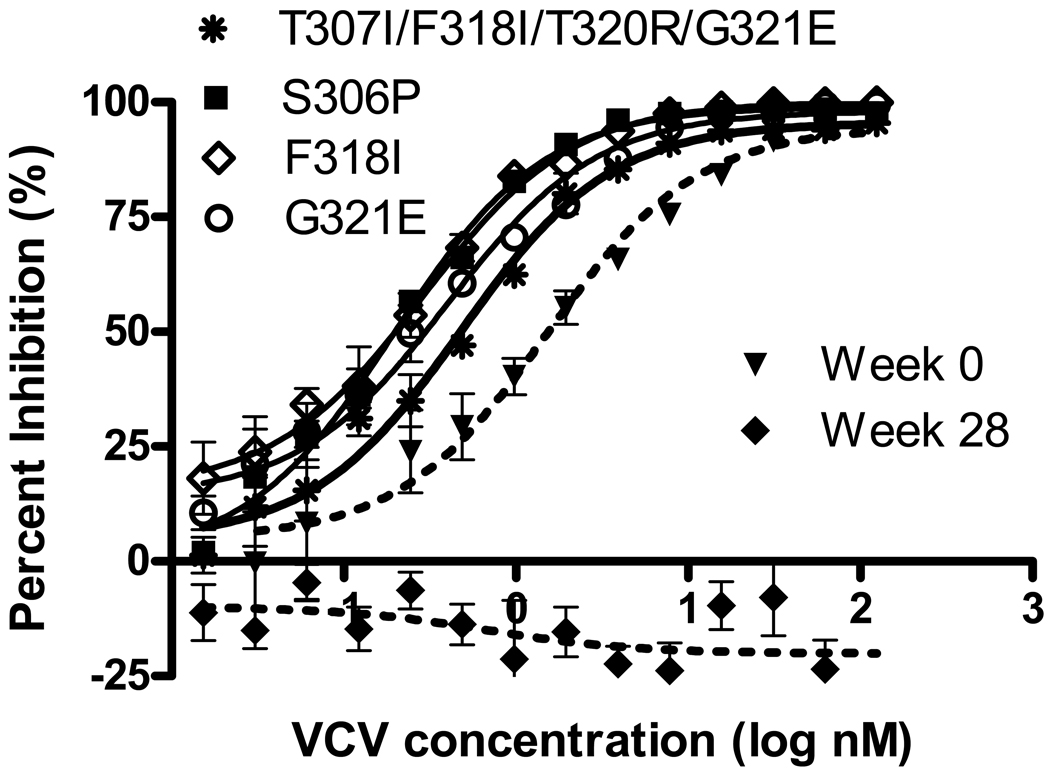
The effect on VCV susceptibility of other V3 loop mutations from 07J-W28, singly and in combination, was also studied (Table 1 and Figure 2). When present by itself, the K305R mutation produced no significant change in IC50 for the CCR5 antagonists tested. Likewise, the double mutant K305R/T307I had no effect on the IC50 for MVC and TAK-779 (VCV not tested). By contrast, the S306P, F318I and G321E mutations each sensitized virus to TAK-779 (Table 1) and HGS004. The S306P mutation also sensitized virus to MVC and VCV (Table 1). The T307I/F318I/T320R, T307I/F318I/T320R/G321E and K305R/T307I/F318I/T320R/H330Y mutants had significantly reduced IC50s for TAK-779 and VCV. Overall, more mutants demonstrated decreased IC50s for VCV and TAK-779 than for MVC (Table 1). None of the combinations of mutations significantly increased the IC50 for any of the drugs tested.
FIG 2.
Inhibition curves for recombinant viruses that demonstrated significant decreases in IC50 compared with 07J-W0 in the presence of VCV. The mutations indicated were inserted into the 07J-W0 envelope. Data shown are the means and standard errors of duplicate assays. VCV susceptibility of recombinant viruses expressing full-length env from 07J-W0 (week 0) and 07J-W28 (week 28) is shown for reference.
