Abstract
Background
Enterotoxigenic Escherichia coli (ETEC) infections result in large economic losses in the swine industry worldwide. ETEC infections cause pro-inflammatory responses in intestinal epithelial cells and subsequent diarrhea in pigs, leading to reduced growth rate and mortality. Administration of probiotics as feed additives displayed health benefits against intestinal infections. Saccharomyces cerevisiae (Sc) is non-commensal and non-pathogenic yeast used as probiotic in gastrointestinal diseases. However, the immuno-modulatory effects of Sc in differentiated porcine intestinal epithelial cells exposed to ETEC were not investigated.
Methodology/Principal Findings
We reported that the yeast Sc (strain CNCM I-3856) modulates transcript and protein expressions involved in inflammation, recruitment and activation of immune cells in differentiated porcine intestinal epithelial IPEC-1 cells. We demonstrated that viable Sc inhibits the ETEC-induced expression of pro-inflammatory transcripts (IL-6, IL-8, CCL20, CXCL2, CXCL10) and proteins (IL-6, IL-8). This inhibition was associated to a decrease of ERK1/2 and p38 MAPK phosphorylation, an agglutination of ETEC by Sc and an increase of the anti-inflammatory PPAR-γ nuclear receptor mRNA level. In addition, Sc up-regulates the mRNA levels of both IL-12p35 and CCL25. However, measurement of transepithelial electrical resistance displayed that Sc failed to maintain the barrier integrity in monolayer exposed to ETEC suggesting that Sc does not inhibit ETEC enterotoxin activity.
Conclusions
Sc (strain CNCM I-3856) displays multiple immuno-modulatory effects at the molecular level in IPEC-1 cells suggesting that Sc may influence intestinal inflammatory reaction.
Introduction
Enterotoxigenic Escherichia coli (ETEC) is a major cause of intestinal infection in piglets inducing diarrhea, reduced growth rate and mortality leading to economic loss [1]. Pathogeny of ETEC is characterized by its adhesion to the intestinal epithelial cells (IEC) through adhesins which interact with their specific receptors localized on the brush border membrane [2], [3], [4]. Following jejunal and ileal mucosa colonization, ETEC strains secrete several enterotoxins, including the heat-labile enterotoxin (LT), the heat-stable enterotoxin (STa and/or STb), and the enteroaggregative E. coli heat-stable enterotoxin 1 (EAST1) [5], [6]. These enterotoxins cause perturbation of hydroelectrolytic secretions in the small intestine resulting in diarrhea [5].
ETEC strains expressing the F4 fimbriae are involved in neonatal and post-weaning diarrhea [1]. ETEC F4+ infections mainly occur during the first week after weaning in piglets expressing the F4 receptor on the intestinal brush border [7]. The weaning-related stress, the dietary changes and the immaturity of the immune system are several factors contributing to the disease severity [1]. ETEC F4+ strains represent the most prevalent form of bacterial infection in piglets [1], [8] and an increase in incidence of ETEC-associated diarrhea was observed worldwide [1]. Furthermore, antibiotic growth promoters were prohibited in the European Union since 2006 (IP/05/1687) and antibiotic-multiresistant ETEC isolates have been identified [9], [10], [11]. Consequently, new prophylactic and/or therapeutic strategies should be developed to protect piglets from ETEC infection. The interest in using probiotic microorganisms such as live yeasts to prevent gastrointestinal diseases in farm animals has increased significantly in the last decade worldwide. Saccharomyces cerevisiae variety boulardii (Sb) has been shown to provide intestinal protection against various enteric pathogens [12]. Indeed, Sb protected the host through multiple mechanisms such as inhibition of pathogen adhesion [13], neutralization of bacterial virulence factors [14], maintenance of epithelial barrier integrity [15], decrease of pathogen-associated inflammation [16] and stimulation of the immune system [17]. Regarding Sb effects on pathogen-associated inflammation, this yeast has been shown to modulate pro-inflammatory signaling pathways leading to the inhibition of mitogen-activated protein kinases (MAPK) and nuclear factor NF-κB activition in IEC [16], [18]. Because IEC play a key role in regulating innate and adaptive immune responses of the gut [19], several studies have evaluated yeast probiotic effects on these cells [15], [16], [18], [20], [21], [22], [23]. Epithelial cells protect the intestine through different mechanisms such as barrier function, mucus secretion, antibacterial peptide synthesis, cytokine and chemokine secretions [19]. IEC detect pathogen-associated molecular patterns (PAMPs) through their pathogen recognition receptors (PRR) and then secrete cytokines and chemokines that activate pro-inflammatory signaling pathways and direct the migration of various effector cells involved in innate and adaptive immunity [24]. However, inflammatory responses induced by enteric pathogens can lead to dysregulation of IEC signaling, disruption of membrane barrier integrity, enhancement of pathogen translocation and disease [25]. With their pivotal role in the gut homeostasis, IEC are particularly relevant to assess yeast immuno-regulatory effects.
