Abstract
P6 has been a vaccine candidate for nontypable Haemophilus influenzae (NTHi) based on its location on the outer membrane and immunogenicity. Because P6 is attached to the inner peptidoglycan layer of NTHi, and is putatively surface exposed, it must be a transmembrane protein. We examined the P6 structure using computational modeling, a P6 modified by site-directed mutagenesis to study monoclonal antibody attachment to P6, and nuclear magnetic resonance spectroscopy. We found that P6 cannot be a transmembrane protein, and therefore may not be surface exposed. We conclude that there may be another protein on the surface of NTHi that has epitopes similar if not identical to P6.
Keywords: nontypable Haemophilus influenzae, P6 protein, outer membrane protein
1. Introduction
For over twenty years, the outer membrane protein (OMP) P6 has been a leading vaccine candidate against nontypable Haemophilus influenzae (NTHi) [1–3]. NTHi is an important cause of acute otitis media (AOM), sinusitis, acute exacerbations of chronic bronchitis and pneumonia (in developing countries). The incidence of ear infections alone in the United States (75% of children experience at least one episode of AOM by their 3rd birthday) points to the urgent need for a vaccine against NTHi [4]. However, one of the major barriers to NTHi vaccine development has been identification of antibody targets that are conserved among all or virtually all strains of the bacteria. P6 has been shown to be conserved among all of the tested strains of NTHi [1, 21].
Although studies have demonstrated that monoclonal antibodies interact with P6 on the surface of the bacterial cell [5, 6] and that P6 is the target of bactericidal antibodies [7–10], our analysis of a recent protein structure of P6 [11] suggests that P6 may not be a surface exposed OMP. In addition, studies on Pal, the homologue to P6 in Escherichia coli (E. coli), mark Pal as a non-surface exposed protein which interacts with the peptidoglycan layer and various other proteins inside of the cell [12–14]. These observations led us to pursue new experimental evidence to corroborate or refute the physical location and orientation of P6. Our findings, presented here, strongly suggest that it is unlikely for P6 to exist as a transmembrane protein in the outer membrane of NTHi.
2. Materials and Methods
2.1. Computational modeling of P6 as a transmembrane protein
Currently, there are several computational tools available on the Expert Protein Analysis System (ExPASy, Swiss Institute of bioinformatics) server [15] for predicting the topology of transmembrane proteins. The topology prediction programs required an input of the P6 primary sequence, minus the first 19 amino acid leader sequence which gets cleaved from the protein and is replaced by the lipid moiety. The programs yielded an output of the predicted transmembrane regions and/or the likelihood that certain regions within the protein are contained within the membrane.
2.2 Protein expression and purification
The NTHi P6 gene in the pET28-a (Kanamycin resistant) vector was a generous gift from Dr. John Orban (University of Maryland Biotechnology Institute). The P6 protein is non-lipidated (the 19 residue N-terminal signal sequence was removed) and contains a 6-Histidine tag at its N-terminus. The protein was expressed and purified as described in the literature with some modifications [11]. Briefly, the protein was expressed in E. coli BL21 (DE3) cells in LB for the ELISA experiments and 15N-labeled minimal media with [15N, 99%]NH4Cl (Cambridge Isotope Laboratories, Inc.) as the sole nitrogen source for the nuclear magnetic resonance (NMR) experiments, and induced with 1 mM IPTG. The cells were harvested by centrifugation at 5000g for 15 minutes and the pellets were frozen overnight. The thawed cells were lysed via sonication and centrifuged at 20,000g for 25 minutes. The supernatant was then purified via TALON resin beads (Clontech) according to the manufacturer's instructions. The protein was eluted in imidazole buffer and exchanged into 50 mM NaPi, 50 mM NaCl pH 7.0 with a PD-10 column desalting column (GE Healthcare). The estimated protein concentration was determined using a BCA assay (Pierce) and using an extinction coefficient (280 nm) of 14,350 cm−1M−1.
2. 3. Site-directed mutagenesis
The P6 D59N mutant was prepared using the QuikChange II site-directed mutagenesis kit (Stratagene/Agilent Technologies) according to the manufacturer's instructions. P6 D59N is a P6 variant where the aspartic acid at position 59 was substituted with an asparagine. The non-lipidated P6 gene in pET28-a was used as a template for the mutagenesis, and the following forward and backward mutant primers were purchased from Integrated DNA Technologies. Forward: 5'-gttacaataccgtttatttcggttttgataaatataacattactggtgaatacg-3' Backward: 5'-cgtattcaccagtaatgttatatttatcaaaaccgaaataaacggtattgtaac-3' The P6 D59N protein was expressed in E. coli and purified as described above for wild-type P6.