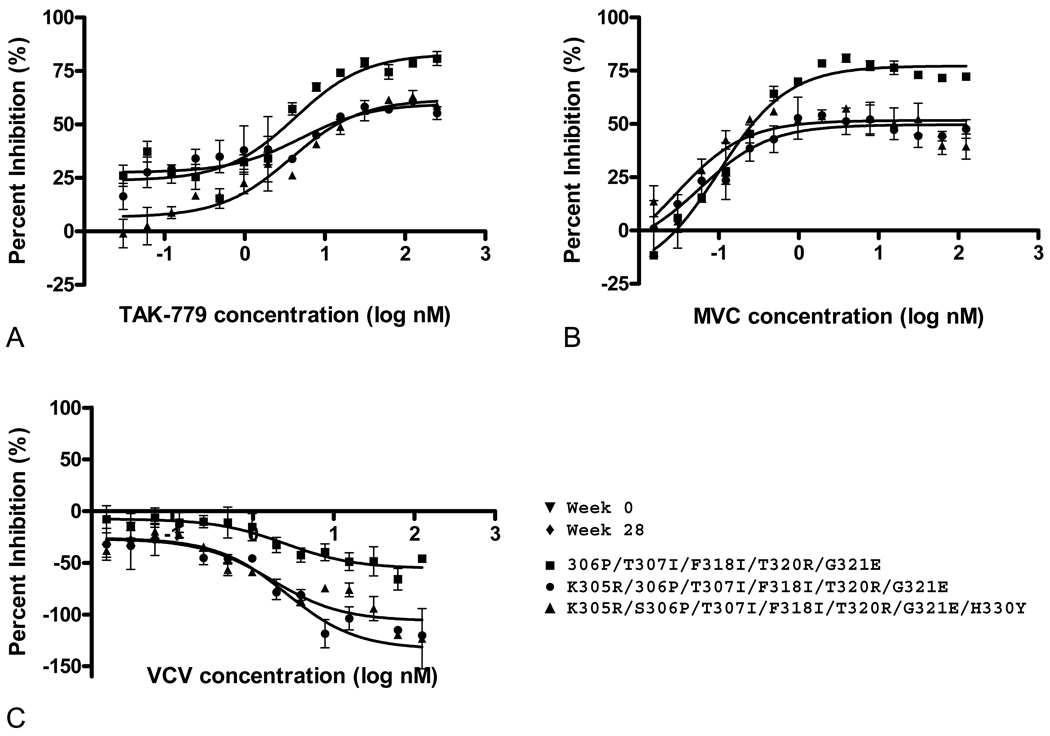
The introduction of six or more V3 mutations into the 07J-W0 env led to significant reductions in the PMIs for TAK-779, MVC and VCV. Although the K305R and S306P mutations had no effect on VCV susceptibility on their own, the addition of S306P to the T307I/ F318I/ T320R/G321E mutant significantly reduced the PMI of all three drugs; further reduction in PMI was observed by the additional incorporation of K305R (Table 2, Figure 3). Each of these mutants showed evidence of cross-resistance to TAK-779 and MVC, but enhanced infection (negative PMI) was observed only with VCV. Addition of the H330Y and S365V mutations did not have a significant effect on drug susceptibility (Table 2).
TABLE 2.
Susceptibility of recombinant viruses showing resistance to small-molecule CCR5 antagonists
| V3 loop sequences of recombinant viruses | TAK-779 | MVC | VCV |
|---|---|---|---|
| PMI (95% CI) | PMI (95% CI) | PMI (95% CI) | |
| 07J-W0 backbone | |||
| CTRPGNNTRKSTRIGPGQTFFATGDIIGDIRQAHC* | 106.7 (96.8, 116.6) | 101.1 (96.2, 106.0) | 94.6 (86.8, 102.6) |
| S306P/T307I/F318I/T320R/G321E | 83.0 (76.3, 89.7) | 77.2 (73.7, 80.8) | −55.5 (−66.7, −44.4) |
| K305R/S306P/T307I/F318I/T320R/G321E | 59.7 (52.2, 67.2) | 49.7 (45.3, 54.0) | −133.5 (−150, −116.6) |
| K305R/S306P/T307I/F318I/T320R/G321E/H330Y | 61.7 (55.4, 68.0) | 51.7 (46.6, 56.7) | −106.2 (−119.7, −92.7) |
| K305R/S306P/T307I/F318I/T320R/G321E/S365V | 68.4 (62.6, 74.1) | 59.2 (55.1, 63.2) | −69.6 (−82.8, −56.5) |
| K305R/S306P/T307I/F318I/T320R/G321E/H330Y/S365V | 49.1 (41.2, 57.0) | 46.4 (42.7, 50.2) | −116.3 (−131.8, −100.8) |
| 07J-W28 backbone | |||
| CTRPGNNTRRPIRIGPGQTFIAREDIIGDIRQAYC† | −10.3 (−3.5, 15.4) | 13.4 (8.0, 18.9) | −9.9 (−19.0, −0.7) |
| 305R→K | 32.9 (21.4, 44.5) | 42.0 (38.2, 45.8) | 14.7 (9.5, 19.8) |
| 306P→S | 66.9 (60.2, 73.6) | 75.9 (73.6, 78.2) | 36.6 (30.9, 42.3) |
Note: PMI = percent maximal inhibition, CI = confidence intervals. Data shown are means of at least two determinations.
