Abstract
Adaptor proteins with multimodular structures can participate in the regulation of various cellular functions. We have cloned a novel adaptor protein, XB130, which binds the p85α subunit of phosphatidyl inositol 3-kinase and subsequently mediates signaling through RET/PTC in TPC-1 thyroid cancer cells. In the present study, we sought to determine the role of XB130 in the tumorigenesis in vivo and in related molecular mechanisms. In WRO thyroid cancer cells, knockdown of XB130 using small interfering RNA inhibited G1-S phase progression, induced spontaneous apoptosis, and enhanced intrinsic and extrinsic apoptotic stimulus-induced cell death. Growth of tumors in nude mice formed from XB130 shRNA stably transfected WRO cells were significantly reduced, with decreased cell proliferation and increased apoptosis. Microarray analysis identified 246 genes significantly changed in XB130 shRNA transfected cells. Among them, 57 genes are related to cell proliferation or survival, including many transcription regulators. Ingenuity Pathway Analysis showed that the top-ranked disease related to XB130 is cancer, and the top molecular and cellular functions are cellular growth and proliferation and cell cycle. A human thyroid tissue microarray study identified expression of XB130 in normal thyroid tissue as well as in human thyroid carcinomas. These observations suggest that the expression of XB130 in these cancer cells may affect cell proliferation and survival by controlling the expression of multiple genes, especially transcription regulators.
Adaptor proteins are molecules of modular structures without enzymatic activity. They are composed of multiple protein-protein and/or protein-lipid interacting domains, through which they link signaling components to form macromolecular complexes and propagate cellular signals.1,2 Depending on the functional role of the interacting partner, adaptor proteins can participate in the regulation of various signaling pathways. A number of adaptor proteins have been demonstrated to regulate tumorigenesis. For example, actin filament associated protein (AFAP) is required for actin stress fiber formation and cell adhesion and is critical for tumorigenic growth in prostate cancer.3,4 Tyrosine kinase substrate 5 is a scaffolding adaptor protein with five Src homology 3 (SH3) domains, co-localizes to podosomes, and regulates migration and invasion of various human cancer cells.5,6 These findings support a broader investigation of the effects of adaptor proteins on tumorigenesis and their potential as diagnostic biomarkers and therapeutic targets of cancer.
Recently, we have discovered the human xb130 gene during the cloning process of human afap gene.7–9 The human xb130 gene encodes 818 amino acids and has an apparent molecular size of approximately 130 kDa. As an adaptor protein, the overall structure of XB130 shares similarity with AFAP, so it is also known as actin filament associated protein 1-like 2 (AFAP1L2). The N-terminal region of XB130 includes several tyrosine phosphorylation sites and a proline-rich sequence that can potentially interact with SH2 and SH3 domain-containing proteins, respectively. The middle portion harbors two pleckstrin-homology domains that may target proteins to cellular membranes through interactions with specific phospholipids such as phosphatidylinositol 3,4,5-triphosphate. The C-terminal region contains a coiled-coil domain, which might be involved in protein oligomerization and DNA binding.9 Our studies have implicated XB130 as a likely substrate and regulator of tyrosine kinase-mediated signaling. Down-regulation of endogenous XB130 with small interfering RNA (siRNA) reduced c-Src activity, IL-8 production, and phosphorylation of Akt in human lung epithelial cells.9
Our recent study revealed expression of XB130 in human thyroid tissue, and we found that XB130 is a downstream mediator of the signaling cascade propagated by RET/PTC, a genetically rearranged, constitutively active, thyroid-cancer-specific tyrosine kinase.10 RET/PTC plays a pathogenic role and exhibits transforming ability by exerting its effects on differentiation and on mitogenic and metastatic potential in papillary thyroid cancer.11,12 XB130 couples RET/PTC signaling to the phosphatidylinositol 3-kinase (PI3K)/Akt signaling through a specific binding site to the p85α subunit of PI3K, thereby regulating cell survival and proliferation.10 We further found that XB130 has a strong affinity with the lamellipodial F-actin meshwork and is involved in the motility and invasiveness of tumor cells.13
The objectives of the present study were to investigate whether XB130 is involved in tumor growth in vivo and to identify the pathogenetic molecular mechanisms. We established XB130 short hairpin RNA (shRNA) stably transfected thyroid cancer cells. Using an in vivo xenograft model, we observed diminished tumor growth due to reduced cell proliferation and enhanced cell death. Further, microarray analyses of XB130 shRNA stably transfected cells showed that many genes are related to cancer mainly through their functions in cell proliferation and cell survival. To confirm the significance of the results in human disease, we used a human thyroid tissue microarray to validate the expression of XB130 in thyroid carcinoma. Our results indicate the important role of XB130 in tumor progression.
Materials and Methods
Cell Lines, Antibodies, and Other Reagents
Human thyroid carcinoma WRO cells were maintained in RPMI 1640, supplemented with 10% FBS, 1 mmol/L pyruvate and nonessential amino acids (GIBCO-BRL, Gaithersburg, MD), and 1% penicillin-streptomycin. Cells were cultured in a standard humidified incubator at 37°C with 5% CO2.
Monoclonal XB130 antibody was generated as described previously.9 Antibody for GAPDH came from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-PCNA antibody came from Abcam (Cambridge, MA). Anti-Ki-67 (clone Ki-S5) came from Chemicon (Temecula, CA). Fas antibody (clone CH11) came from Upstate Biotechnology Inc. (Lake Placid, NY). Staurosporine came from Sigma-Aldrich (St. Louis, MO).
Protein Studies
Immunoblotting experiments were performed according to procedures described previously.7,8 Briefly, cells were lysed with modified radioimmune precipitation assay buffer (50 mmol/L Tris-HCl, pH 7.5; 150 mmol/L NaCl; 2 mmol/L EGTA; 2 mmol/L EDTA; and 1% Triton X-100) containing 10 μg/ml each aprotinin, leupeptin, pepstatin, 1 mmol/L phenylmethylsulfonyl fluoride, 1 mmol/L Na3VO4, and 10 mmol/L NaF. Protein concentration was measured by a modified Bradford assay (Bio-Rad, Hercules, CA). Cell lysates containing equal amounts of total proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and then transferred onto nitrocellulose membranes. Membranes were then probed with the indicated antibodies. Proteins were revealed by an enhanced chemiluminescence detection kit (Thermo, Rockford, IL). Band densities were quantified using ImageJ software (http://rsb.info.nih.gov/ij/).
siRNA Transfection
Two siRNAs were designed according to the XB130 sequence, as described previously.9,10 Cells were transfected with 50 nmol/L pooled XB130 siRNAs using the oligofectamine reagent (Invitrogen, San Diego, CA), as described previously.9,10,14 The medium containing siRNA was replaced with fresh medium after 24 hours. The siSTABLE V2 nontargeting siRNA#1 from Dharmacon (Lafayette, CO) was used as a negative control.
Cell Cycle Analysis
Cell cycle distribution was analyzed by flow cytometry.15 Briefly, cells were harvested in phosphate-buffered saline (PBS) and fixed in 70% cold ethanol overnight. Fixed cells were washed twice in PBS and permeabilized with 0.1% Triton X-100 and 2 mg/ml RNase A in PBS for 30 minutes. They were then washed once in PBS and stained with 50 μg/ml of propidium iodide (PI) (Sigma-Aldrich). Stained cells were analyzed by the Cytomix FC500 flow cytometry system (Beckman Coulter, Fullerton, CA).
Cell Proliferation
Cells were seeded onto 6-well plates at a density of 1.0 × 105 cells per well and incubated at 37°C with 5% CO2. At various time points, cells were detached from the flasks in a trypsin-EDTA solution and counted by a hemocytometer.