Saccharomyces cerevisiae (Sc) is non-commensal and non-pathogenic yeast used in food industry as brewer and baker's yeast. Sc and Sb are members of the same yeast species [26] but they present some genetical, metabolical and physiological differences [27], [28]. Sc (strain CNCM I-3856) is a probiotic yeast studied for its beneficial effects on animal growth, host immune function and inhibition of Salmonella spp. adhesion [29], [30]. Furthermore, Sc (strain CNCM I-3856) has been shown to decrease inflammation in a mouse model of chemically-induced colitis [31], to reduce digestive discomfort and abdominal pain in IBS patients [32] and to exert in vitro antagonist effect against E. coli O157:H7 [33]. In the current study, we use an in vitro model of differentiated porcine intestinal epithelial IPEC-1 cells co-cultured with Sc (strain CNCM I-3856) and F4+ ETEC (strain GIS26). IPEC-1 cells provide a relevant model since F4+ ETEC has been shown to bind IPEC-1 cells which express cytokines and chemokines after ETEC stimulation [34], [35]. In addition, the ETEC strain GIS26 has been shown to infect newly weaned piglets (Verdonck et al. 2002). Few data are available regarding yeast immuno-modulatory effects in porcine IEC exposed to ETEC. These data showed that Sc and Sb inhibited IL-1α transcript expression in non-differentiated IPEC-J2 cell line [36]. Consequently, using differentiated IPEC-1 cells, we investigated in this report whether Sc modulates gene expressions and signaling pathways involved in inflammation, recruitment and activation of immune cells. Then, we assessed whether Sc could prevent the disruption of the membrane barrier integrity induced by ETEC.
Results
Saccharomyces cerevisiae (strain CNCM I-3856) and ETEC (strain GIS26) modulate differently immune gene expression in differentiated IPEC-1 cells
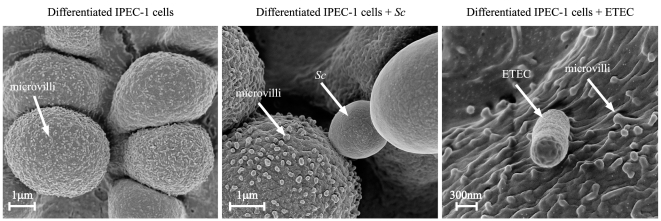
In this work, we aimed to compare the effects of Sc and ETEC on IPEC-1 gene expressions involved in inflammation, innate and adaptive immunity. As illustrated by scanning electron microscopy, differentiated IPEC-1 cells displayed microvilli and both Sc and ETEC interact with these cells (Figure 1). Characterization of Sc and ETEC immuno-modulatory effects was first assessed by analysis of transcript expressions. The analysed genes are presented in Table 1. ETEC increased significantly the mRNA expression (p<0.01) of the pro-inflammatory cytokines TNF-α (x 3075.6), IL1-α (x 46.9), IL-6 (x 7.8), the pro-Th2 cytokine IL-5 (x 2.6), and the chemokines CCL20 (x 5726.1), CXCL2 (x 857.4), IL-8 (x 670.2), CXCL10 (x 7.1) and CXCL12 (x 3) (Table 2). In contrast, Sc did not significantly up-regulate these transcripts but increased the expression (p<0.01) of the pro-Th1 cytokine IL-12p35 (x 10.7), the chemokine CCL25 (x 2.7), the anti-inflammatory nuclear receptor PPAR-γ (x 2.6), the mucine MUC1 (x 2.21) and decreased the mRNA expression of the pro-Th2 cytokine BAFF (÷ 2.5, p<0.01) (Table 2). Thus, this result shows that Sc and ETEC display different modulatory effects on transcripts involved in both inflammatory and immune responses.
Figure 1. Interaction of Saccharomyces cerevisiae or ETEC with differentiated IPEC-1 cells.
Differentiated IPEC-1 cells were cultured overnight with Sc (3×106 yeasts/well) or exposed to ETEC (3×107 CFU/well) for 45 min. IPEC-1 cells were then assessed to scanning electron microscopy. (A) Differentiated IPEC-1 cells expressed microvilli on their surface, (B) Sc interacts with differentiated IPEC-1 cells, (C) ETEC interacts with differentiated IPEC-1 cells.