Full-length 07J-W0 envelope; amino acid V3 sequence shown (starting at HXB2 position 296)
Full-length envelope from 017-W28; amino acid V3 sequence shown
FIG 3.
Inhibition curves for representative recombinant viruses carrying multiple V3 mutations in an 07J-W0 backbone that demonstrate a significantly decreased PMI in the presence of TAK-779 (A), MVC (B) and VCV (C). Data shown are the means and standard errors of duplicate assays.
Effect of V3 Loop Back-Mutations on CCR5 Antagonist Susceptibility
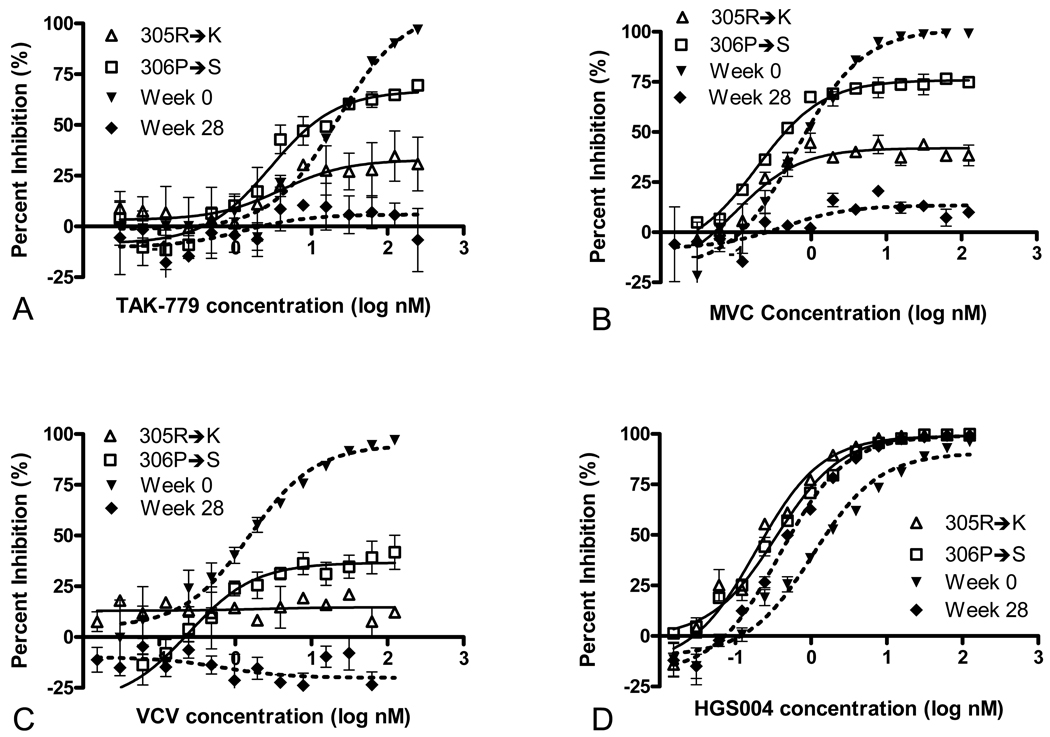
To further explore the contributions of specific amino acid substations to VCV resistance, two reversion mutants were generated: a 306P→S mutant, which recapitulated the dominant V3 loop present at a time of sub-maximal VCV resistance (week 19 of VCV treatment, PMI ~40%), and a 305R→K mutant. Partial restoration of susceptibility to TAK-779, MVC and VCV was observed with both of these reversion mutants (Table 2 and Figure 4).Reversion of S306P to wild-type had a greater effect on PMI than did reversion of K305R.
FIG 4.