Analysis of Apoptotic Cells
After being treated with staurosporine or FasAb for 24 hours, cells were harvested and stained with fluorescein isothiocyanate-conjugated annexin V and phosphatidylinositol using the annexin V kit (Beckman Coulter, Brea, CA) according to the manufacturer's protocols and analyzed by the Cytomix FC500 flow cytometry system.
shRNA Plasmid Transfection and Establishment of Stably Transfected Cell Lines
A SureSilencing shRNA kit for XB130 was purchased from SuperArray (Frederick, MD). Four shRNA plasmids for XB130 were included in the kit. Negative control shRNA plasmid was a scrambled artificial sequence that does not match any human gene (see Supplementary Figure S1A at http://ajp.amjpathol.org). WRO cells were transfected with shRNA plasmids using Lipofectamine 2000 (Invitrogen). Transfected cells were selected with 0.5 mg/ml G418 (Roche, Indianapolis, IN) for 14 days, and antibiotic-resistant pools were collected. To screen out the best gene ablation efficiency, the expression levels of XB130 protein from antibiotic-resistant pools were determined by Western blotting. To establish single-cell colonies, cell pools were diluted to obtain 1 cell/200 μL and plated onto 96-well plates. Cell clones were expanded under continuous selection using 0.5 mg/ml G418. Single colonies were chosen by Western blotting and used for further experiments.
Xenograft Models in Nude Mice
The animal study was approved by the Animal Use and Care Committee of the University Health Network. Suspensions of 2.0 × 106 XB130 shRNA transfected WRO cells in 200 μL of PBS were injected subcutaneously into one side of the lower flanks of 6-week-old female nude mice, and the same amount of NC shRNA-transfected cells were injected into the opposite side. Two independent experiments were repeated with five and four animals per group. At regular intervals, the tumor diameters were measured with a caliper and tumor volumes were calculated using the formula16:
At defined time points, all mice were sacrificed, and the weights of resected tumors were measured. Tumors were fixed in 10% formaldehyde in PBS, paraffin embedded, and stained with hematoxylin and eosin.
Immunohistochemical Analysis
Tissue sections (5 μm) from the xenografts underwent immunohistochemical staining. Formalin-fixed and paraffin-embedded tissue sections were deparaffinized in xylene, rehydrated in a graded alcohol series, and transferred to PBS. The sections were immersed in 10 mmol/L citrate buffer (pH 6.0) and heated in a microwave for 15 minutes. The microwave-irradiated sections were cooled to room temperature. The slides were then blocked for endogenous peroxidase and biotin with 1.5% H2O2 in methanol. Nonspecific binding was blocked by incubating the slides with 5% horse serum albumin in a humid chamber for 60 minutes. Incubation for the primary antibody was performed overnight at 4°C with the monoclonal XB130 antibody or the mouse monoclonal anti-Ki-67 antibody diluted 1:200 in PBS with 0.3% Triton X-100/5% horse serum albumin. The secondary antibody was a biotinylated anti-mouse IgG (1:200 in PBS with 0.3% Triton X-100/5% horse serum albumin for 1 hour in a humidified chamber). Detection was made using a Vecstatin ABC kit (Vector Laboratories, Burlingame, CA) with 3-3′-diaminobenzidine as chromogen; slides were counterstained with hematoxylin. Isotype-matched mouse IgG was used as negative control antibodies with the same dilution as the primary antibodies mentioned earlier. To evaluate the proliferation index, Ki-67-stained cells were quantified in five randomly selected fields at 400× magnification. The percentage of cells that showed positive nuclear staining for the Ki-67 antigen was considered to be the proliferation index,17 which was calculated by analyzing five different tumors from each group.
A tissue microarray was built using cases of human thyroid tissues as described previously.18,19 The following tissue cores were included (total = 251): normal thyroid, n = 23; benign lesions, n = 71; follicular adenomas, n = 35; chronic thyroiditis, n = 18; Hürthle cell adenoma, n = 7; infarcted/degenerative/reactive follicular lesion, n = 6; nodular hyperplasia, n = 4; follicular neoplasm, n = 1; malignant cores, n = 157; follicular carcinoma, n = 7; papillary carcinoma, n = 141; Hürthle cell carcinoma, n = 5; anaplastic/insular, n = 4).
The tissue microarray was stained with an anti-XB130 monoclonal antibody as described earlier. XB130-positive staining was scored on the basis of the intensity of cytoplasmic signal using the following criteria: the cores were scored as 0 (no signal); 1 (weak); 2 (moderate); or 3 (strong) and then averaged for each sample (three cores). In samples in which one of the three cores was not interpretable (2.8%) because of either insufficient numbers of cells or loss of the core during array slide processing, the intensity score was based on the average of the remaining two cores.
Terminal Deoxynucleotidyl Transferase-Mediated Deoxyuridine Triphosphate Biotin Nick-End Labeling Staining
An in situ cell death detection kit (Roche, Laval, QC) was used to assess cell death in 5-μm sections obtained from tumors according to a procedure described previously.20 After deparaffinization, dehydration, and permeabilization with 20 μg/ml proteinase K in 10 mmol/L Tris/HCl (pH 7.4 to 8) for 15 minutes, tetramethylrhodamine red-labeled nucleotides were detected under a fluorescence microscope. The nuclei were counterstained with Hoechst 33258 (Pierce, Rockford, IL) at a dilution of 1:5000 for 10 minutes. Label solution without terminal transferase was used as negative control. Slides for positive control were pretreated with DNase (90 U/ml in 40 mmol/L Tris-HCl, pH 7.5, 1 mg/ml BSA). All slides were subjected to identical exposure times. For an evaluation of the apoptotic index in each section, terminal deoxynucleotidyl transferase-mediated deoxyuridine triphosphate biotin nick-end labeling (TUNEL)-stained cells were quantified in five randomly selected fields at 400× magnification, and the apoptotic index was calculated as the number of positive cells/total number of cells × 100%,17 from five tumors per group.
Microarray and Data Analysis
Total RNA was extracted using RNeasy kit (Qiagen, Valencia, CA). Equal amounts of RNA from four single clones in each group (NC or XB130 shRNA transfected cells) were used for microarray. cDNA was synthesized using high-capacity cDNA reverse transcription kits (Applied Biosystems, Foster City, CA). The RNA integrity number determined by the Agilent Bioanalyzer 2100 (Agilent Technologies, Inc., Santa Clara, CA) was used as a measure of the quality of the RNA. Human gene ST 1.0 chips (28,132 spotted genes) from Affymetrix (Santa Clara, CA) were used. Affymetrix CEL files were imported into Partek software (Partek Inc., St. Louis, MO) using the default Partek normalization parameters. Probe-level data were preprocessed, including background correction, normalization, and summarization, using robust multiarray average analysis. Data normalization was performed across all arrays using quantile normalization. The background-adjusted, normalized values were then compiled, or summarized, using the median polish technique, to generate a single measure of expression. As a preliminary analysis, principle component analysis was performed using Partek. Differential expression analysis was performed using Significance Analysis of Microarray (SAM).21 Statistical software JMP version 5 (SAS Institute Inc., Cary, NC) was used for hierarchical cluster analysis. Signal transduction network was analyzed with Ingenuity Pathway Analysis (IPA) (Ingenuity Systems, Inc., Redwood City, CA).
Real-Time Quantitative RT-PCR
The primers used for quantitative RT-PCR for selected genes are shown in Table 1. Total RNA was extracted using RNeasy kit (Qiagen), and cDNA was synthesized from total RNA using MuLV Reverse Transcriptase (Applied Biosystems).22 Quantitative RT-PCR was performed using SYBR Green I Master PCR kit on LightCycler480 (Roche).22 Each assay included a standard curve of five serial dilutions and a no-template negative control. All assays were performed in triplicate. Gene expression levels were normalized to the level of SDHA, a housekeeping gene.
Table 1.
The Primers of the Selected Genes for Quantitative RT-PCR
| Gene name | Forward primer | Reverse primer |
|---|---|---|
| HSPA1A | 5′-AAGAACCAGGTGGCGCTGAA-3′ | 5′-TTGTCTCCGTCGTTGATCAC-3′ |
| BHLHE40 | 5′-TAATTGATCAGCAGCAGCAG-3′ | 5′-GGCCAGATACTGAAGCACCT-3′ |
| TOB1 | 5′-CTCCAGTACAGTAATGCCTT-3′ | 5′-GCTGGTTAGAATACTGCATG-3′ |
| DDIT3 | 5′-GCGTATCATGTTAAAGATGAG-3′ | 5′-ATGTATGAAGATACACTTCC-3′ |
| SLC7A11 | 5′-TGACTGGAGTCCCTGCGTATT-3′ | 5′-TCTTCTGGTACAACTTCCAG-3′ |
| MYC | 5′-TCTCCTTGCAGCTGCTTAGA-3′ | 5′-GAGTCGTAGTCGAGGTCATA-3′ |
| SDHA | 5′-CGGCATTCCCACCAACTAC-3′ | 5′-GGCCGGGCACAATCTG-3′ |
Statistical Analysis
Statistical analysis was carried out using Student's t-test (for two-group comparisons) or Tukey-Kramer HSD test (for multiple comparisons). Differences were considered significant when the P value was less than 0.05. Statistical analyses were performed using JMP version 5.