2. 4. ELISA
50 ng of purified recombinant P6 or P6 D59N protein were added to each well of an Apogent medium binding plate (Nunc), incubated at room temperature for approximately 3 hours and then refrigerated overnight. The plate was washed 3 times with PBS with 0.1% TWEEN-20. The plate was blocked with PBS/3% skim milk/ 0.1% TWEEN-20 (200 μl/well) for 1 hour at 37°C. After the plate was washed 3 times, 100 μl of the unpurified 7F3 and 4G4 monoclonal antibodies (kindly provided by Dr. Timothy Murphy, University at Buffalo) were added to each well at different dilutions (10 fold, 20 fold, 40 fold and 80 fold in PBS/3% skim milk/0.1% TWEEN-20) and allowed to incubate for 1 hour at room temperature. The plate was washed again; the goat anti-mouse IgG with HRP (1:10,000 dilution in PBS/3% skim milk/ 0.1% TWEEN-20, 100 μl/well) was added to the wells and allowed to incubate at room temperature for 1 hour. After washing, 100μl of TMB substrate (KPL) was placed in each well and allowed to develop for 30 minutes at room temperature, and the reaction was stopped by adding 100μl of 1M phosphoric acid to each well. The plates were read using an automated ELISA reader at 450nm.
2. 5. NMR Spectroscopy
The NMR data were collected on a Varian INOVA 500 MHz spectrometer (operating at 499.839 MHz for 1H) at 299 K. A 1H-15N HSQC spectrum (8 scans, 1024 × 128 points) was collected for both the purified recombinant wild-type P6 and P6 D59N (~1 mM protein concentration, pH 7.0). The NMR spectra were processed using NMRpipe [16] and visualized (the two spectra were overlaid) using Sparky [17]. The residue chemical shift assignments for wild-type P6 have been published in the literature [11].
3. Results
3.1. Computational modeling of P6 as a transmembrane protein
Seven protein topology programs were used for prediction analyses of the location of P6 in NTHi. None of the programs predicted membrane spanning regions contained within P6 (Table 1).
Table 1.
Summary of outputs from the transmembrane prediction tools (19 amino acid leader sequence not included in analysis).
| Topology Prediction Tool | Output |
|---|---|
| TMHMM Server v. 2.024 | No predicted transmembrane |
| Dense Alignment Surface25 | No predicted transmembrane |
| TMpred26 | No predicted transmembrane |
| SOSUI27 | No predicted transmembrane |
| HMMTOP28 | No predicted transmembrane |
| PHDhtm28 | No predicted transmembrane |
| Phobius30 | No predicted transmembrane |
3.2. Immunologic assays to detect P6
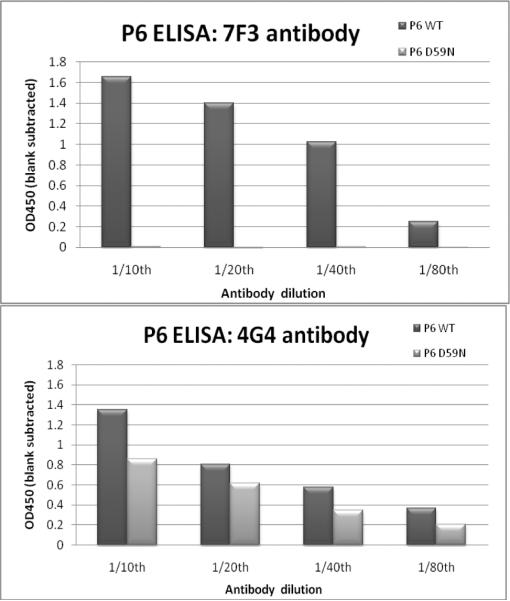
ELISA was performed on purified recombinant wild-type P6 and P6 D59N. P6 D59N did not show any binding to the 7F3 monoclonal antibody. The 4G4 monoclonal antibody did bind to P6 D59N at approximately half the binding seen to wild-type P6 (Fig. 1).
Fig. 1.
ELISA data on purified recombinant P6 wild-type (WT) and P6 D59N proteins against 7F3 (left) and 4G4 (right) monoclonal antibodies.