Table 1. Primer sequences, annealing temperatures of primer sets (°C), expected PCR fragment sizes (bp) and accession numbers or references.
| Primer name | Primer sequence | Annealing temperature(s) (°C) | PCR product (bp) | Accession number or reference |
| APRIL/TNFSF13 | S: TGCTCACCCGTAAACAGAAGAS: TAAACTCCAGCATCCCAGAC | 60 | 172 | Meurens et al., 2009 |
| BAFF/TNFSF13B | S: GAGAGCAGCTCCATTCAAAGAS: GCATGCCACTGTCTGCAATC | 60 | 103 | Meurens et al., 2009 |
| CCL2/MCP-1 | S: GTCACCAGCAGCAAGTGTCAS: CCAGGTGGCTTATGGAGTC | 60 | 112 | Meurens et al., 2009 |
| CCL17/TARC | S: TGCTGCTCCTGGTTGCTCTCAS: ATGGCGTCCCTGGTACACTC | 67 | 169 | Bruel et al., 2009 |
| CCL20/MIP3 alpha | S: GCTCCTGGCTGCTTTGATGTCAS: CATTGGCGAGCTGCTGTGTG | 66 | 146 | Meurens et al., 2009 |
| CCL22/MDC | S: GTCCTCCTTGCTGTGATACAS: CTCGGTCCCTCAAGGTTAG | 60 | 184 | DB798783 |
| CCL25/TECK | S: ACCTGCCTGCTGTGATATTCAS: TCCGATTGTCCAGGATCTTC | 62 | 105 | NM_001025214 |
| Cdx-1 | S: ACAGCCGCTATATCACCATCAS: GTTCACTTTGCGCTCCTTTG | 60 | 116 | ENSFM00600000921619 http://ensembl.org |
| Cdx-2 | S: CAGTCGCTACATCACCATTCAS: GCTGTTGCTGCAACTTCTTC | 60 | 137 | GU_17420 |
| CXCL2/GRO beta | S: TGCTGCTCCTGCTTCTAGTGAS: TGGCTATGACTTCCGTTTGG | 60 | 171 | Meurens et al., 2009 |
| CXCL10/IP-10 | S: CCCACATGTTGAGATCATTGC AS: CATCCTTATCAGTAGTGCCG | 60 | 168 | Meurens et al., 2009 |
| CXCL12/SDF-1 | S: TGCCTCAGCGATGAGAAACAS: GGGTCAATGCACACTTGTC | 58 | 173 | AY312066 |
| HMBS2 | S: AGGATGGGCAACTCTACCTGAS: GATGGTGGCCTGCATAGTCT | 58 | 83 | Nygard et al., 2007 |
| HPRT-1 | S: GGACTTGAATCATGTTTGTGAS: CAGATGTTTCCAAACTCAAC | 60 | 91 | Nygard et al., 2007 |
| IFN gamma | A: GCTCTGGGAAACTGAATGACAS: TCTCTGGCCTTGGAACATAG | 60 | 167 | Meurens et al., 2009 |
| IL-1 alpha/LAF | S: CCCGTCAGGTCAATACCTCAS: GCAACACGGGTTCGTCTTC | 60 | 170 | NM_214029 |
| IL-4/BCGF | S: CAACCCTGGTCTGCTTACTGAS: CTTCTCCGTCGTGTTCTCTG | 65 | 173 | Meurens et al., 2009 |
| IL-5/EDF | S: TGGAGCTGCCTACGTTAGTGAS: TCGCCTATCAGCAGAGTTCG | 64 | 105 | Meurens et al., 2009 |
| IL-6/IFN beta 2 | S: ATCAGGAGACCTGCTTGATGAS: TGGTGGCTTTGTCTGGATTC | 62 | 177 | Meurens et al., 2009 |
| IL-8/CXCL-8 | S: TCCTGCTTTCTGCAGCTCTCAS: GGGTGGAAAGGTGTGGAATG | 62 | 100 | Meurens et al., 2009 |
| IL-10/B-TCGF | S: GGTTGCCAAGCCTTGTCAGAS: AGGCACTCTTCACCTCCTC | 60 | 202 | NM_214041 |
| IL-12p35 | S: GGCCTGCTTACCACTTGAACAS: GCATTCATGGCCTGGAACTC | 64 | 180 | Meurens et al., 2009 |
| IL-13 | A: TGGCGCTCTGGTTGACTCTGAS: CCATGCTGCCGTTGCATAGG | 67 | 159 | Meurens et al., 2009 |
| IL-17A/CTLA-8 | A: CCAGACGGCCCTCAGATTACAS: CACTTGGCCTCCCAGATCAC | 66 | 103 | Meurens et al., 2009 |
| IL-23p19 | S: CTCCTTCTCCGCCTCAAGATCC AS:TTGCTGCTCCATGGGCGAAGAC | 70 | 82 | Meurens et al., 2009 |