Results
XB130 Controls Cell Cycle Progression of Thyroid Cancer Cells
To investigate the role of XB130 in cancer cell cycle progression, we conducted knockdown experiments with XB130 siRNA in human thyroid carcinoma WRO cells, and we analyzed the effects using flow cytometry. XB130 siRNA effectively reduced XB130 protein levels (Figure 1A). Down-regulation of XB130 partially reduced cell cycle progression from G1 to S phase (Figure 1B). The expression of cell proliferation markers Ki-67 and PCNA were also reduced in XB130 siRNA treated cells (Figure 1C). These results suggest that XB130 plays important roles in the regulation of thyroid cancer cell proliferation.
Figure 1.
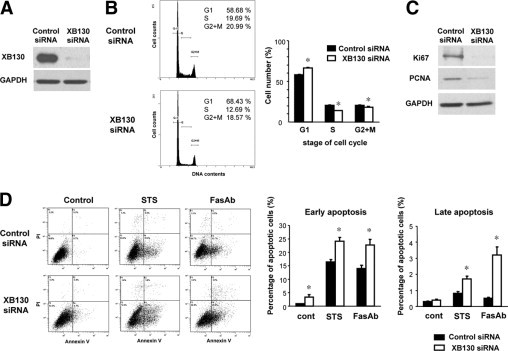
XB130 controls cell cycle progression and survival of WRO thyroid cancer cells. A: XB130 siRNA effectively reduced XB130 protein levels in WRO cells 48 hours after transfection. B: Down-regulation of XB130 inhibited G1-S phase progression of WRO cells. Cells transfected with control or XB130 siRNA were stained with propidium iodide (PI) and analyzed by flow cytometry. Mean ± SEM n = 6. *P < 0.05 (compared with control siRNA). C: Reduced Ki-67 and PCNA levels in XB130 siRNA treated cells. D: Down-regulation of XB130 enhanced spontaneous and induced cell death. Cells transfected with control or XB130 siRNA were treated with staurosporine (STS, 200 nmol/L) or with Fas antibody (FasAb, clone CH11, 100 ng/ml) for 24 hours. Apoptosis was determined by flow cytometry using PI/annexin V double staining. n = 6. Mean ± SEM *P < 0.05 (compared with control siRNA).
XB130 Controls Survival of Thyroid Cancer Cells
To determine the role of XB130 in the survival of cancer cells, we treated WRO cells with XB130 siRNA and analyzed apoptosis. Cells can undergo apoptosis through two major pathways: extrinsic (death receptor), and intrinsic (mitochondrial).23 Down-regulation of XB130 induced early apoptosis (annexin V positive/PI negative) in WRO cells at 48 hours after transfection of siRNA (Figure 1D). Further, XB130 siRNA enhanced extrinsic (FasAb, clone CH11, 100 ng/ml) or intrinsic (staurosporine, 200 nmol/L) apoptotic stimulus-induced early and late apoptosis (annexin V/PI double positive) (Figure 1D). These findings indicate that XB130 expression levels affected cell survival in WRO cells.
Diminished Proliferation in XB130 shRNA Stably Transfected WRO Cell Lines
To determine the roles of XB130 in vivo, we established XB130 shRNA stably transfected WRO cell lines. Four shRNA plasmids (C1, C2, C3, and C4) targeting XB130 and one negative control (NC) shRNA plasmid were transfected to WRO cells (see Supplemental Figure S1A at http://ajp.amjpathol.org). Following antibiotic selection with G418, Western blotting revealed the effective reduction of XB130, especially in C3 or C4 plasmid transfected cells (see Supplemental Figure S1B at http://ajp.amjpathol.org). To select cells with the most optimal gene ablation efficiency, we established single-cell colonies. From the C3 or C4 cell pool, 12 colonies were established, and XB130 expressions were determined by Western blotting (Figure 2A). Four XB130 shRNA transfected colonies (C3-1, C3-4, C4-3, and C4-11) and two NC shRNA transfected colonies (NC-1 and NC-12) were further analyzed for cell proliferation. The numbers of XB130 shRNA transfected cells were significantly lower than those of NC shRNA transfected cells at 72 hours after plating (Figure 2B). NC-1, C3-1, and C4-3 were further studied as representatives of each shRNA plasmid transfected WRO cells in subsequent in vitro and in vivo studies. At 24 hours and 48 hours after plating, cell counts of C3-1 and C4-3 cells were significantly lower than those of NC-1 cells (Figure 2C). Flow cytometry showed that in C3-1 and C4-3 cells, the populations of cells in the G0/G1 phase had increased, and cells in the S phase had decreased in comparison with NC-1 cells at 48 hours after plating (Figure 2D).
Figure 2.
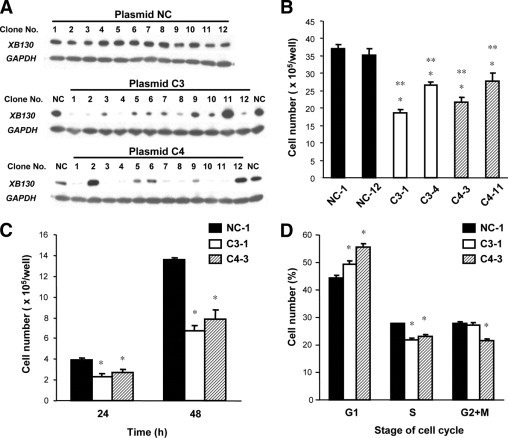
Cell proliferation was diminished in XB130 shRNA stably transfected WRO cells. A: Expression of XB130 in shRNA stably transfected WRO cells as determined by Western blotting. shRNA plasmids (C3 and C4) targeting XB130 and a negative control (NC) shRNA plasmid were transfected in WRO cells. Twelve single colonies were established from each transfected cell pool. B: Cell numbers of XB130 shRNA transfected cells (C3-1, C3-4, C4-3, and C4-11) were significantly lower than those of NC shRNA transfected cells (NC-1 and NC-12) at 72 hours after plating. n = 4. Mean ± SEM *P < 0.05 (compared with NC-1). **P < 0.05 (compared with NC-12). C: Cell numbers of C3-1 and C4-3 cells were significantly lower than those of NC-1 cells at 24 hours and 48 hours after plating. n = 4. Mean ± SEM *P < 0.05 (compared with NC-1). D: G1-S phase progression was significantly diminished in C3-1 and C4-3 cells in comparison with NC-1 cells at 48 hours after plating. Analysis of cell cycle was performed by flow cytometry using PI staining. n = 4. Mean ± SEM *P < 0.05 (compared with NC-1).
Reduced Growth of Tumors Formed from XB130 shRNA Transfected Cells
To determine the role of XB130 in tumor growth in vivo, a xenograft model in nude mice was used.19,24,25 Injection of shRNA stably-transfected WRO cells (2.0 × 106 cells, clones C3-1 and C4-3) subcutaneously into nude mice resulted in significantly smaller tumor volumes than those of the negative control shRNA transfected cells (NC-1) (Figure 3, A and B). The average weight of C3 tumors was reduced by 50.9%, and the average weight of C4 tumors was reduced by 81.0% in comparison with NC tumors (Figure 3B). These results indicate that XB130 has an essential role in the growth of thyroid cancer cells in vivo.
Figure 3.