Susceptibility to TAK-779 (A), MVC (B), VCV (C) and HGS004 (D) of recombinant viruses carrying R305K and P306S reversion mutations in V3 of the 07J-W28 envelope. Data shown are the means and standard errors of duplicate assays (for some data points, error bars are not visible due to very low interassay variation.) The susceptibilities of recombinant viruses expressing full-length env from 07J-W0 (week 0) and 07J-W28 (week 28) are shown for reference
Susceptibility to HGS004
None of the V3-loop mutants demonstrated significant resistance to the CCR5 monoclonal antibody HGS004. Viruses tested in the presence of HGS004 demonstrated either a decrease or no change in IC50 as compared to 07J-W0 or 07J-W28. In addition to the S306P, F318I and G321E single mutants, 07J-W28 and the 305R→K/306P→S reversion mutants all demonstrated decreased IC50s for HGS004 as compared to 07J-W0 (See Figure 4 and Table 1, Supplemental Digital Content).
DISCUSSION
We constructed and tested a series of 19 gp120 mutants that incorporated amino acid substitutions from the V3 loop of a VCV-resistant HIV-1 subtype C isolate, and from variants previously identified by deep sequencing and clonal analysis of plasma virus obtained at serial time points during the development of VCV resistance.34 Three patterns of CCR5 antagonist susceptibility were observed: 1) sigmoid inhibition curves with IC50s similar to 07J-W0; 2) significant decreases in IC50 with leftward shifts of the inhibition curves compared with 07J-W0; and 3) flattening of the inhibition curves with decreased or negative PMI. None of the mutants showed an increase in IC50 or rightward shift in the VCV inhibition curve. A majority of the mutants demonstrated sigmoid dose-response curves to MVC, TAK-779 and VCV that were similar to those obtained with recombinant virus that incorporated the full-length 07J-W0 env; all mutants showed a sigmoid dose-response to the CCR5 mAb HGS004. Viruses manifesting resistance to the CCR5 antagonists all had reduced PMI (< 90%).
A previous report based on the in vitro selection of a VCV-resistant isolate suggested that the first steps in the evolution of resistance involves a competitive mechanism, whereby gp120 develops an increased affinity for the drug-free form of CCR5, resulting in a modest increase in IC50.23 Accumulation of additional mutations subsequently leads to development of a non-competitive form of resistance, in which the virus utilizes the drug-bound receptor.23 The data presented here, together with our earlier findings, suggest that the evolution of VCV resistance in this subtype C isolate may be characterized by the progressive loss of inhibition through a non-competitive mechanism due to the selection of pre-existing minority variants that already carried VCV resistance-associated mutations in V3.16,34 The IC50s of small molecule CCR5 antagonists increase over the natural course of HIV-1 infection.35,38–40 Whether clinical isolates with higher pre-treatment CCR5 antagonist IC50s are more likely to develop clinical resistance to these drugs remains an open question.
In earlier work we identified V3 sequences present as minority variants at week 0 that contained some but not all of the V3 modifications present in 07J-W28. Several of these variants showed evidence of positive selection during VCV treatment.34 Surprisingly, reconstruction of those sequences by introducing the relevant mutations into the pretreatment (07J-W0) envelope resulted in envelopes that showed no resistance or increased susceptibility to VCV. For example, the T307I/F318I/T320R/H330Y mutant, which was present at 0.002% of the population at week 0, increased to 15% of the population by week 12.34 However, a variant carrying these mutations in a week 0 backbone showed a fold-change in VCV IC50 of 0.18 compared to the wild-type. Similarly, the K305R/ T307I/F318I/T320R/H330Y mutant, which was present at 0.2% of the starting population and increased to 25% at week 12, showed a fold-change of 0.24 in VCV IC50. These results suggest that sequences outside of V3 in the week 12 envelope may confer a significant selective advantage for the viruses carrying V3 sequences identified by deep sequencing, either by contributing to VCV resistance or by improving overall viral fitness.
The finding that certain V3 loop mutations associated with VCV resistance in the 07J-W28 envelope increased susceptibility to VCV when present individually could help explain the relatively infrequent observation of CCR5 antagonist resistance in clinical trials to date, as sequential accumulation of such mutations would be highly unfavorable in the setting of VCV treatment. Mutations in the V3 loop could increase susceptibility to small-molecule CCR5 antagonists if these mutations reduce affinity of gp120 for CCR5, slow viral entry, or both. The in vivo significance of increased VCV susceptibility conferred by these mutations in certain envelope backbones requires further study.