| IL-33 | S: AGCTTCGCTCTGGCCTTATCAS: GCTGACAGGCAGCAAGTACC | 63 | 126 | Meurens et al., 2009 |
| MUC1 | S: TAAAGAAGACGGGCTTCTGGAS: CCGCTTTAAGCCGATCAAAC | 60 | 134 | XM_001926883 |
| MUC2 | S: ACCCGCACTACGTCACCTTCAS: GGCAGGACACCTGGTCATTG | 62 | 150 | Bruel et al., 2009 |
| MUC4 | S: CTGCTCTTGGGCACTATATGAS: CCTGTGACTGCAGAATCAAC | 60 | 133 | DQ848681 |
| PBD-1 | S: ACCGCCTCCTCCTTGTATTCAS: CACAGGTGCCGATCTGTTTC | 62 | 150 | Meurens et al., 2009 |
| PBD-2 | S: TTGCTGCTGCTGACTGTCTGAS: CTTGGCCTTGCCACTGTAAC | 62 | 180 | Meurens et al., 2009 |
| PPAR gamma | S: AAGACGGGGTCCTCATCTCCAS: CGCCAGGTCGCTGTCATCT | 62 | 149 | Bassaganya-Riera et al., 2006 |
| RPL-19 | S: AACTCCCGTCAGCAGATCCAS: AGTACCCTTCCGCTTACCG | 60 | 147 | Meurens et al., 2009 |
| Secretory component | S: ACTGGTGTCGCTGGGAAGAGAS: GACCGTGAAGGTGCCATTGC | 64 | 131 | CJ025705 |
| TGF beta | S: GAAGCGCATCGAGGCCATTCAS: GGCTCCGGTTCGACACTTTC | 64 | 162 | Meurens et al., 2009 |
| TNF alpha/TNFSF2 | S: CCAATGGCAGAGTGGGTATGAS: TGAAGAGGACCTGGGAGTAG | 62 | 116 | Meurens et al., 2009 |
| TSLP | S: AGGGCTTGTGCTAACCTACAS: ATCCGGCCTATCATCACAG | 58 | 164 | Meurens et al., 2009 |
Reference genes are underlined.
Table 2. Effects of Saccharomyces cerevisiae or ETEC on transcript expressions in IPEC-1 cells (fold changes in comparison to controls).
| Transcripts | Expression level | S. cerevisiae | ETEC |
| APRIL | moderate | 0.60 | 1.13 |
| BAFF | low | 0.40 ** | 0.74 |
| CCL2 | moderate | 1.32 | 6.83 |
| CCL17 | not detected | - | - |
| CCL20 | moderate | 3.61 | 5726.08 ** |
| CCL22 | not detected | - | - |
| CCL25 | moderate | 2.73** | 1.11 |
| CXCL2 | moderate | 1.65 | 857.44 ** |
| CXCL10 | moderate | 1.18 | 7.05 ** |
| CXCL12 | moderate | 1.50 | 2.98 ** |
| IFN-γ | not detected | - | - |
| IL-1α | moderate | 7.37 | 46.93 ** |
| IL-4 | not detected | - | - |
| IL-5 | moderate | 0.97 | 2.56 ** |
| IL-6 | high | 0.39 | 7.80 ** |
| IL-8 | moderate | 5.08 | 670.15 ** |
| IL-10 | low | 1.07 | 2.00 |
| IL-12p35 | moderate | 10.69 ** | 0.85 |
| IL-13 | low | 0.93 | 0.93 |
| IL-17A | not detected | - | - |
| IL-23p19 | high | 0.47 | 1.66 |
| IL-33 | not detected | - | - |
| MUC1 | high | 2.21 ** | 1.30 |
| MUC2 | low | 1.46 | 0.74 |
| MUC4 | moderate | 0.9 | 0.77 |
| PBD-1 | low | 0.5 | 1.07 |
| PBD-2 | not detected | - | - |
| PPAR-γ | high | 2.59 ** | 1.21 |
| Secretory component | high | 0.66 | 0.78 |
| TGF-β | high | 1.64 | 0.62 |
| TNF-α | low | 5.12 | 3075.63 ** |
| TSLP | moderate | 1.17 | 1.36 |
Level of mRNA expressions in untreated cells are expressed in the second column (high: Amplification around 17–24 cq (cycle quantification), moderate: Around 25–29 cq, low: Around 30–33 cq, not detected: More than 33 cq). Asterisks
denote p<0.01