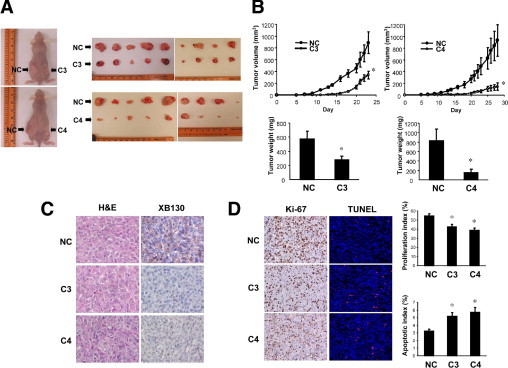
Reduced tumor growth of XB130 shRNA transfected WRO cells in vivo. Suspensions of 2.0 × 106 XB130 shRNA transfected WRO cells (C3-1 or C4-3 cells) in 200 μL of PBS were injected subcutaneously into one side of the lower flanks of 6-week-old female nude mice, and the same amount of NC shRNA-transfected cells (NC-1 cells) were injected into the opposite side. A: Photos of tumors from shRNA transfected WRO cells in mice. C3 or C4 tumors were smaller than NC tumors. B: Tumor growths were significantly diminished and weights of resected tumors were significantly lower in C3 or C4 tumors in comparison with NC tumors. n = 9. Mean ± SEM *P < 0.05 (compared with NC). C: XB130 expression was decreased in C3 and C4 tumors in comparison with NC tumors (magnification, 400×). D: The proliferation index (percentage of Ki-67 positive cells) was decreased, and the apoptosis index (percentage of TUNEL positive cells) was increased in C3 and C4 tumors in comparison with NC tumors. n = 5. Mean ± SEM *P < 0.05 (compared with NC).
Reduced Proliferation and Increased Apoptosis in Tumors from XB130 shRNA Transfected Cells
Immunohistochemical analysis using a monoclonal XB130 antibody showed detectable expression of XB130 in NC tumors but not in C3 and C4 tumors under the same staining conditions (Figure 3C). To determine the cause of the attenuated growth of C3 and C4 tumors, we checked the appearance of cell proliferation and apoptosis markers in the tumor tissue. We found a clear reduction of Ki-67-stained cells in C3 and C4 tumors in comparison with NC tumors (Figure 3D). Further, the percentage of TUNEL-positive cells was significantly higher in C3 and C4 tumors than in NC tumors (Figure 3D). These results indicate that the diminished growth of tumors formed from XB130 shRNA transfected cells was the result of decreased cell proliferation and increased apoptosis.
Gene Expression Profile in XB130 shRNA Transfected Cells
To determine the molecular mechanisms by which XB130 regulates cellular functions, we analyzed gene expression profiles in XB130 shRNA transfected cells by microarray and bioinformatics studies. Four negative control shRNA transfected colonies (NC-1, 2, 9, and 12) and four XB130 shRNA transfected colonies (C3-1, C3-4, C4-3, and C4-11) were analyzed for microarray after the confirmation of XB130 expression using Western blotting (Figure 4A). Principal component analysis results indicated a clear separation of samples based on the expression of XB130 (see Supplemental Figure S2A at http://ajp.amjpathol.org). Further, SAM analysis showed that 246 genes were significantly changed by the stable knockdown of XB130 in WRO cells with a false discovery rate q value <6.66% and fold change >1.3 (see Supplemental Figure S2B at http://ajp.amjpathol.org). Of the genes, 86 were up-regulated, and 160 were down-regulated in XB130 shRNA transfected cells. XB130 was one of the top 10 genes down-regulated in XB130 shRNA transfected cells (fold of change: −2.749, q value <0.001). In the hierarchical cluster analysis of 246 genes, XB130 shRNA transfected cells were grouped together and separated from NC shRNA transfected cells (Figure 4B). Ingenuity pathway analysis showed that the top-ranked disease related to XB130 is cancer, followed by cardiovascular disease and endocrine system disorders (see Supplementary Tables S1 at http://ajp.amjpathol.org). The top molecular and cellular functions related to XB130 are cellular growth and proliferation and cell cycles. Among 246 significantly changed genes, 57 genes showed cell proliferation or survival-related functions (cellular growth and proliferation, cell cycle, cell death, and organismal survival revealed by ingenuity pathway analysis) (Table 2). It is interesting that among these genes, only 18 were up-regulated, and the other 39 genes were down-regulated (Table 2). We then checked signal transduction networks induced by the knockdown of XB130 expression. The top-ranked signal network is cardiovascular disease, cell death, endocrine system disorders (Figure 4C). Among the 28 genes included in this network, 21 genes can be found in Table 2. These results indicate that the expression level of XB130 influences genes related to cellular growth and proliferation, cell cycle, cell death, and organismal survival.
Figure 4.
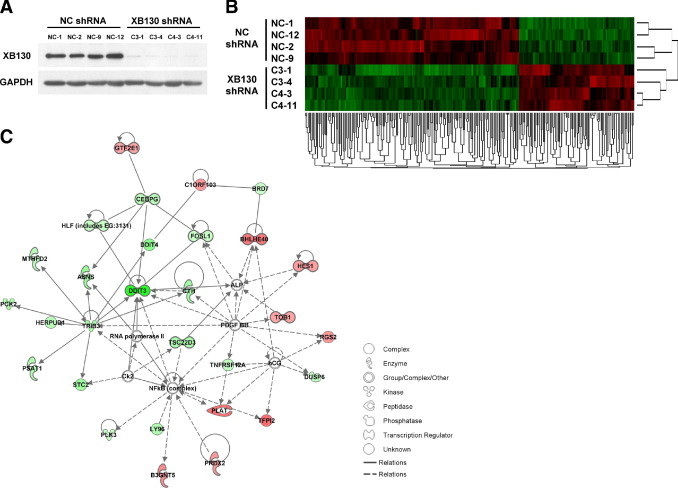
Analysis of gene expression change in XB130 shRNA transfected WRO cells. A: Four NC shRNA transfected colonies (NC-1, 2, 9, and 12) and four XB130 shRNA transfected colonies (C3-1, C3-4, C4-3, and C4-11) were used for microarray studies. XB130 protein levels were effectively reduced in XB130 shRNA transfected cells shown by Western blotting. B: Hierarchical clustering analysis demonstrates gene expression patterns. Each row corresponds to an individual sample, and each column corresponds to a particular gene. There are 246 genes that were significantly regulated. The matrix represents the expression level of an individual gene in each sample, with red and green indicating gene expression levels above or below, respectively, compared to that in the reference RNA. C: The top signaling network related to XB130 down-regulation analyzed using ingenuity pathway analysis. The network is named Cardiovascular Disease, Cell Death, Endocrine System Disorders.
Table 2.