Two mutations that increased susceptibility to VCV (306P and 318I) are rare or previously unreported in env sequences from subtype B and C viruses (Los Alamos HIV Sequence Database).41–42 In contrast, the K305R substitution is found in 16% of subtype B and 11% of subtype C sequences.42 This mutation had no effect on CCR5 antagonist susceptibility and was retained in all the env clones obtained from this subject after cessation of drug.16 Interestingly, another study found that K305R decreased PMI when present by itself, underscoring the concept that the phenotypic effect of V3 mutations in env is context dependent.15
Further evidence of the influence of the envelope backbone on phenotypic expression of V3 mutations is provided by the observation that the 305R/S306P/T307I/F318I/ T320R/G321E/H330Y mutant in the context of the 07J-W0 backbone exhibited a more negative PMI with VCV than the same mutations in the context of the week 28 backbone (−106.2 versus −9.9, respectively). This finding is counterintuitive, as one might expect the envelope showing the greatest resistance (most negative PMI) to emerge as the dominant species. One possible explanation is that changes in the env backbone required to allow emergence of the intermediate V3 mutants in which the 306P eventually arose moderated the effect of the V3 mutations on VCV susceptibility. It is also possible that a virus carrying the 07J-W28 backbone has greater overall fitness than the week 0 virus, a question that can be resolved by performing growth competition experiments.
Prior characterization of a VCV-resistant subtype D clinical isolate demonstrated that at least 3 mutations were required in order to produce detectable reduction in PMI.26 Clonal analysis of V3 sequences from the pre-VCV sample demonstrated the presence of F309L, L317F, and D320N (analogous to position 321 in our V3 alignment) mutations in 2 of 12 clones; the frequency of these mutations in the population increased in response to drug pressure and in association with the development of resistance.26 This finding supports our model that pre-existing variants act as a scaffold on which further novel mutations lead to complete VCV resistance. Detailed analysis of individual VCV-resistant HIV-1 isolates obtained in vitro and in vivo suggests that V3 mutations associated with VCV resistance significantly alter interactions of V3 with the second extracellular loop of CCR5 (ECL2) and enhance the interaction of gp120 with the N-terminus of co-receptor.25–26 Further study is required to determine the generalizability of V3 loop and regional CCR5 interactions to other VCV-resistant clinical isolates across subtypes.
We previously reported that recombinant viruses containing the 07J-W28 envelope are cross resistant to TAK-779;16 here we extend those findings to MVC. Cross-resistance to multiple small-molecule CCR5 antagonists is not surprising, given that these inhibitors are believed to bind in a common pocket formed by the CCR5 transmembrane domains.21,43 However, for any given mutant, the nature and magnitude of resistance to VCV, MVC and TAK-779 varied, suggesting that these antagonists differ in their interactions with CCR5. In general, alterations in PMI were much greater for VCV than for MVC or TAK-779. Maximal resistance to TAK-779 and MVC was not observed even when the full complement of mutations from the 07J-W28 V3 loop was inserted into the 07J-W0 backbone, suggesting again the likely contribution of additional mutations in other parts of envelope to CCR5 antagonist resistance.
A limitation of our current study is that it focused on a single VCV-resistant subtype C isolate; other VCV-resistant viruses may have different characteristics. Despite this limitation, our findings extend our understanding of VCV resistance, as most information to date comes from the detailed study of a small number of isolates selected in vitro. It remains unclear whether resistance-associated mutations that are derived in vitro will be similar to those that arise during the development of clinical resistance. The availability of deep sequencing data on minority and emerging V3 loop variants allowed us to relate phenotypic data from the mutants we studied to the changing structure of the virus population in response to VCV selection.
In summary, the results presented here provide further evidence of the complexity of resistance to CCR5 antagonists, which remains incompletely understood. The emergence of VCV resistance most likely requires a complex scaffold to which mutations that have a major effect on VCV susceptibility can be added. The variable effect of these mutations on MVC and TAK 779 susceptibilities suggest that these small molecule CCR5 antagonists can have different interactions with CCR5 even though they occupy a similar binding site.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by a National Research Service Award (T32) AI07387-19 to TJH, and by NIH grants K08 AI081547 (to AMNT), R37 AI553537, K24 RR016482 (to DRK), and the Harvard University Center for AIDS Research (P30 AI060354). We also thank the AIDS Clinical Trials Group A5211 protocol team for providing the 07J-W0 and 07J-W28 viruses; M. Subramanian for providing HGS004; B.M. Baroudy for providing vicriviroc; and the NIH AIDS Research and Reference Reagent Program for providing maraviroc and TAK779.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Presented in part at the 17th Conference on Retroviruses and Opportunistic Infections, 2010, San Francisco, CA (abstract 524).
