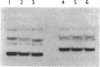
Abstract
To predict alterations in single-strand DNA mobility in non-denaturing electrophoretic gels, Zuker's RNA folding program was modified. Energy files utilized by the LRNA RNA folding algorithm were modified to emulate folding of single-strand DNA. Energy files were modified to disallow G-T base pairing. Stacking energies were corrected for DNA thermodynamics. Constraints on loop nucleotide sequences were removed. The LRNA RNA folding algorithm using the DNA fold energy files was applied to predict folding of PCR generated single-strand DNA molecules from polymorphic human ALDH2 and TPH alleles. The DNA-Fold version 1.0 program was used to design primers to create and abolish SSCP mobility shifts. Primers were made that add a 5' tag sequence or alter complementarity to an internal sequence. Differences in DNA secondary structure were assessed by SSCP analysis and compared to single-strand DNA secondary structure predictions. Results demonstrate that alterations in single-strand DNA conformation may be predicted using DNA-Fold 1.0.
Full text
PDF
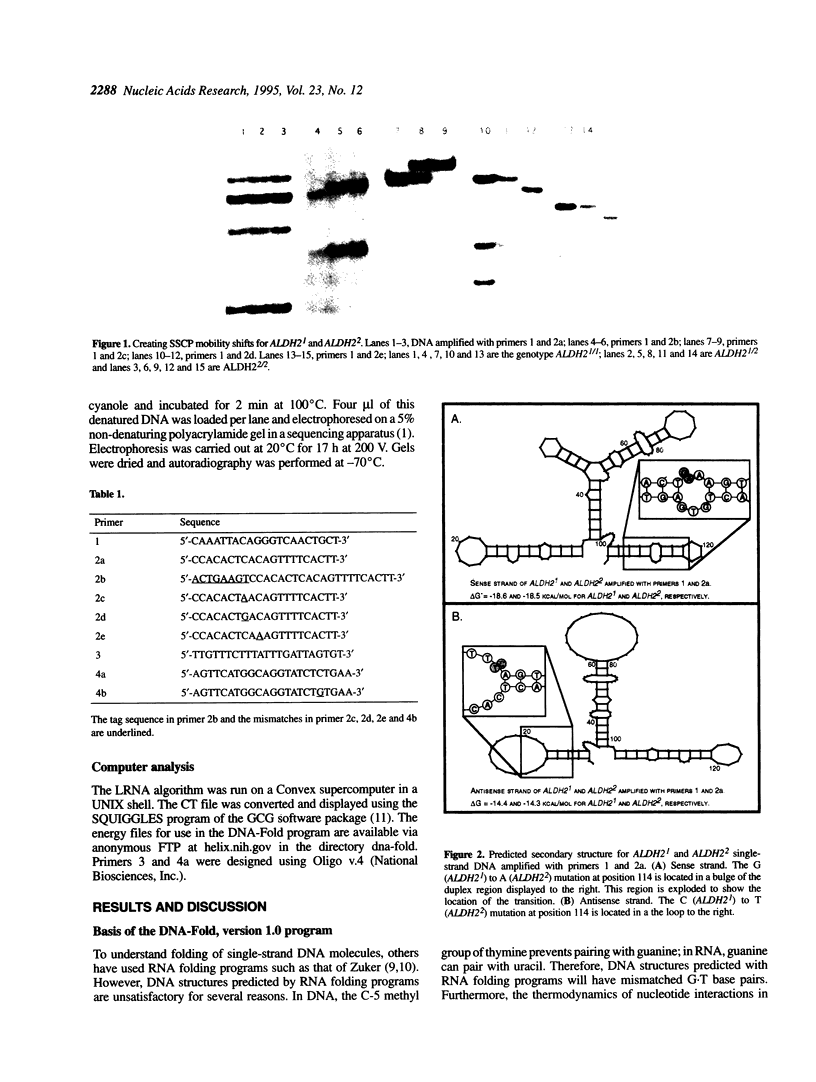
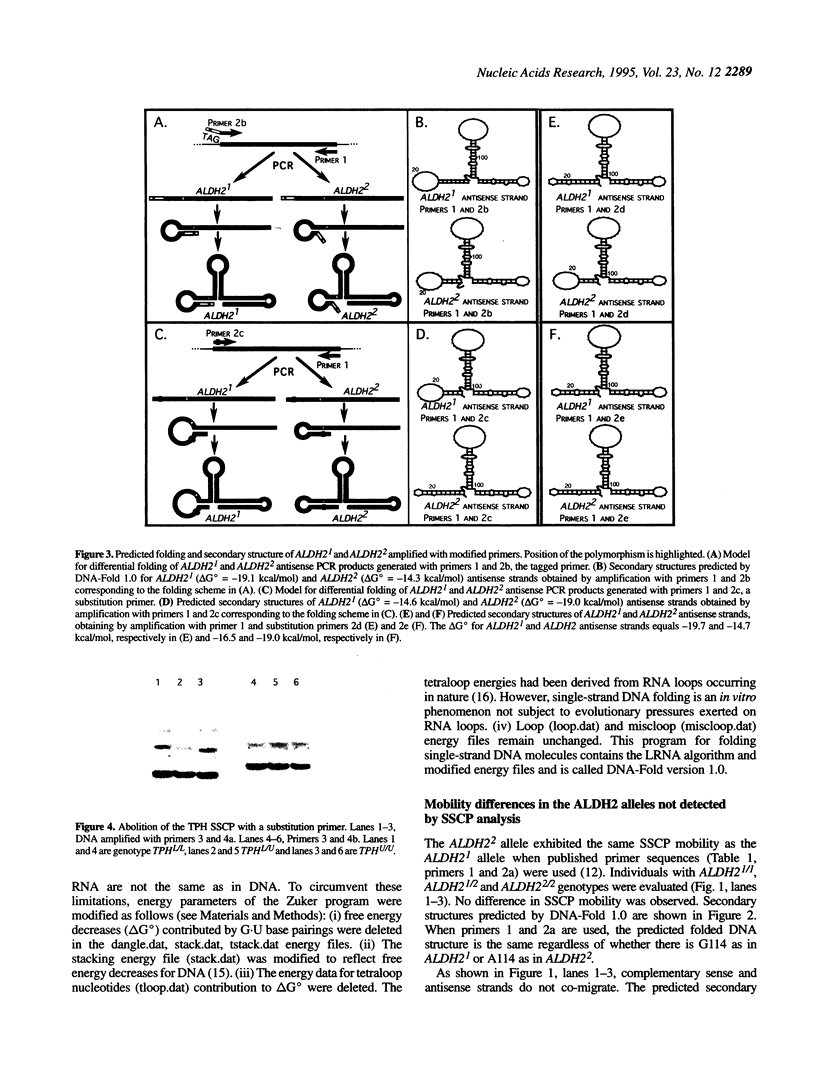
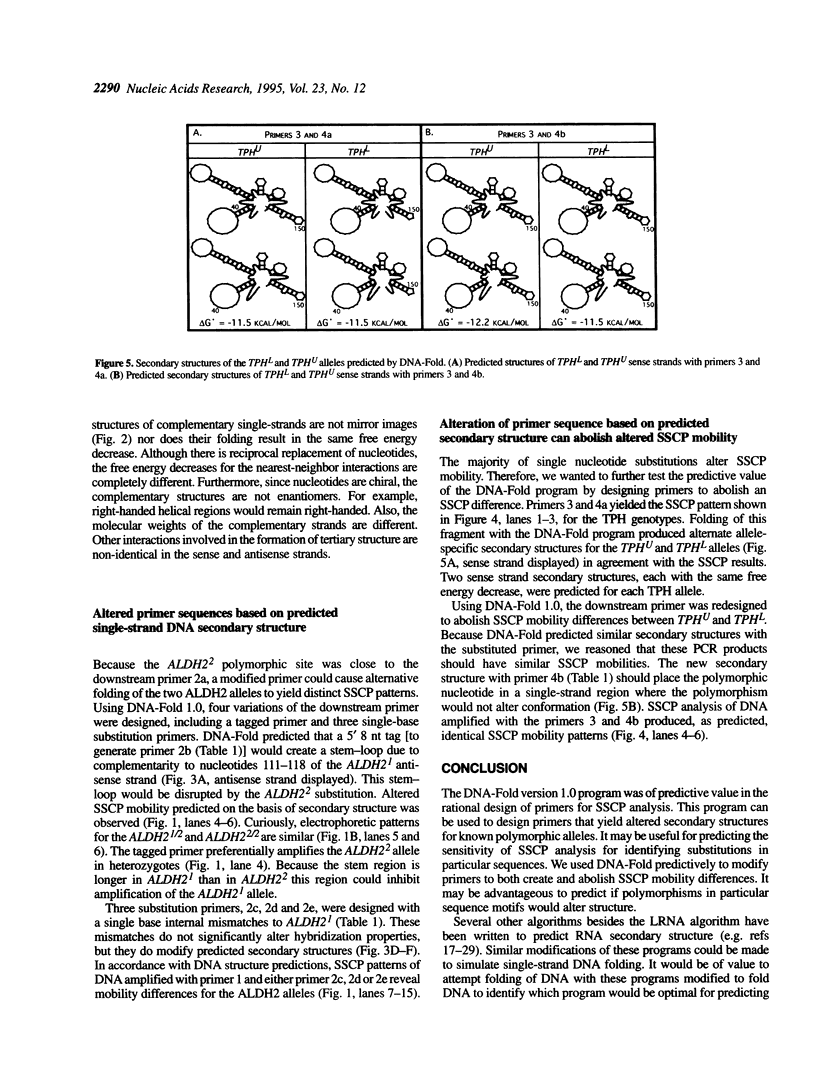

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abrahams J. P., van den Berg M., van Batenburg E., Pleij C. Prediction of RNA secondary structure, including pseudoknotting, by computer simulation. Nucleic Acids Res. 1990 May 25;18(10):3035–3044. doi: 10.1093/nar/18.10.3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breslauer K. J., Frank R., Blöcker H., Marky L. A. Predicting DNA duplex stability from the base sequence. Proc Natl Acad Sci U S A. 1986 Jun;83(11):3746–3750. doi: 10.1073/pnas.83.11.3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J. H., Le S. Y., Maizel J. V. A procedure for RNA pseudoknot prediction. Comput Appl Biosci. 1992 Jun;8(3):243–248. doi: 10.1093/bioinformatics/8.3.