Saccharomyces cerevisiae modulates the ETEC-induced transcript expression in differentiated IPEC-1 cells
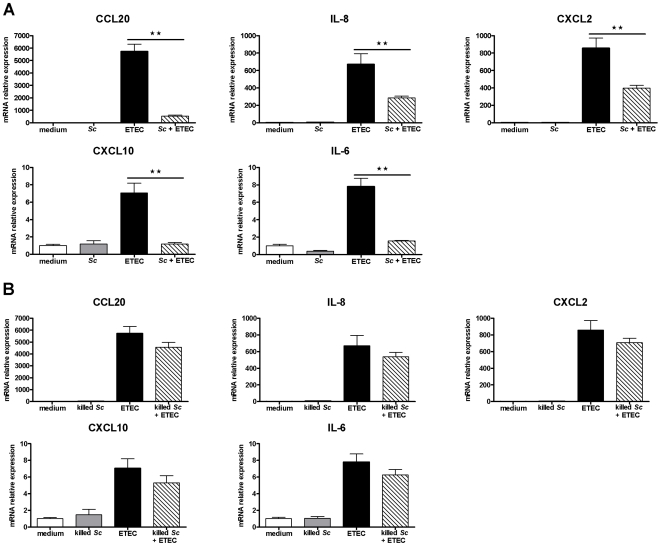
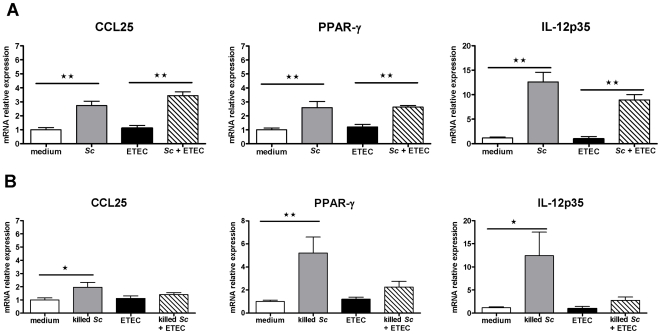
Overnight pre-incubation of IPEC-1 cells with viable Sc before ETEC exposure for 3 h inhibited significantly CCL20 (÷ 10.7), CXCL10 (÷ 6), IL-6 (÷ 5), IL-8 (÷ 2.3), and CXCL2 (÷ 2.2) mRNA expressions (p<0.01) (Figure 2.A). In contrast, Sc did not decrease significantly the mRNA expressions of TNF-α, IL-1α, IL-5 and CXCL12 which were up-regulated by ETEC. In addition, Sc inhibitory effect on BAFF mRNA expression was not conserved in presence of ETEC (data not shown). However, when Sc was killed, no inhibitory effects were observed suggesting that yeast-secreted factors are essential to inhibit the ETEC-induced gene expressions (Figure 2.B). Regarding Sc stimulatory effects, overnight pre-incubation of IPEC-1 cells with viable yeasts still up-regulated IL-12p35, CCL25 and PPAR-γ mRNA expressions (p<0.05) in IPEC-1 cells exposed to ETEC for 3 h (respectively x 8.9, x 3.1 and x 2.2) (Figure 3.A). The MUC1 mRNA was still up-regulated in presence of ETEC as observed with Sc alone (x 2.0, p<0.01, data not shown). However, when the yeast was killed, no stimulatory effects were observed in presence of ETEC and only IPEC-1 cells cultured with killed Sc alone showed this pattern of up-regulated mRNA expression (Figure 3.B). Because Sc up-regulated CCL25 mRNA levels, we also analysed whether IPEC-1 cells expressed the homeobox transcription factors Cdx-1 and -2, which are involved in CCL25 transcription. Analysis displayed that in these conditions of co-culture, Cdx-1 and -2 mRNA were not expressed in differentiated IPEC-1 cells (data not shown).
Figure 2. Viable Saccharomyces cerevisiae down-regulates cytokine and chemokine mRNA relative expressions induced by ETEC.
Differentiated IPEC-1 cells were cultured overnight with (A) viable Sc or (B) killed Sc (3×106 yeasts/well) and then ETEC (3×107 CFU/well) was added to the co-culture for 3 h. Gene expression was analysed by RT-qPCR. Data are presented as means ± SEM (n = 6), asterisks denote: ** (P<0.01). (A) Data are representative of three independent experiments.
Figure 3. Saccharomyces cerevisiae up-regulates CCL25, PPAR-γ and IL-12p35 mRNA relative expressions.
Differentiated IPEC-1 cells were cultured overnight with (A) viable Sc or (B) killed Sc (3×106 yeasts/well) and then ETEC (3×107 CFU/well) was added to the co-culture for 3 h. Gene expression was analysed by RT-qPCR. Data are presented as means ± SEM (n = 6), asterisks denote: * (P<0.05), ** (P<0.01). (A) Data are representative of three independent experiments.