REFERENCES
- 1.Tsibris AM, Kuritzkes DR. Chemokine antagonists as therapeutics: focus on HIV-1. Annual review of medicine. 2007;58:445–459. doi: 10.1146/annurev.med.58.080105.102908. [DOI] [PubMed] [Google Scholar]
- 2.Gulick RM, Lalezari J, Goodrich J, et al. Maraviroc for previously treated patients with R5 HIV-1 infection. The New England journal of medicine. 359:1429–1441. doi: 10.1056/NEJMoa0803152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gulick RM, Su Z, Flexner C, et al. Phase 2 study of the safety and efficacy of vicriviroc, a CCR5 inhibitor, in HIV-1-Infected, treatment-experienced patients: AIDS clinical trials group 5211. The Journal of infectious diseases. 2007;196:304–312. doi: 10.1086/518797. [DOI] [PubMed] [Google Scholar]
- 4.Landovitz RJ, Angel JB, Hoffmann C, et al. Phase II study of vicriviroc versus efavirenz (both with zidovudine/lamivudine) in treatment-naive subjects with HIV-1 infection. The Journal of infectious diseases. 2008;198:1113–1122. doi: 10.1086/592052. [DOI] [PubMed] [Google Scholar]
- 5.Strizki JM, Tremblay C, Xu S, et al. Discovery and characterization of vicriviroc (SCH 417690), a CCR5 antagonist with potent activity against human immunodeficiency virus type 1. Antimicrobial agents and chemotherapy. 2005;49:4911–4919. doi: 10.1128/AAC.49.12.4911-4919.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dorr P, Westby M, Dobbs S, et al. Maraviroc (UK-427,857), a potent, orally bioavailable, and selective small-molecule inhibitor of chemokine receptor CCR5 with broad-spectrum anti-human immunodeficiency virus type 1 activity. Antimicrobial agents and chemotherapy. 2005;49:4721–4732. doi: 10.1128/AAC.49.11.4721-4732.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schurmann D, Fatkenheuer G, Reynes J, et al. Antiviral activity, pharmacokinetics and safety of vicriviroc, an oral CCR5 antagonist, during 14-day monotherapy in HIV-infected adults. AIDS. 2007;21:1293–1299. doi: 10.1097/QAD.0b013e3280f00f9f. [DOI] [PubMed] [Google Scholar]
- 8.Ketas TJ, Kuhmann SE, Palmer A, et al. Cell surface expression of CCR5 and other host factors influence the inhibition of HIV-1 infection of human lymphocytes by CCR5 ligands. Virology. 2007;364:281–290. doi: 10.1016/j.virol.2007.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ketas TJDV, Lam E, Maddon PJ, Olson WC. PRO 140, a Humanized CCR5 Monoclonal Antibody, is Active Against Genotypically Diverse and Enfuvirtide-Resistant Strains of HIV-1. Abstract WEPEA093. Presented at: 4th International AIDS Society Conference on HIV Pathogenesis, Treatment, and Prevention; Sydney, Australia. 2007. [Google Scholar]
- 10.Lalezari J, Yadavalli GK, Para M, et al. Safety, pharmacokinetics, and antiviral activity of HGS004, a novel fully human IgG4 monoclonal antibody against CCR5, in HIV-1-infected patients. The Journal of infectious diseases. 2008;197:721–727. doi: 10.1086/527327. [DOI] [PubMed] [Google Scholar]
- 11.Murga JD, Franti M, Pevear DC, Maddon PJ, Olson WC. Potent antiviral synergy between monoclonal antibody and small-molecule CCR5 inhibitors of human immunodeficiency virus type 1. Antimicrobial agents and chemotherapy. 2006;50:3289–3296. doi: 10.1128/AAC.00699-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hartley O, Gaertner H, Wilken J, et al. Medicinal chemistry applied to a synthetic protein: development of highly potent HIV entry inhibitors. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:16460–16465. doi: 10.1073/pnas.0404802101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kuhmann SE, Pugach P, Kunstman KJ, et al. Genetic and phenotypic analyses of human immunodeficiency virus type 1 escape from a small-molecule CCR5 inhibitor. Journal of virology. 2004;78:2790–2807. doi: 10.1128/JVI.78.6.2790-2807.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marozsan AJ, Kuhmann SE, Morgan T, et al. Generation and properties of a human immunodeficiency virus type 1 isolate resistant to the small molecule CCR5 inhibitor, SCH-417690 (SCH-D) Virology. 2005;338:182–199. doi: 10.1016/j.virol.2005.04.035. [DOI] [PubMed] [Google Scholar]
- 15.Ogert RA, Wojcik L, Buontempo C, et al. Mapping resistance to the CCR5 co-receptor antagonist vicriviroc using heterologous chimeric HIV-1 envelope genes reveals key determinants in the C2-V5 domain of gp120. Virology. 2008;373:387–399. doi: 10.1016/j.virol.2007.12.009. [DOI] [PubMed] [Google Scholar]
- 16.Tsibris AM, Sagar M, Gulick RM, et al. In vivo emergence of vicriviroc resistance in a human immunodeficiency virus type 1 subtype C-infected subject. Journal of virology. 2008;82:8210–8214. doi: 10.1128/JVI.00444-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Westby M, Smith-Burchnell C, Mori J, et al. Reduced maximal inhibition in phenotypic susceptibility assays indicates that viral strains resistant to the CCR5 antagonist maraviroc utilize inhibitor-bound receptor for entry. Journal of virology. 2007;81:2359–2371. doi: 10.1128/JVI.02006-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.MARAVIROC Tablets NDA 22–128 ANTIVIRAL DRUGS ADVISORY COMMITTEE (AVDAC) BRIEFING DOCUMENT. 2007:104. www.fda.gov/OHRMS/DOCKETS/AC/07/briefing/2007-4283b1-01-Pfizer.pdf.