3.3. Nuclear magnetic resonance
NMR spectroscopy was utilized to confirm that the D59N mutation did not alter the protein structure from that of wild-type P6. The 1H-15N HSQC “fingerprint” spectra for both proteins, confirmed that the structure of P6 D59N is highly similar to that of wild-type P6, with only minor chemical shift differences between the two spectra (data not shown).
3.4. Protein structure analysis
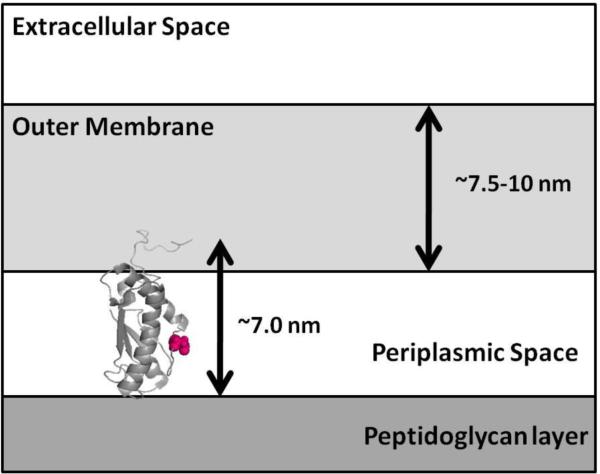
When the P6 protein structure is oriented such that its longest measurement is vertical, the protein is about ~7 nm in length. The OM of NTHi has been measured to be 7.5–10 nm thick [18]. Residue 59 is located in the center of the protein (Fig. 2). Monoclonal antibodies 7F3 and 4G4 interact with P6 residue 59. Therefore, P6 structure analysis shows it is impossible for P6 to interact with the peptidoglycan layer in the interior of the bacterial cell and span the OM to become surface-exposed and have residue 59 interact with monoclonal antibodies on the cell surface.
Fig. 2.
A cartoon representation of the backbone of NTHi P6 (PDB ID 2AIZ) is oriented vertically, with residue 59 highlighted in space-fill (left). Because the OM is measured between 7.5–10 nm thick, and P6 is proposed to interact with monoclonal antibodies on the cell surface via residue 59 (pink), it is unlikely that P6 is long enough to span the OM such that it can also interact with the peptidoglycan layer.
4. Discussion
P6 has been reported to comprise 1–5% (by weight) of the total protein in the outer membrane of NTHi [5]. Several experimental approaches have been utilized to demonstrate the surface exposure of P6. Monoclonal and polyclonal antibodies to P6 have been shown to stain the entire organism by immunofluorescence and immunoelectron microscopy [1, 5, 6, 19]. NTHi strain 3524 was radio-labeled with 125I, and then after disruption of the bacterial outer membrane, the OMPs were dispersed on an agarose gel. The presence of a band at ~16 kDa that was extrinsically labeled suggested that P6 was surface exposed [1]. Adsorbed antiserum was shown to immunoprecipitate P6 [19], P6 was shown to be accessible to bactericidal antibodies [7–10, 20] and the anti-P6 antibody was shown to be eluted from the surface of intact NTHi [5]. It is also generally accepted that NTHi P6 interacts with the peptidoglycan layer in the periplasmic space of the bacterial cell [11, 21].
There have been several key immunological experiments involving monoclonal antibodies to P6 that have suggested that P6 is surface exposed [5–10]. Apicella et al. performed immunoelectron microscope experiments that utilized the P6 monoclonal antibodies 3B9 and 4G4 [5, 6]. The presence of gold-labeled secondary antibodies on NTHi cells (visualized via an electron microscope) suggested that both monoclonal antibodies bound to P6 on the surface of the cells [5, 6]. A third monoclonal antibody, 7F3, was proposed to bind to the same or a closely related epitope on P6 as the 4G4 antibody [6]. Pre-incubation of NTHi with monoclonal antibody 7F3 inhibited human serum bactericidal killing, suggesting that P6 was indeed surface exposed and the target of bactericidal activity [20]. While the specific amino acids that make up the epitope for 7F3 and 4G4 have not been fully identified, aspartic acid at position 59 has been implicated in antibody binding to P6 [21]. The results of our experiments demonstrate that D59 does indeed participate in the binding of P6 to both 7F3 and 4G4 monoclonal antibodies. The OM typically measures between 7.5 and 10 nm thick. At its longest measurement, P6 is only ~7 nm long, making it unlikely that P6 would be able to interact with monoclonal antibodies on the cell surface via residue 59 (which is located in the middle of the protein) and span the OM such that it can also interact with the peptidoglycan layer (Fig. 2).