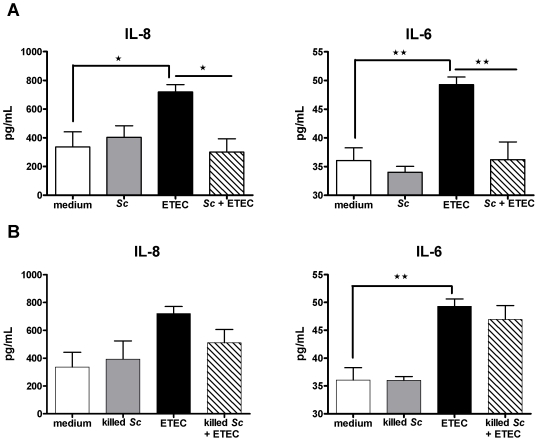
Saccharomyces cerevisiae decreases the ETEC-induced IL-6 and IL-8 secretions in differentiated IPEC-1 cells
At the protein level, overnight pre-incubation of IPEC-1 cells with viable or killed Sc did not modify apical IL-6 and IL-8 secretions in comparison to untreated cells (Figure 4.A, 4.B) whereas exposure with ETEC for 30 min up-regulated significantly the secretion of IL-6 (49.29±1.32 versus 36.06±2.21 pg/mL, p<0.05) and IL-8 (720±50.87 versus 335.8±105.8 pg/mL, p<0.05) (Figure 4.A, 4.B). In contrast, neither Sc nor ETEC induced the basolateral secretion of IL-6 and IL-8. The basolateral concentration of these cytokines was below the detection threshold of ELISA kits (data not shown). Overnight pre-incubation of IPEC-1 cells with viable Sc before ETEC exposure for 30 min inhibited significantly the apical secretions of IL-6 (36.21±3.10 versus 49.29±1.32 pg/mL, p<0.01) and IL-8 (300±92.21 versus 720±50.87 pg/mL, p<0.05) (Figure 4.A, 4.B). However, killed Sc did not inhibit significantly the ETEC-induced apical secretions of IL-6 and IL-8 (Figure 4.A, 4.B). These results are in accordance with those observed for the modulation of IL-6 and IL-8 transcripts indicating that Sc display anti-inflammatory properties at the molecular level.
Figure 4. Saccharomyces cerevisiae decreases IL-6 and IL-8 secretions induced by ETEC.
Differentiated IPEC-1 cells were cultured overnight with (A) viable or (B) killed Sc (3×106 yeasts/well) and then ETEC (3×107 CFU/well) was added to the co-culture for 30 min. Differentiated IPEC-1 cells were then washed and cultured for another 24 h before the assessment of apical IL-6 and IL-8 secretions by ELISA. Data are presented as means of cytokine concentration ± SEM (n = 3-4), asterisks denote: * (P<0.05), ** (P<0.01). Data are representative of two independent experiments.
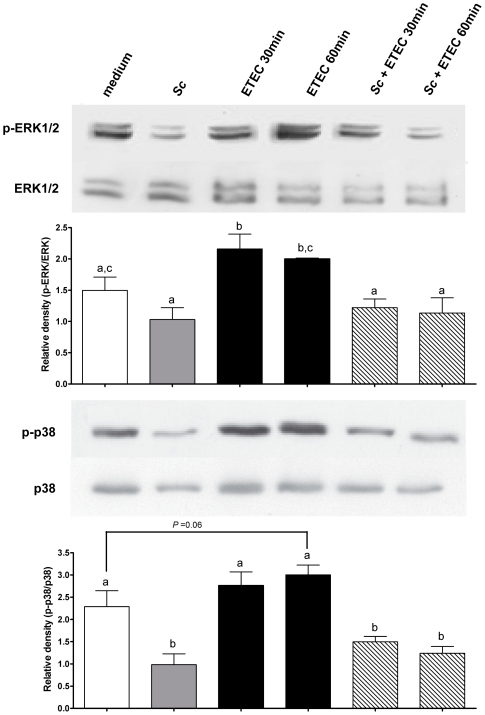
Saccharomyces cerevisiae decreases ERK1/2 and p38 MAPK phosphorylation in differentiated IPEC-1 cells
We next examined the effect of Sc on the activation of different protein kinases which are involved in pro-inflammatory gene expressions. Thus, we analysed Sc modulatory effects on MAPK (ERK1/2, p38, JNK), Akt and AMP-activated protein kinase (AMPK) phosphorylation using phospho-specific antibodies. As shown in Figure 5, Sc decreases the phosphorylation of ERK1/2 and p38. In control cells, the phosphorylated active-forms of ERK1/2 and p38 are detectable. Cells pre-incubated with Sc alone showed a significant decrease in p38 phosphorylation (p<0.05) but not in ERK1/2 (Figure 5). In contrast, ETEC exposure for 30 min enhanced the phosphorylation level of ERK1/2 (p<0.05) and ETEC exposure for 60 min enhanced the phosphorylation of p38 (p = 0.06) (Figure 5). When IPEC-1 cells were pre-incubated with Sc and then infected with ETEC, the active form of ERK1/2 was reduced at the level of control cells (p<0.05) whereas the active form of p38 was lower than control cells (p<0.05) (Figure 5). No effect was observed on Akt and AMPK activation, and JNK detection level was too low to display any regulatory effect of Sc (data not shown). These results indicated that Sc inhibitory effects on cytokine and chemokine mRNA expressions are associated with a decrease of ERK1/2 and p38 phosphorylation in IPEC-1 cells.