- 19.Yusa K, Maeda Y, Fujioka A, Monde K, Harada S. Isolation of TAK-779-resistant HIV-1 from an R5 HIV-1 GP120 V3 loop library. The Journal of biological chemistry. 2005;280:30083–30090. doi: 10.1074/jbc.M414360200. [DOI] [PubMed] [Google Scholar]
- 20.Ogert RA, Ba L, Hou Y, et al. Structure-function analysis of human immunodeficiency virus type 1 gp120 amino acid mutations associated with resistance to the CCR5 coreceptor antagonist vicriviroc. Journal of virology. 2009;83:12151–12163. doi: 10.1128/JVI.01351-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Watson C, Jenkinson S, Kazmierski W, Kenakin T. The CCR5 receptor-based mechanism of action of 873140, a potent allosteric noncompetitive HIV entry inhibitor. Molecular pharmacology. 2005;67:1268–1282. doi: 10.1124/mol.104.008565. [DOI] [PubMed] [Google Scholar]
- 22.Pugach P, Ketas TJ, Michael E, Moore JP. Neutralizing antibody and anti-retroviral drug sensitivities of HIV-1 isolates resistant to small molecule CCR5 inhibitors. Virology. 2008;377:401–407. doi: 10.1016/j.virol.2008.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pugach P, Marozsan AJ, Ketas TJ, Landes EL, Moore JP, Kuhmann SE. HIV-1 clones resistant to a small molecule CCR5 inhibitor use the inhibitor-bound form of CCR5 for entry. Virology. 2007;361:212–228. doi: 10.1016/j.virol.2006.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Trkola A, Kuhmann SE, Strizki JM, et al. HIV-1 escape from a small molecule, CCR5-specific entry inhibitor does not involve CXCR4 use. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:395–400. doi: 10.1073/pnas.012519099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Berro R, Sanders RW, Lu M, Klasse PJ, Moore JP. Two HIV-1 variants resistant to small molecule CCR5 inhibitors differ in how they use CCR5 for entry. PLoS pathogens. 2009;5:e1000548. doi: 10.1371/journal.ppat.1000548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ogert RA, Hou Y, Ba L, et al. Clinical resistance to vicriviroc through adaptive V3 loop mutations in HIV-1 subtype D gp120 that alter interactions with the N-terminus and ECL2 of CCR5. Virology. 2010 doi: 10.1016/j.virol.2010.01.037. epublished ahead of print. [DOI] [PubMed] [Google Scholar]
- 27.Colonno R, Rose R, McLaren C, Thiry A, Parkin N, Friborg J. Identification of I50L as the signature atazanavir (ATV)-resistance mutation in treatment-naive HIV-1-infected patients receiving ATV-containing regimens. The Journal of infectious diseases. 2004;189:1802–1810. doi: 10.1086/386291. [DOI] [PubMed] [Google Scholar]
- 28.Demeter LM, Meehan PM, Morse G, et al. HIV-1 drug susceptibilities and reverse transcriptase mutations in patients receiving combination therapy with didanosine and delavirdine. J Acquir Immune Defic Syndr Hum Retrovirol. 1997;14:136–144. doi: 10.1097/00042560-199702010-00006. [DOI] [PubMed] [Google Scholar]
- 29.Dueweke TJ, Pushkarskaya T, Poppe SM, et al. A mutation in reverse transcriptase of bis(heteroaryl)piperazine-resistant human immunodeficiency virus type 1 that confers increased sensitivity to other nonnucleoside inhibitors. Proceedings of the National Academy of Sciences of the United States of America. 1993;90:4713–4717. doi: 10.1073/pnas.90.10.4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gong YF, Robinson BS, Rose RE, et al. In vitro resistance profile of the human immunodeficiency virus type 1 protease inhibitor BMS-232632. Antimicrobial agents and chemotherapy. 2000;44:2319–2326. doi: 10.1128/aac.44.9.2319-2326.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rimsky LT, Shugars DC, Matthews TJ. Determinants of human immunodeficiency virus type 1 resistance to gp41-derived inhibitory peptides. Journal of virology. 