Transmembrane proteins typically demonstrate a pattern of hydrophobic/hydrophilic amino acids which can be used to predict the probable “membrane spanning” regions. In the case of NTHi P6, seven out of seven transmembrane prediction programs (available on the ExPASy server) did not recognize any membrane spanning regions within the P6 amino acid sequence (Table 1). These results, along with our protein structure analysis of P6 and the NTHi OM, suggest that P6 cannot possibly span the OM in order to interact with monoclonal antibodies (at residue 59) on the cell surface and with the peptidoglycan layer in the interior of the bacterial cell.
It is important to mention that the experiments presented in this work were performed on recombinant non-lipidated P6 protein in which the leader sequence containing the first 19 amino acids was removed. During in vivo processing of P6, the N-terminal leader sequence is removed and replaced by a lipid moiety. The ELISA and NMR experiments were attempted on the lipidated version of P6; however, purified lipidated P6 unfolds and/or aggregates such that NMR spectroscopy cannot be performed on the protein (data not shown). The lipid moiety of a bacterial lipoprotein anchors it to the membrane by insertion into the inner or outer leaflet of the lipid bilayer. Since P6 has to interact with antibodies and peptidoglycan through its amino acids (and not through its lipid moiety), using the non-lipidated version of P6 does not prevent us from making conclusions about the length of native P6 or the location of its epitopes.
The structural similarity between NTHi P6 and E. coli Pal is notable. The NMR structure of NTHi P6 was determined in 2006 [11]. It showed a high degree of similarity between NTHi P6 and E. coli Pal [12]. The N-terminally truncated 108-residue Pal crystal structure can be overlaid onto the NTHi P6 NMR structure (Fig. 3). The overall folds of the proteins are highly similar, suggesting the potential for similar binding partners and/or similar functions in vivo. Although the functions of Pal in E. coli are not definitive, Pal is known to be anchored to the inner leaflet (i.e., the inner lipid layer) of the OM of E. coli via its lipid moiety and is proposed to interact with the peptidoglycan layer as well as several Tol proteins and outer membrane protein A (OmpA) [12–13, 22–23]. Importantly, there is no controversy that E. coli Pal is attached to the OM via its lipid moiety, but is otherwise defined as non-membrane spanning protein. In other words, with the exception of the N-terminal lipid, Pal does not span either layer of the OM and faces in towards the intermembrane (i.e., periplasmic) space of E. coli. Although there is no definitive evidence demonstrating a similar orientation for P6 in the NTHi OM, the remarkable similarity between the structures of NTHi P6 and E. coli Pal points to the possibility of similar membrane orientations.
Fig. 3.
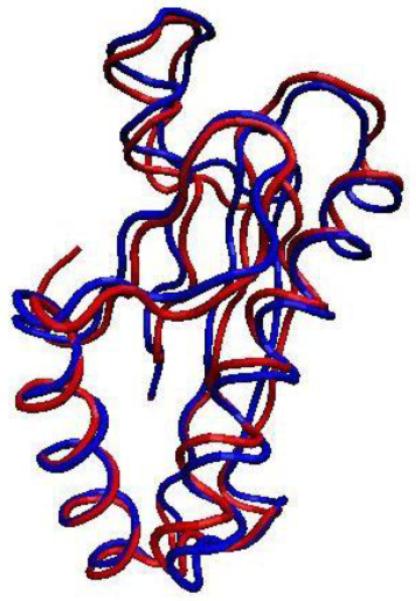
Backbone representations of truncated P6 from NTHi (blue, PDB ID 2AIZ) and Pal from E. coli (red, PDB ID 1OAP) overlay with an RMSD of ~1.2 Å. The graphical representation was prepared using the Visual Molecular Dynamics program [31].
In conclusion, our results show that NTHi P6 is not a transmembrane protein since it is not of sufficient length to be surface-exposed and interact with the peptidoglycan layer on the inside of the bacterial cell [11, 21]. In order for P6 to interact with the peptidoglycan layer and interact with monoclonal/bactericidal antibodies on the cell surface (as shown by immunological experiments), then P6 must either be inserted into the OM via two distinct “flipped” orientations or there is another OMP on the surface of NTHi that expresses an epitope similar to P6. Since the former has never been described for an OMP of any bacterial species, we are now pursuing studies to identify the OMP of NTHi that has structural epitopes similar to P6 but that is not P6.