Figure 5. Saccharomyces cerevisiae decreases the MAP Kinase (ERK1/2, p38) phosphorylation in differentiated IPEC-1 cells.
Differentiated IPEC-1 cells were exposed for 30 and 60 min with ETEC (3×107 CFU/well) in the presence and absence of Sc (3×106 yeasts/well). Western blots for phospho-ERK1/2 (p-ERK1/2) and phospho-p38 (p-p38) were performed. Total ERK1/2 and total p38 are shown as loading controls and did not change with each condition over time. Data are presented as means ± SEM, (n = 3) and the different letters represent significant differences between the treatments (P<0.05). Results are representative of three independent experiments.
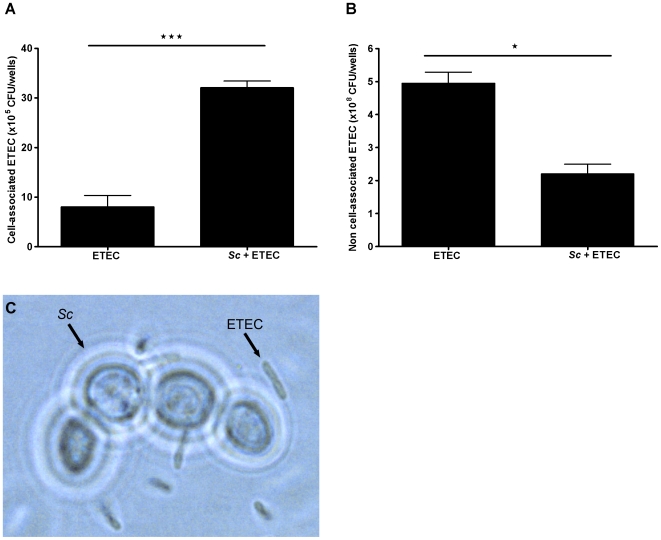
Saccharomyces cerevisiae agglutinates ETEC
We investigated whether Sc modulates ETEC growth and adhesion on IPEC-1 cells. Overnight pre-incubation of IPEC-1 cells with Sc before ETEC exposure increased the number of cell-associated ETEC (8.02±2.31×105 CFU/wells in ETEC group versus 32.08±1.33×105 CFU/wells in Sc+ETEC group) and decreased the number of non cell-associated ETEC (4.95±0.34×108 CFU/wells in ETEC group versus 2.2±0.29×108 CFU/wells in Sc+ETEC group) (Fig. 6.A, 6.B). Consequently, we assessed whether adherent yeasts on IPEC-1 cells could interact with ETEC thus forming agglutinates on the surface monolayer. Scanning electron microscopy of IPEC-1 cells exposed with ETEC did not demonstrate ETEC agglutination by Sc on the surface monolayer. In contrast, a physical interaction between yeasts and ETEC isolated from IPEC-1 cells culture supernatant was observed by phase-contrast microscopy (Fig. 6.C).
Figure 6. Saccharomyces cerevisiae agglutinates ETEC.
(A) IPEC-1 cells were exposed to ETEC (3×107 CFU/well) for 30 min in the presence or absence of Sc (3×106 yeasts/well). Cells were then lysed and cell lysates were diluted and plated on agar in order to quantify the cell-associated bacteria, (n = 4). (B) IPEC-1 cells were exposed to ETEC (3×107 CFU/well) for 3 h in the presence or absence of Sc (3×106 yeasts/well) and 100 µl of culture supernatant were then harvested from the apical compartment, diluted and plated on agar in order to quantify the non cell-associated bacteria, (n = 4). (C) IPEC-1 cells were overnight pre-incubated with Sc (3×106 yeasts/well) and then exposed to ETEC (3×107 CFU/well) for 3 h. Apical IPEC-1 cell culture supernatant was harvested and physical interaction between Sc and ETEC was observed by phase contrast microscopy (x 1000). Data are presented as means ± SEM, asterisks denote: * (P<0.05), *** (P<0.001). (A, B) Data are representatives of two independent experiments.