Significantly Changed Cell Growth or Survival-Related Genes in XB130 shRNA Transfected Cells
| Up-regulated | |||||||
|---|---|---|---|---|---|---|---|
| Bio functions |
|||||||
| Gene symbol | Gene name | Cellular growth and proliferation | Cell cycle | Cell death | Organismal survival | Fold change | q-value |
| HSPA1B | Heat shock 70kDa protein 1B | ● | 3.094 | 0.016 | |||
| TFPI2 | Tissue factor pathway inhibitor 2 | ● | 2.199 | 0.061 | |||
| HSPA1A | Heat shock 70kDa protein 1A | ● | ● | ● | 2.919 | 0.016 | |
| PLAT | Plasminogen activator, tissue | ● | ● | ● | 2.145 | 0.027 | |
| BHLHE40 | Basic helix-loop-helix domain containing, class B, 2 | ● | ● | 2.051 | 0.028 | ||
| DNAJB4 | DnaJ (Hsp40) homolog, subfamily B, member 4 | ● | ● | 1.792 | 0.027 | ||
| DNAJB1 | DnaJ (Hsp40) homolog, subfamily B, member 1 | ● | ● | ● | 1.660 | 0.061 | |
| PTGES | Prostaglandin E synthase | ● | ● | ● | 1.659 | 0.016 | |
| AKAP12 | A kinase (PRKA) anchor protein 12 | ● | ● | ● | 1.639 | 0.028 | |
| EDNRA | Endothelin receptor type A | ● | 1.566 | 0.028 | |||
| KITLG | KIT ligand | ● | ● | ● | 1.557 | 0.061 | |
| DICER1 | Dicer 1, ribonuclease type III | ● | ● | ● | ● | 1.549 | 0.036 |
| RGS2 | Regulator of G-protein signaling 2, 24kDa | ● | 1.541 | 0.016 | |||
| PRDX2 | Peroxiredoxin 2 | ● | ● | 1.527 | 0.027 | ||
| SERTAD3 | SERTA domain containing 3 | ● | 1.500 | 0.028 | |||
| HES1 | Hairy and enhancer of split 1, (Drosophila) | ● | ● | ● | 1.464 | 0.061 | |
| ITGA5 | Integrin, alpha 5 (fibronectin receptor, alpha polypeptide) | ● | ● | ● | ● | 1.435 | 0.052 |
| TOB1 | Transducer of ERBB2, 1 | ● | 1.403 | 0.036 | |||
| Down-Regulated | |||||||
|---|---|---|---|---|---|---|---|
| Bio functions |
|||||||
| Gene symbol | Gene name | Cellular growth and proliferation | Cell cycle | Cell death | Organismal survival | Fold change | q-value |
| DDIT3 | DNA-damage-inducible transcript 3 | ● | ● | ● | −5.223 | <0.001 | |
| SLC7A11 | Solute carrier family 7, (cationic amino acid transporter, y+ system) member 11 | ● | ● | −3.011 | <0.001 | ||
| DDIT4 | DNA-damage-inducible transcript 4 | ● | −2.766 | <0.001 | |||
| CYR61 | Cysteine-rich, angiogenic inducer, 61 | ● | ● | ● | ● | −2.753 | <0.001 |
| AFAP1L2 | Actin filament associated protein 1-like 2 | ● | −2.749 | <0.001 | |||
| ASNS | Asparagine synthetase | ● | −2.529 | <0.001 | |||
| TIE1 | Tyrosine kinase with immunoglobulin-like and EGF-like domains 1 | ● | ● | −2.434 | 0.052 | ||
| TRIB3 | Tribbles homolog 3 (Drosophila) | ● | −2.278 | 0.036 | |||
| CTH | Cystathionase (cystathionine gamma-lyase) | ● | ● | −2.219 | 0.061 | ||
| EGR1 | Early growth response 1 | ● | ● | ● | −2.163 | 0.041 | |
| NUPR1 | Nuclear protein 1 | ● | ● | −2.081 | 0.026 | ||
| GADD45A | Growth arrest and DNA-damage-inducible, alpha | ● | ● | ● | ● | −2.045 | <0.001 |
| PPP1R15A | Protein phosphatase 1, regulatory (inhibitor) subunit 15A | ● | ● | ● | −2.039 | 0.026 | |
| TSC22D3 | TSC22 domain family, member 3 | ● | −2.009 | 0.026 | |||
| TUBE1 | Tubulin, epsilon 1 | ● | −1.941 | 0.033 | |||
| MYC | V-myc myelocytomatosis viral oncogene homolog (avian) | ● | ● | ● | ● | −1.938 | <0.001 |
| LY96 | LY96 | ● | ● | −1.886 | 0.033 | ||
| ULBP1 | UL16 binding protein 1 | ● | −1.854 | 0.026 | |||
| SLC3A2 | Solute carrier family 3 (activators of dibasic and neutral amino acid transport), member 2 | ● | −1.694 | 0.027 | |||
| SLC7A5 | Solute carrier family 7 (cationic amino acid transporter, y+ system), member 5 | ● | −1.680 | 0.052 | |||
| CEBPG | CCAAT/enhancer binding protein (C/EBP), gamma | ● | −1.664 | 0.041 | |||
| MTHFD2 | Methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 2, methenyltetrahydrofolate cyclohydrolase | ● | −1.663 | <0.001 | |||
| HERPUD1 | Homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1 | ● | −1.650 | <0.001 | |||
| DUSP6 | Dual specificity phosphatase 6 | ● | ● | −1.618 | 0.061 | ||
| TNFRSF12A | Tumor necrosis factor receptor superfamily, member 12A | ● | ● | −1.455 | 0.038 | ||
| MAFF | V-maf musculoaponeurotic fibrosarcoma oncogene homolog F (avian) | ● | ● | −1.447 | 0.041 | ||
| PLK3 | Polo-like kinase 3 (Drosophila) | ● | ● | −1.435 | 0.036 | ||
| PHGDH | Phosphoglycerate dehydrogenase | ● | −1.418 | 0.033 | |||
| LPAR3 | Lysophosphatidic acid receptor 3 | ● | −1.406 | 0.036 | |||
| EREG | Epiregulin | ● | ● | −1.400 | 0.038 | ||
| HLF | Hepatic leukemia factor | ● | −1.399 | 0.061 | |||
| HIST1H1C | Histone cluster 1, H1c | ● | ● | ● | −1.397 | <0.001 | |
| FOSL1 | FOS-like antigen 1 | ● | ● | ● | ● | −1.378 | 0.033 |
| SIAH1 | Seven in absentia homolog 1 (Drosophila) | ● | ● | ● | ● | −1.373 | 0.052 |
| HIST1H1D | Histone cluster 1, H1 days | ● | ● | −1.353 | 0.067 | ||
| DECR1 | 2,4-dienoyl CoA reductase 1, mitochondrial | ● | ● | −1.339 | 0.061 | ||
| DCLRE1A | DNA cross-link repair 1A (PSO2 homolog, S. cerevisiae) | ● | −1.331 | 0.026 | |||
| SETD8 | SET domain containing (lysine methyltransferase) 8 | ● | −1.315 | 0.067 | |||
| HIST1H1E | Histone cluster 1, H1e | ● | ● | −1.302 | 0.033 | ||
Verification of Gene Expression by Real-Time Quantitative RT-PCR
In our previous microarray studies using Affymetrix (Santa Clara, CA) GeneChips, the microarray results showed very good correlation with that of quantitative RT-PCR.26,27 In the present study, six genes (HSPA1A, BHLHE40, TOB1, DDIT3, SLC7A11, and MYC) were further tested using quantitative RT-PCR. The expression levels of HSPA1A, BHLHE40, and TOB1 mRNA were significantly increased, and DDIT3 and MYC were significantly decreased in XB130 shRNA transfected cells compared to NC shRNA transfected cell (Figure 5). The expression levels of SLC7A11 tended to be decreased in XB130 shRNA transfected cells although the difference was not statistically significant (Figure 5). These changes are in agreement with the microarray results.
Figure 5.
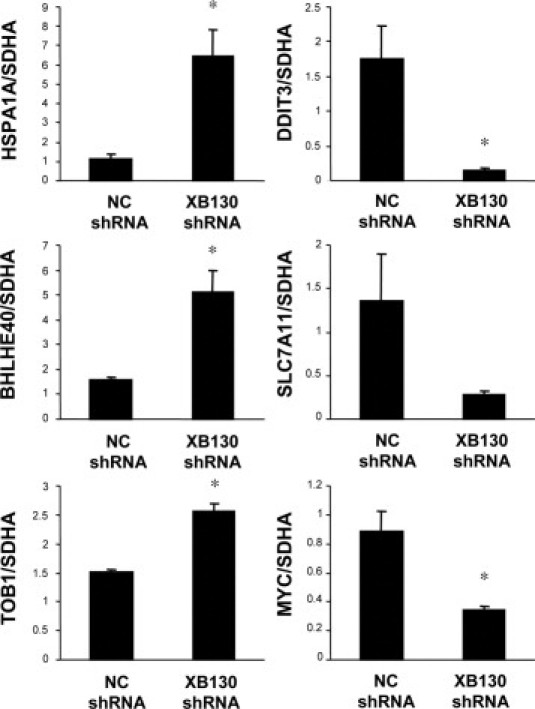
Verification of gene expression by real-time quantitative RT-PCR. The expressions of six selected genes (HSPA1A, BHLHE40, TOB1, DDIT3, SLC7A11, and MYC) in XB130 shRNA transfected cells were compared with NC shRNA transfected cells using real-time quantitative RT-PCR. Gene expression levels were normalized to the level of SDHA. n = 4. Mean ± SEM *P < 0.05 (compared with NC shRNA transfected cell).
XB130 Expression in Human Thyroid Carcinomas
To explore the relevance of our findings to the behavior of clinical thyroid cancer, we examined the expression of XB130 in a tissue microarray. XB130 protein was identified to be expressed in the cytoplasm of thyroid follicular cells in normal thyroid tissue (Figure 6A), as shown previously.10 The respective staining also confirmed the presence of XB130 in papillary as well as insular forms of thyroid carcinoma (Figure 6A). Semiquantitative analysis of cytoplasmic intensity scores demonstrated that the XB130 intensity in papillary thyroid carcinoma and anaplastic/insular carcinoma was significantly lower than that in normal and benign lesion (P < 0.001), and the intensity in the Hürthle cell carcinoma group was significantly lower than that in the normal group (P < 0.01) (Figure 6B). The reduced expression of XB130 in tumor cells may be due to the disruption of tissue structures and the loss of differentiation characteristics of thyroid cells.