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean M., White M. B., Amos J., Gerrard B., Stewart C., Khaw K. T., Leppert M. Multiple mutations in highly conserved residues are found in mildly affected cystic fibrosis patients. Cell. 1990 Jun 1;61(5):863–870. doi: 10.1016/0092-8674(90)90196-l. [DOI] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goedde H. W., Agarwal D. P., Harada S., Rothhammer F., Whittaker J. O., Lisker R. Aldehyde dehydrogenase polymorphism in North American, South American, and Mexican Indian populations. Am J Hum Genet. 1986 Mar;38(3):395–399. [PMC free article] [PubMed] [Google Scholar]
- Gultyaev A. P. The computer simulation of RNA folding involving pseudoknot formation. Nucleic Acids Res. 1991 May 11;19(9):2489–2494. doi: 10.1093/nar/19.9.2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogeweg P., Hesper B. Energy directed folding of RNA sequences. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):67–74. doi: 10.1093/nar/12.1part1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu L. C., Tani K., Fujiyoshi T., Kurachi K., Yoshida A. Cloning of cDNAs for human aldehyde dehydrogenases 1 and 2. Proc Natl Acad Sci U S A. 1985 Jun;82(11):3771–3775. doi: 10.1073/pnas.82.11.3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Häner R., Dervan P. B. Single-strand DNA triple-helix formation. Biochemistry. 1990 Oct 23;29(42):9761–9765. doi: 10.1021/bi00494a001. [DOI] [PubMed] [Google Scholar]
- Impraim C., Wang G., Yoshida A. Structural mutation in a major human aldehyde dehydrogenase gene results in loss of enzyme activity. Am J Hum Genet. 1982 Nov;34(6):837–841. [PMC free article] [PubMed] [Google Scholar]
- Jacobson A. B., Good L., Simonetti J., Zuker M. Some simple computational methods to improve the folding of large RNAs. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):45–52. doi: 10.1093/nar/12.1part1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeger J. A., Turner D. H., Zuker M. Improved predictions of secondary structures for RNA. Proc Natl Acad Sci U S A. 1989 Oct;86(20):7706–7710. doi: 10.1073/pnas.86.20.7706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeger J. A., Turner D. H., Zuker M. Predicting optimal and suboptimal secondary structure for RNA. Methods Enzymol. 