Acknowledgements
This study was supported by NIH NIDCD RO1 08671 (to MEP) and the Rochester Institute of Technology. We would like to thank Dr. Timothy Murphy (University at Buffalo, SUNY) for the generous gift of the monoclonal antibodies 7F3 and 4G4. We would also like to thank Dr. John Orban (University of Maryland Biotechnology Institute) for the generous gift of the non-lipidated P6 plasmid and Dr. Lisa Parsons for her advice on working with the P6 protein. We would also like to thank Dr. Kara Bren and Dr. Sarah Bowman for their help with the NMR spectroscopy.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Murphy TF, Nelson MB, Dudas KC, Mylotte JM, Apicella MA. Identification of a specific epitope of Haemophilus influenzae on a 16,600-dalton outer membrane protein. J Infect Dis. 1985;152:1300–07. doi: 10.1093/infdis/152.6.1300. [DOI] [PubMed] [Google Scholar]
- [2].Murphy TF. Vaccine development for non-typeable Haemophilus influenzae and Moraxella catarrhalis: progress and challenges. Expert Rev Vaccines. 2005;4:843–53. doi: 10.1586/14760584.4.6.843. [DOI] [PubMed] [Google Scholar]
- [3].Murphy TF, Bakaletz LO, Kyd JM, Watson B, Klein DL. Vaccines for otitis media: proposals for overcoming obstacles to progress. Vaccine. 2005;23:2696–702. doi: 10.1016/j.vaccine.2004.12.014. [DOI] [PubMed] [Google Scholar]
- [4].National Institute on Deafness and Other Communication Disorders: Health Info, Hearing, Ear, Infection and Deafness, Otitis Media (Ear Infection) 2002 NIH Pub No 97-4216. [Google Scholar]
- [5].Nelson MB, Murphy TF, vanKeulen H, Rekosh D, Apicella MA. Studies on P6, an important outer-membrane protein antigen of Haemophilus influenzae. Rev Infect Dis. 1988;10:S331–36. doi: 10.1093/cid/10.supplement_2.s331. [DOI] [PubMed] [Google Scholar]
- [6].Bogdan JA, Apicella MA. Mapping of a surface-exposed, conformational epitope of the P6 protein of Haemophilus influenzae. Infect Immun. 1995;63:4395–401. doi: 10.1128/iai.63.11.4395-4401.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].DeMaria TF, Murwin DM, Leake ER. Immunization with outer membrane protein P6 from nontypeable Haemophilus influenzae induces bactericidal antibody and affords protection in the chinchilla model of otitis media. Infect Immun. 1996;64:5187–92. doi: 10.1128/iai.64.12.5187-5192.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Green BA, Quinn-Dey T, Zlotnik GW. Biologic activities of antibody to a peptidoglycan-associated lipoprotein of Haemophilus influenzae against multiple clinical isolates of H. influenzae type b. Infect Immun. 1987;55:2878–83. doi: 10.1128/iai.55.12.2878-2883.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Kyd JM, Dunkley ML, Cripps AW. Enhanced respiratory clearance of nontypeable Haemophilus influenzae following mucosal immunization with P6 in a rat model. Infect Immun. 1995;63:2931–40. doi: 10.1128/iai.63.8.2931-2940.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Sabirov A, Casey JR, Murphy TF, Pichichero ME. Breast feeding is associated with a reduced frequency of acute otitis media and high serum antibody levels against NTHi and outer membrane protein vaccine antigen candidate P6. Pediatric Res. 2009;66:565–70. doi: 10.1203/PDR.0b013e3181b4f8a6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Parsons LM, Lin F, Orban J. Peptidoglycan recognition by Pal, an outer membrane lipoprotein. Biochemistry. 2006;45:2122–28. doi: 10.1021/bi052227i. [DOI] [PubMed] [Google Scholar]
- [12].Godlewska R, Wiśniewska K, Pietras Z, Jagusztyn-Kyrnicka EK. Peptidoglycan-associated lipoprotein (Pal) of Gram-negative bacteria: function, structure, role in pathogenesis and potential application in immunoprophylaxis. FEMS Microbiol Letters. 2009;298:1–11. doi: 10.1111/j.1574-6968.2009.01659.x. [DOI] [PubMed] [Google Scholar]
- [13].Bonsor DA, Grishkovskaya I, Dodson EJ, Kleanthous C. Moleclar mimcry enables competitive recruitment by a natively disordered protein. JACS. 2007;129:4800–7. doi: 10.1021/ja070153n. [DOI] [PubMed] [Google Scholar]