Figure 6.
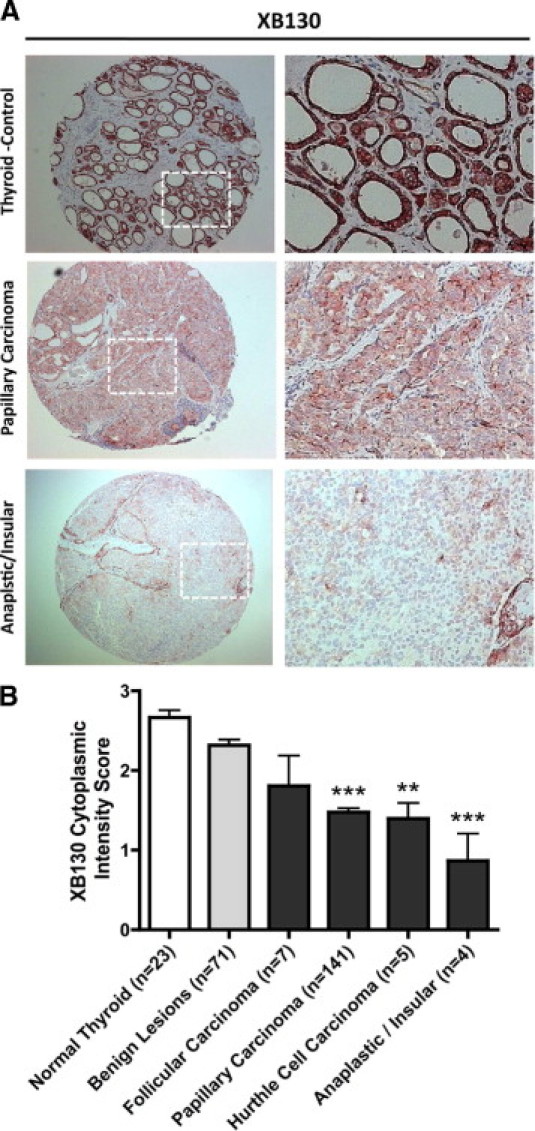
XB130 protein expression analysis with a thyroid tissue microarray. A: Immunohistochemistry staining with XB130 monoclonal antibody for normal thyroid, papillary carcinoma, and anaplastic/insular carcinoma (10×). Enlargement for selected areas as indicated is shown in the right column. Predominantly cytoplasmic staining was observed in normal thyroid tissue localized to thyroid follicular cells. B: Semiquantitative analysis of thyroid tissue microarray showed expression of XB130 in carcinoma samples at reduced levels. **P < 0.01 as compared to the normal group, ***P < 0.001 as compared to both the normal and benign lesion groups.
Discussion
The incidence of thyroid malignancy is increasing by 5% to 6% per year, and among these neoplasms, differentiated thyroid cancer, predominantly papillary thyroid carcinomas, constitute approximately 90% of the lesions.28,29 The treatment of differentiated thyroid cancer typically involves surgical resection, followed by radioactive iodine ablation and long-term thyroid hormone replacement. This protocol provides an excellent prognosis for the majority of patients with these cancers.28–30 However, recurrence develops in 20% to 40% of patients, and with the rare occurrence of distance metastasis or extensive local invasion, the prognosis is significantly poorer.28,31 Understanding the specific mechanisms involved in the deregulation of cell growth and cell death in thyroid cancer is critical for developing new treatments.
In a recent study, we demonstrated that XB130 is expressed in several thyroid cancer cell lines and that it is a substrate and binding partner of RET/PTC, an oncogenic protein tyrosine kinase found in thyroid carcinoma.10 Down-regulation of XB130 expression in the TPC1 cell line derived from papillary thyroid carcinoma that carries the RET/PTC-1 rearrangement reduced proliferation and promoted anoikis.10 The WRO cell line used in the present study also carries a RET/PTC-1 rearrangement (data not shown). Using the in vivo xenograft model we demonstrated that constitutive down-regulation of XB130 reduces growth of tumors derived from WRO cells. Furthermore, we performed XB130 immunohistochemistry staining on human thyroid tumors and confirmed expression of XB130 in multiple types of thyroid cancers. Although XB130 appears to be reduced (not increased, as might be expected) in tumors, and the reduced expression seems to parallel loss of tumor differentiation (Figure 6), it is found in follicular and papillary thyroid carcinoma with recognizable expression and may contribute to the tumorigenesis in these tumors. Taken together, these results suggest that XB130 is not only involved in signal transduction of cultured cells but also plays a crucial role in the growth of thyroid cancer cells that carry the expression of XB130.
In addition to TPC-1 and WRO thyroid cancer cells, we observed similar effects of XB130 down-regulation on the proliferation and survival of A549 cells, a human lung carcinoma cell line (data not shown). It is interesting that a study by Emaduddin and colleagues32 investigating the implications of Src tyrosine kinases in certain colorectal cancers distinguished XB130 from SW629 colorectal cancer cells as being one of the tyrosine phosphorylated proteins binding to the Lck-SH2 domain. Lck is an Src family member that is not detectable in normal colonic epithelium but becomes aberrantly expressed in a subset of colorectal carcinomas. Furthermore, Cunha and colleagues performed gene expression profiling using 102 soft tissue tumor samples and found XB130 to be one of the genes strongly related to local aggressiveness.33 Therefore, in addition to being a factor in thyroid cancer, XB130 may also play important roles in other neoplasms.
RNA interference (RNAi), especially siRNA, has become a powerful tool for investigating gene functions. There is rapidly growing interest in extending its application to in vivo systems, such as in animal models of disease and in human therapeutics.34,35 Knockdown of XB130 with siRNA transiently inhibited tumor cell proliferation and promoted spontaneous as well as inducing cell death. We used shRNA to develop stably transfected cell lines, in which XB130 expression was persistently down-regulated. Using an in vivo xenograft model, we found not only impedance of tumor growth but also evidence showing that both proliferation and survival are reduced in the formed tumors. RNAi-mediated suppression of XB130 could be a specific gene-targeting therapy that would suppress the progression of thyroid cancers that expresses XB130.
Oncoproteins may enhance tumor growth by promoting cell proliferation and/or inhibiting cell death. To determine how down-regulation of XB130 can affect both cell proliferation and survival, we used a bioinformatics approach to analyze the genome-wide consequence of XB130 knockdown. Indeed, microarray analysis data showed that among the 246 significantly changed genes, 57 genes have cell proliferation or survival related functions (Table 2). Two thirds of these genes are down-regulated in XB130 shRNA stably transfected cells, suggesting that XB130-related signaling may help to maintain the expression of these genes. Moreover, many of these genes are well connected in the top signaling network (Figure 4C), indicating that these genes are not only functionally related but are also regulated together at expression levels by XB130-related signal transduction pathways. The top function of XB130-related genes is connected to cancer, and the top molecular and cellular functions of these genes are related to cell proliferation and survival.
We noted that four transcription regulators (GTF2E1, BHLHE40, HES1, TOB1) are up-regulated in the signal network (Figure 4C). Both BHLHE40 and HES1 belong to the basic helix-loop-helix family of transcription factors. HES1 is a transcriptional repressor of genes that requires a bHLH protein for their transcription.36 TOB1 is a member of the tob/btg1 family of anti-proliferative proteins.37 Up-regulation of TOB1 may contribute to the reduced proliferation of WRO cells. Many down-regulated genes are directly connected in the top signal network. Among them are a group of transcription regulators, including DDIT3, CEBPG, FOSL1, HLF, and TSC22D3. The most down-regulated gene is DDIT3, which encodes a protein named DNA damage-inducible transcript 3, appears to be a hub of multiple down-regulated genes. It has been shown that DDIT3 can interact with multiple proteins, especially transcription factors, such as JunD, C-jun and C-Fos, ATF3, and CEBPB.38 We have found that XB130 specifically mediates AP-1 and SRE transcription activation when co-expressed with c-Src.9 Down-regulation of transcription factors such as DDIT3, CEBPB, and FOSL1 may impact cell proliferation and survival. Knocking down XB130 also reduced the gene expression of MYC; c-Myc-encoded protein functions in cell proliferation, differentiation, and neoplastic disease. Amplification of the c-Myc gene has been found in several types of human tumors, including lung, breast, and colon carcinomas.39,40
In summary, we identified the role of XB130 in tumor growth by promoting cell proliferation and survival in thyroid cancer cells. Microarray data suggest that XB130 has profound effects on the expression of genes related to cell proliferation and survival. These results suggest that XB130 could be a novel oncoprotein in thyroid cancers. Similar roles may also be present in other cancer cells. A deeper understanding of these mechanisms may lead to the discovery that XB130 is an important mediator in tumor development and may be a novel therapeutic target for cancer.