1990;183:281–306. doi: 10.1016/0076-6879(90)83019-6. [DOI] [PubMed] [Google Scholar]
- Jordan B. R. Computer generation of pairing schemes for RNA molecules. J Theor Biol. 1972 Feb;34(2):363–378. doi: 10.1016/0022-5193(72)90168-3. [DOI] [PubMed] [Google Scholar]
- Le S. Y., Chen J. H., Braun M. J., Gonda M. A., Maizel J. V. Stability of RNA stem-loop structure and distribution of non-random structure in the human immunodeficiency virus (HIV-I). Nucleic Acids Res. 1988 Jun 10;16(11):5153–5168. doi: 10.1093/nar/16.11.5153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez H. M. An RNA folding rule. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):323–334. doi: 10.1093/nar/12.1part1.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mironov A. A., D'iakonova L. P., Kister A. E. Predskazanie ansamblei vtorichnykh struktur RNK. Kineticheskii analiz samoorganizatsii. Mol Biol (Mosk) 1984 Nov-Dec;18(6):1686–1694. [PubMed] [Google Scholar]
- Nielsen D. A., Dean M., Goldman D. Genetic mapping of the human tryptophan hydroxylase gene on chromosome 11, using an intronic conformational polymorphism. Am J Hum Genet. 1992 Dec;51(6):1366–1371. [PMC free article] [PubMed] [Google Scholar]
- Nielsen D. A., Goldman D., Virkkunen M., Tokola R., Rawlings R., Linnoila M. Suicidality and 5-hydroxyindoleacetic acid concentration associated with a tryptophan hydroxylase polymorphism. Arch Gen Psychiatry. 1994 Jan;51(1):34–38. doi: 10.1001/archpsyc.1994.03950010034005. [DOI] [PubMed] [Google Scholar]
- Nussinov R., Jacobson A. B. Fast algorithm for predicting the secondary structure of single-stranded RNA. Proc Natl Acad Sci U S A. 1980 Nov;77(11):6309–6313. doi: 10.1073/pnas.77.11.6309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nussinov R., Pieczenik G. Structural and combinatorial constraints on base pairing in large nucleotide sequences. J Theor Biol. 1984 Feb 7;106(3):245–259. doi: 10.1016/0022-5193(84)90029-8. [DOI] [PubMed] [Google Scholar]