- [14].Bonsor DA, Hecht O, Vankemmelbeke M, Sharma A, Krachler AM, Housden NG, et al. Allosteric □-propeller signaling in TolB and its manipulation by translocating colicins. EMBO J. 2009;28:2846–57. doi: 10.1038/emboj.2009.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, Bairoch A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nuc Acids Res. 2003;31:3784–8. doi: 10.1093/nar/gkg563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–93. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- [17].Goddard D, Kneller DG. SPARKY 3. University of California; San Francisco: [Google Scholar]
- [18].Salton MRJ, Kim K-S. Bacteriology, Structure. In: Baron S, Davis CP, Niesel DW, Woods GW, editors. Medical Microbiology. University of Texas Medical Branch at Galveston; Galveston, TX: 1996. [Google Scholar]
- [19].Munson RS, Jr., Granoff DM. Purification and partial characterization of outer membrane proteins P5 and P6 from Haemophilus influenzae type b. Infect Immun. 1985;49:544–9. doi: 10.1128/iai.49.3.544-549.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Murphy TF, Bartos LC, Rice PA, Nelson MB, Dudas KC, Apicella MA. Identification of a 16,600-dalton outer membrane protein on nontypeable Haemophilus influenzae as a target for human serum bactericidal antibody. J Clin Invest. 1986;78:1020–7. doi: 10.1172/JCI112656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Murphy TF, Kirkham C, Sikkema DJ. Neonatal, urogenital isolates of biotype 4 nontypeable Haemophilus influenzae express a variant P6 outer membrane protein molecule. Infect Immun. 1992;60:2016–22. doi: 10.1128/iai.60.5.2016-2022.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Bouveret E, Bénédetti H, Rigal A, Loret E, Lazdunski C. In vitro characterization of peptidoglycan-associated lipoprotein (PAL)-peptidoglycan and PAL-TolB interactions. J of Bacteriol. 1999;181:6306–11. doi: 10.1128/jb.181.20.6306-6311.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Cascales E, Lloubés R. Deletion analyses of the peptidoglycan-associated lipoprotein Pal reveals three independent binding sequences including a TolA box. Mol Microbiol. 2004;51:873–85. doi: 10.1046/j.1365-2958.2003.03881.x. [DOI] [PubMed] [Google Scholar]
- [24].Krogh A, Larsson B, von Heijne G, Sonnhammer ELL. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J Mol Biol. 2001;305:567–80. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- [25].Cserzo M, Wallin E, Simon I, von Heijne G, Elofsson A. Prediction of transmembrane alpha-helices in procariotic membrane proteins: the Dense Alignment Surface method. Prot Eng. 1997;10:673–76. doi: 10.1093/protein/10.6.673. [DOI] [PubMed] [Google Scholar]
- [26].Hofmann K, Stoffel W. TMbase - A database of membrane spanning proteins segments. Biol Chem. 1993;374:166. [Google Scholar]
- [27].Hirokawa T, Boon-Chieng S, Mitaku S. SOSUI: Classification and secondary structure prediction system for membrane proteins. Bioinformatics. 1998;14:378–9. doi: 10.1093/bioinformatics/14.4.378. [DOI] [PubMed] [Google Scholar]
- [28].Tusnády GE, Simon I. Principles governing amino acid composition of integral membrane proteins: Applications to topology prediction. J Mol Biol. 1998;283:489–506. doi: 10.1006/jmbi.1998.2107. [DOI] [PubMed] [Google Scholar]
- [29].Combet C, Blanchet C, Geourjon C, Deléage G. NPS@: Network Protein Sequence Analysis. TIBS. 2000;25:147–50. doi: 10.1016/s0968-0004(99)01540-6. [DOI] [PubMed] [Google Scholar]
- [30].Kall L, Krogh A, Sonnhammer ELL. A combined transmembrane topology and signal peptide prediction method. J Mol Biol. 2004;338:1027–36. doi: 10.1016/j.jmb.2004.03.016. [DOI] [PubMed] [Google Scholar]
- [31].Humphrey W, Dalke A, Schulten K. VMD - Visual Molecular Dynamics. J Molec Graphics. 1996;14:33–8. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]