Acknowledgments
We are grateful to Dr. Bing Han, Mr. Matthew Rubacha, and Ms. Roli Bawa for technical assistance.
Footnotes
Supported by operating grants (MOP-13270 and MOP-42546) from the Canadian Institutes of Health Research and by Research Fellowship Awards from Uehara Memorial Foundation and International Society of Heart and Lung Transplantation (A.S.).
Supplemental material for this article can be found at http://ajp.amjpathol.org or at doi:10.1016/j.ajpath.2010.11.024.
Supplementary data
Establishment of XB130 shRNA stably transfected WRO cell lines. (A) shRNA plasmids for XB130 stable transfection. (B) The C3 or C4 shRNA transfection was more effective to reduce the expression of XB130. shRNA transfected WRO cells were selected with 0.5 mg/ml G418, and the expression levels of XB130 protein from antibiotic-resistant pools were determined by western blotting.
Principle Component Analysis (PCA) and Significance Analysis of Microarray (SAM). (A) Principle Component Analysis (PCA) was performed using Partek software. PCA results indicated a significant separation of samples based on the expression of XB130. (B) Differential expression analysis was performed using Significance Analysis of Microarray (SAM). SAM analysis showed that 246 genes were significantly changed by the stable knock down of XB130 in WRO cells with criteria Fold Discovery Rate (FDR) q value < 6.66% and fold change > 1.3.
Top Bio Functions and Networks of XB130 analyzed by Ingenuity Pathway analysis
References
- 1.Flynn D.C. Adaptor proteins. Oncogene. 2001;20:6270–6272. doi: 10.1038/sj.onc.1204769. [DOI] [PubMed] [Google Scholar]
- 2.Csiszar A. Structural and functional diversity of adaptor proteins involved in tyrosine kinase signaling. Bioessays. 2006;28:465–479. doi: 10.1002/bies.20411. [DOI] [PubMed] [Google Scholar]
- 3.Dorfleutner A., Stehlik C., Zhang J., Gallick G.E., Flynn D.C. AFAP-110 is required for actin stress fiber formation and cell adhesion in MDA-MB-231 breast cancer cells. J Cell Physiol. 2007;213:740–749. doi: 10.1002/jcp.21143. [DOI] [PubMed] [Google Scholar]
- 4.Zhang J., Park S.I., Artime M.C., Summy J.M., Shah A.N., Bomser J.A., Dorfleutner A., Flynn D.C., Gallick G.E. AFAP-110 is overexpressed in prostate cancer and contributes to tumorigenic growth by regulating focal contacts. J Clin Invest. 2007;117:2962–2973. doi: 10.1172/JCI30710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Seals D.F., Azucena E.F., Jr, Pass I., Tesfay L., Gordon R., Woodrow M., Resau J.H., Courtneidge S.A. The adaptor protein Tks5/Fish is required for podosome formation and function, and for the protease-driven invasion of cancer cells. Cancer Cell. 2005;7:155–165. doi: 10.1016/j.ccr.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 6.Blouw B., Seals D.F., Pass I., Diaz B., Courtneidge S.A. A role for the podosome/invadopodia scaffold protein Tks5 in tumor growth in vivo. Eur J Cell Biol. 2008;87:555–567. doi: 10.1016/j.ejcb.2008.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lodyga M., Bai X.H., Mourgeon E., Han B., Keshavjee S., Liu M. Molecular cloning of actin filament-associated protein: a putative adaptor in stretch-induced Src activation. Am J Physiol Lung Cell Mol Physiol. 2002;283:L265–L274. doi: 10.1152/ajplung.00492.2001. [DOI] [PubMed] [Google Scholar]
- 8.Han B., Bai X.H., Lodyga M., Xu J., Yang B.B., Keshavjee S., Post M., Liu M. Conversion of mechanical force into biochemical signaling. J Biol Chem. 2004;279:54793–54801. doi: 10.1074/jbc.M406880200. [DOI] [PubMed] [Google Scholar]
- 9.Xu J., Bai X.H., Lodyga M., Han B., Xiao H., Keshavjee S., Hu J., Zhang H., Yang B.B., Liu M. XB130, a novel adaptor protein for signal transduction. J Biol Chem. 2007;282:16401–16412. doi: 10.1074/jbc.M701684200. [DOI] [PubMed] [Google Scholar]
- 10.Lodyga M., De Falco V., Bai X.H., Kapus A., Melillo R.M., Santoro M., Liu M. XB130, a tissue-specific adaptor protein that couples the RET/PTC oncogenic kinase to PI 3-kinase pathway. Oncogene. 2009;28:937–949. doi: 10.1038/onc.2008.447. [DOI] [PubMed] [Google Scholar]
- 11.Grieco M., Santoro M., Berlingieri M.T., Melillo R.M., Donghi R., Bongarzone I., Pierotti M.A., Della Porta G., Fusco A., Vecchio G. PTC is a novel rearranged form of the ret proto-oncogene and is frequently detected in vivo in human thyroid papillary carcinomas. Cell. 1990;60:557–563. doi: 10.1016/0092-8674(90)90659-3. [DOI] [PubMed] [Google Scholar]
- 12.Kondo T., Ezzat S., Asa S.L. Pathogenetic mechanisms in thyroid follicular-cell neoplasia. Nat Rev Cancer. 2006;6:292–306. doi: 10.1038/nrc1836. [DOI] [PubMed] [Google Scholar]
- 13.Lodyga M., Bai X.H., Kapus A., Liu M. Novel adaptor protein XB130 is a Rac-controlled component of lamellipodia, which regulates cell motility and invasion. J Cell Sci. 2010 doi: 10.1242/jcs.071050. In press. [DOI] [PubMed] [Google Scholar]
- 14.Mura M., Han B., Andrade C.F., Seth R., Hwang D., Waddell T.K., Keshavjee S., Liu M. The early responses of VEGF and its receptors during acute lung injury: implication of VEGF in alveolar epithelial cell survival. Crit Care. 2006;10:R130. doi: 10.1186/cc5042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Noguchi P.D., Johnson J.B., Browne W. Measurement of DNA synthesis by flow cytometry. Cytometry. 1981;1:390–393. doi: 10.1002/cyto.990010607. [DOI] [PubMed] [Google Scholar]
- 16.Takei Y., Kadomatsu K., Yuzawa Y., Matsuo S., Muramatsu T. A small interfering RNA targeting vascular endothelial growth factor as cancer therapeutics. Cancer Res. 2004;64:3365–3370. doi: 10.1158/0008-5472.CAN-03-2682. [DOI] [PubMed] [Google Scholar]
- 17.Nozawa H., Tadakuma T., Ono T., Sato M., Hiroi S., Masumoto K., Sato Y. Small interfering RNA targeting epidermal growth factor receptor enhances chemosensitivity to cisplatin, 5-fluorouracil and docetaxel in head and neck squamous cell carcinoma. Cancer Sci. 2006;97:1115–1124. doi: 10.1111/j.1349-7006.2006.00287.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Briese J, Cheng S, Ezzat S, Liu W, Winer D, Wagener C, Bamberger AM, Asa SL: Osteopontin (OPN) expression in thyroid carcinoma. Anticancer Res 30:1681–1688 [PubMed]
- 19.Liu W., Wei W., Winer D., Bamberger A.M., Bamberger C., Wagener C., Ezzat S., Asa S.L. CEACAM1 impedes thyroid cancer growth but promotes invasiveness: a putative mechanism for early metastases. Oncogene. 2007;26:2747–2758. doi: 10.1038/sj.onc.1210077. [DOI] [PubMed] [Google Scholar]
- 20.Mura M., Andrade C.F., Han B., Seth R., Zhang Y., Bai X.H., Waddell T.K., Hwang D., Keshavjee S., Liu M. Intestinal ischemia-reperfusion-induced acute lung injury and oncotic cell death in multiple organs. Shock. 2007;28:227–238. doi: 10.1097/01.shk.0000278497.47041.e3. [DOI] [PubMed] [Google Scholar]