- Orita M., Iwahana H., Kanazawa H., Hayashi K., Sekiya T. Detection of polymorphisms of human DNA by gel electrophoresis as single-strand conformation polymorphisms. Proc Natl Acad Sci U S A. 1989 Apr;86(8):2766–2770. doi: 10.1073/pnas.86.8.2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleij C. W., Rietveld K., Bosch L. A new principle of RNA folding based on pseudoknotting. Nucleic Acids Res. 1985 Mar 11;13(5):1717–1731. doi: 10.1093/nar/13.5.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiki R. K., Gelfand D. H., Stoffel S., Scharf S. J., Higuchi R., Horn G. T., Mullis K. B., Erlich H. A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988 Jan 29;239(4839):487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- Solovyov V. V., Zharkikh A. A., Kolchanov N. A., Ratner V. A. The template RNAs of RNA polymerases can have compact secondary structure, formed by long double helices with partial violations of the complementarity. FEBS Lett. 1984 Jan 2;165(1):72–78. doi: 10.1016/0014-5793(84)80017-4. [DOI] [PubMed] [Google Scholar]
- Stoll J., Goldman D. Isolation and structural characterization of the murine tryptophan hydroxylase gene. J Neurosci Res. 1991 Apr;28(4):457–465. doi: 10.1002/jnr.490280402. [DOI] [PubMed] [Google Scholar]
- Tuerk C., Gauss P., Thermes C., Groebe D. R., Gayle M., Guild N., Stormo G., d'Aubenton-Carafa Y., Uhlenbeck O. C., Tinoco I., Jr CUUCGG hairpins: extraordinarily stable RNA secondary structures associated with various biochemical processes. Proc Natl Acad Sci U S A. 1988 Mar;85(5):1364–1368. doi: 10.1073/pnas.85.5.1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M., Stiegler P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1981 Jan 10;9(1):133–148. doi: 10.1093/nar/9.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]