1998;72:986–993. doi: 10.1128/jvi.72.2.986-993.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wei X, Decker JM, Liu H, et al. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrobial agents and chemotherapy. 2002;46:1896–1905. doi: 10.1128/AAC.46.6.1896-1905.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cocchi F, DeVico AL, Garzino-Demo A, Cara A, Gallo RC, Lusso P. The V3 domain of the HIV-1 gp120 envelope glycoprotein is critical for chemokine-mediated blockade of infection. Nature medicine. 1996;2:1244–1247. doi: 10.1038/nm1196-1244. [DOI] [PubMed] [Google Scholar]
- 34.Tsibris AM, Korber B, Arnaout R, et al. Quantitative deep sequencing reveals dynamic HIV-1 escape and large population shifts during CCR5 antagonist therapy in vivo. PloS one. 2009;4:e5683. doi: 10.1371/journal.pone.0005683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Etemad B, Fellows A, Kwambana B, et al. Human immunodeficiency virus type 1 V1-to-V5 envelope variants from the chronic phase of infection use CCR5 and fuse more efficiently than those from early after infection. Journal of virology. 2009;83:9694–9708. doi: 10.1128/JVI.00925-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kimpton J, Emerman M. Detection of replication-competent and pseudotyped human immunodeficiency virus with a sensitive cell line on the basis of activation of an integrated beta-galactosidase gene. Journal of virology. 1992;66:2232–2239. doi: 10.1128/jvi.66.4.2232-2239.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sagar M, Wu X, Lee S, Overbaugh J. Human immunodeficiency virus type 1 V1–V2 envelope loop sequences expand and add glycosylation sites over the course of infection, and these modifications affect antibody neutralization sensitivity. Journal of virology. 2006;80:9586–9598. doi: 10.1128/JVI.00141-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jansson M, Popovic M, Karlsson A, et al. Sensitivity to inhibition by beta-chemokines correlates with biological phenotypes of primary HIV-1 isolates. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:15382–15387. doi: 10.1073/pnas.93.26.15382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koning FA, Kwa D, Boeser-Nunnink B, et al. Decreasing sensitivity to RANTES (regulated on activation, normally T cell-expressed and -secreted) neutralization of CC chemokine receptor 5-using, non-syncytium-inducing virus variants in the course of human immunodeficiency virus type 1 infection. The Journal of infectious diseases. 2003;188:864–872. doi: 10.1086/377105. [DOI] [PubMed] [Google Scholar]
- 40.Repits J, Oberg M, Esbjornsson J, et al. Selection of human immunodeficiency virus type 1 R5 variants with augmented replicative capacity and reduced sensitivity to entry inhibitors during severe immunodeficiency. The Journal of general virology. 2005;86:2859–2869. doi: 10.1099/vir.0.81111-0. [DOI] [PubMed] [Google Scholar]
- 41.Felsovalyi K, Nadas A, Zolla-Pazner S, Cardozo T. Distinct sequence patterns characterize the V3 region of HIV type 1 gp120 from subtypes A and C. AIDS research and human retroviruses. 2006;22:703–708. doi: 10.1089/aid.2006.22.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Patel MB, Hoffman NG, Swanstrom R. Subtype-specific conformational differences within the V3 region of subtype B and subtype C human immunodeficiency virus type 1 Env proteins. Journal of virology. 2008;82:903–916. doi: 10.1128/JVI.01444-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kondru R, Zhang J, Ji C, et al. Molecular interactions of CCR5 with major classes of small-molecule anti-HIV CCR5 antagonists. Molecular pharmacology. 2008;73:789–800. doi: 10.1124/mol.107.042101. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.