- 21.Tusher V.G., Tibshirani R., Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaneda H., Waddell T.K., de Perrot M., Bai X.H., Gutierrez C., Arenovich T., Chaparro C., Liu M., Keshavjee S. Pre-implantation multiple cytokine mRNA expression analysis of donor lung grafts predicts survival after lung transplantation in humans. Am J Transplant. 2006;6:544–551. doi: 10.1111/j.1600-6143.2005.01204.x. [DOI] [PubMed] [Google Scholar]
- 23.Tang P.S., Mura M., Seth R., Liu M. Acute lung injury and cell death: how many ways can cells die? Am J Physiol Lung Cell Mol Physiol. 2008;294:L632–L641. doi: 10.1152/ajplung.00262.2007. [DOI] [PubMed] [Google Scholar]
- 24.Liu W., Asa S.L., Ezzat S. 1alpha,25-Dihydroxyvitamin D3 targets PTEN-dependent fibronectin expression to restore thyroid cancer cell adhesiveness. Mol Endocrinol. 2005;19:2349–2357. doi: 10.1210/me.2005-0117. [DOI] [PubMed] [Google Scholar]
- 25.Liu W., Cheng S., Asa S.L., Ezzat S. The melanoma-associated antigen A3 mediates fibronectin-controlled cancer progression and metastasis. Cancer Res. 2008;68:8104–8112. doi: 10.1158/0008-5472.CAN-08-2132. [DOI] [PubMed] [Google Scholar]
- 26.dos Santos C.C., Han B., Andrade C.F., Bai X., Uhlig S., Hubmayr R., Tsang M., Lodyga M., Keshavjee S., Slutsky A.S., Liu M. DNA microarray analysis of gene expression in alveolar epithelial cells in response to TNFalpha, LPS, and cyclic stretch. Physiol Genomics. 2004;19:331–342. doi: 10.1152/physiolgenomics.00153.2004. [DOI] [PubMed] [Google Scholar]
- 27.dos Santos C.C., Okutani D., Hu P., Han B., Crimi E., He X., Keshavjee S., Greenwood C., Slutsky A.S., Zhang H., Liu M. Differential gene profiling in acute lung injury identifies injury-specific gene expression. Crit Care Med. 2008;36:855–865. doi: 10.1097/CCM.0B013E3181659333. [DOI] [PubMed] [Google Scholar]
- 28.Mazzaferri E.L., Jhiang S.M. Long-term impact of initial surgical and medical therapy on papillary and follicular thyroid cancer. Am J Med. 1994;97:418–428. doi: 10.1016/0002-9343(94)90321-2. [DOI] [PubMed] [Google Scholar]
- 29.Sherman S.I., Brierley J.D., Sperling M., Ain K.B., Bigos S.T., Cooper D.S., Haugen B.R., Ho M., Klein I., Ladenson P.W., Robbins J., Ross D.S., Specker B., Taylor T., Maxon H.R., 3rd Prospective multicenter study of thyroid carcinoma treatment: initial analysis of staging and outcome. National Thyroid Cancer Treatment Cooperative Study Registry Group. Cancer. 1998;83:1012–1021. doi: 10.1002/(sici)1097-0142(19980901)83:5<1012::aid-cncr28>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 30.Cooper D.S., Doherty G.M., Haugen B.R., Kloos R.T., Lee S.L., Mandel S.J., Mazzaferri E.L., McIver B., Sherman S.I., Tuttle R.M. Management guidelines for patients with thyroid nodules and differentiated thyroid cancer. Thyroid. 2006;16:109–142. doi: 10.1089/thy.2006.16.109. [DOI] [PubMed] [Google Scholar]
- 31.Durante C., Haddy N., Baudin E., Leboulleux S., Hartl D., Travagli J.P., Caillou B., Ricard M., Lumbroso J.D., De Vathaire F., Schlumberger M. Long-term outcome of 444 patients with distant metastases from papillary and follicular thyroid carcinoma: benefits and limits of radioiodine therapy. J Clin Endocrinol Metab. 2006;91:2892–2899. doi: 10.1210/jc.2005-2838. [DOI] [PubMed] [Google Scholar]
- 32.Emaduddin M., Edelmann M.J., Kessler B.M., Feller S.M. Odin (ANKS1A) is a Src family kinase target in colorectal cancer cells. Cell Commun Signal. 2008;6:7. doi: 10.1186/1478-811X-6-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cunha IW, Carvalho KC, Martins WK, Marques SM, Muto NH, Falzoni R, Rocha RM, Aguiar S, Simoes AC, Fahham L, Neves EJ, Soares FA, Reis LF: Identification of genes associated with local aggressiveness and metastatic behavior in soft tissue tumors. Transl Oncol 3:23–32 [DOI] [PMC free article] [PubMed]
- 34.Behlke M.A. Progress towards in vivo use of siRNAs. Mol Ther. 2006;13:644–670. doi: 10.1016/j.ymthe.2006.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Akhtar S., Benter I.F. Nonviral delivery of synthetic siRNAs in vivo. J Clin Invest. 2007;117:3623–3632. doi: 10.1172/JCI33494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takata T., Ishikawa F. Human Sir2-related protein SIRT1 associates with the bHLH repressors HES1 and HEY2 and is involved in HES1- and HEY2-mediated transcriptional repression. Biochem Biophys Res Commun. 2003;301:250–257. doi: 10.1016/s0006-291x(02)03020-6. [DOI] [PubMed] [Google Scholar]
- 37.Matsuda S., Kawamura-Tsuzuku J., Ohsugi M., Yoshida M., Emi M., Nakamura Y., Onda M., Yoshida Y., Nishiyama A., Yamamoto T. Tob, a novel protein that interacts with p185erbB2, is associated with anti-proliferative activity. Oncogene. 1996;12:705–713. [PubMed] [Google Scholar]
- 38.Ubeda M., Vallejo M., Habener J.F. CHOP enhancement of gene transcription by interactions with Jun/Fos AP-1 complex proteins. Mol Cell Biol. 1999;19:7589–7599. doi: 10.1128/mcb.19.11.7589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alitalo K., Schwab M., Lin C.C., Varmus H.E., Bishop J.M. Homogeneously staining chromosomal regions contain amplified copies of an abundantly expressed cellular oncogene (c-myc) in malignant neuroendocrine cells from a human colon carcinoma. Proc Natl Acad Sci USA. 1983;80:1707–1711. doi: 10.1073/pnas.80.6.1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nau M.M., Brooks B.J., Battey J., Sausville E., Gazdar A.F., Kirsch I.R., McBride O.W., Bertness V., Hollis G.F., Minna J.D. L-myc, a new myc-related gene amplified and expressed in human small cell lung cancer. Nature. 1985;318:69–73. doi: 10.1038/318069a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Establishment of XB130 shRNA stably transfected WRO cell lines. (A) shRNA plasmids for XB130 stable transfection. (B) The C3 or C4 shRNA transfection was more effective to reduce the expression of XB130. shRNA transfected WRO cells were selected with 0.5 mg/ml G418, and the expression levels of XB130 protein from antibiotic-resistant pools were determined by western blotting.
Principle Component Analysis (PCA) and Significance Analysis of Microarray (SAM). (A) Principle Component Analysis (PCA) was performed using Partek software. PCA results indicated a significant separation of samples based on the expression of XB130. (B) Differential expression analysis was performed using Significance Analysis of Microarray (SAM). SAM analysis showed that 246 genes were significantly changed by the stable knock down of XB130 in WRO cells with criteria Fold Discovery Rate (FDR) q value < 6.66% and fold change > 1.3.
Top Bio Functions and Networks of XB130 analyzed by Ingenuity Pathway analysis