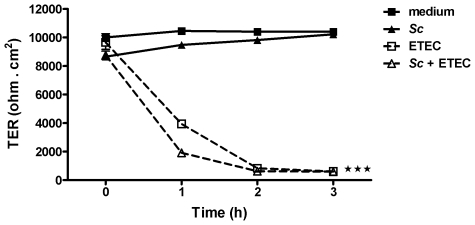
Saccharomyces cerevisiae did not prevent the disruption of membrane barrier integrity induced by ETEC
To assess the protective effect of Sc on the monolayer barrier function, transepithelial electrical resistance (TER) was measured in IPEC-1 cells pre-incubated or not with Sc and then exposed to ETEC for 3 h. Overnight pre-incubation of IPEC-1 cells with Sc alone did not alter the monolayer resistance (TER>8000 ohm.cm2) (Figure 7). In contrast, when IPEC-1 cells were exposed to ETEC, a significant TER decrease was already observed at 1 h of exposure (3934±50 ohm.cm2, p<0.001) (Figure 7). Overnight pre-incubation of IPEC-1 cells with Sc before ETEC exposure did not preserve the barrier function in comparison to both untreated and Sc-treated cells, thus resulting in a significant TER decrease after 1 h of ETEC exposure (1915±246 ohm.cm2, p<0.001) (Figure 7). In presence of Sc, despite a higher TER drop at 1 h of ETEC exposure, no differences were observed at 2 h and 3 h in comparison to monolayers exposed with ETEC alone (Figure 7).
Figure 7. Saccharomyces cerevisiae failed to preserve the barrier function.
Measurement of transepithelial electrical resistance (TER) in differentiated IPEC-1 cells untreated, pre-incubated with Sc (3×106 yeasts/well), exposed to ETEC (3×107 CFU/well) or pre-incubated with Sc and then exposed with ETEC. Data are presented as means ± SEM, (n = 4). Data are representative of two independent experiments, asterisks denote: *** (P<0.001).
Discussion
In pigs, enterotoxigenic Escherichia coli (ETEC) is the most common etiologic agent of enteric diseases in the weaning period [1]. ETEC infection induces pro-inflammatory responses in porcine intestinal epithelial cells [37] and causes diarrhea resulting in reduced growth rate, mortality and economic loss [1], [5]. Since 2006, administration of antibiotics as growth promoters in the piglet diet was prohibited in the European Union (IP/05/1687) and consequently, alternative strategies such as probiotics were developed to prevent intestinal diseases and to maintain health status [38], [39].
In this study, we investigated Saccharomyces cerevisiae (Sc, strain CNCM I-3856) modulatory effects on the expression of genes involved in inflammation, recruitment and activation of immune cells. IEC represent a relevant model to study yeast probiotic effects in the intestinal tract because IEC are involved in several mechanisms allowing protection such as barrier function, mucus layer protection, antibacterial peptide secretion and cytokine and chemokine secretion [19]. Consequently, IEC are involved in innate immunity as well as in the induction of adaptive immunity at the mucosal surface. In this study, we used as model of IEC the porcine IPEC-1 cell line exposed to ETEC. Previous reports suggest the presence of F4 receptor onto the apical surface of IPEC-1 cells since ETEC F4+ can adhere to these cells [34], [35]. In addition, IPEC-1 exposure to ETEC F4+ stimulate pro-inflammatory responses in IEC [35], [37]. Moreover, bacterial constituents like flagellin and lipopolysaccharide have been shown to activate MAPK signaling pathways in IEC [40], [41]. In the current work, we showed that only viable Sc decreased the ETEC-induced mRNA expressions of pro-inflammatory IL-6, IL-8, CCL20, CXCL2 and CXCL10. In addition, viable Sc decreased the apical secretions of IL-6 and IL-8 whereas no secretions were observed at the basolateral level. This result indicates a polarization of IL-6 and IL-8 secretions as described previously in both polarized epithelial and endothelial cells [42], [43], [44]. Taken together, these results indicate that Sc viability is essential and confirm the presence and the action of a soluble secreted factor as demonstrated in previous studies [45], [46]. In addition, a recent study reported that Sc (strain CNCM I-3856) exerts antagonist effect on E. coli O157:H7 probably through ethanol production [33], thus highlighting another potential anti-inflammatory effect used by the yeast. In our study, inhibition of pro-inflammatory gene expressions was correlated with a decrease of the MAPK ERK1/2 and p38 phosphorylation. These results are reminiscent of other studies showing that Sb inhibits IL-8 expression and MAPK phosphorylation induced by enteric pathogens such as enterohemorrhagic E. coli [21], Clostridium difficile [20], S. enterica Serovar Typhimurium [18] and Shigella flexneri [16]. Inhibition of pro-inflammatory IL-6, CCL20 and CXCL10 transcript expressions could also be associated with the inhibition of ERK1/2 and p38 MAPK signaling pathways since the activation of these protein kinases has been shown to correlate with IL-6, CCL20 and CXCL10 expressions in different cell types [47], [48], [49], [50]. In contrast, detection level of JNK in untreated and ETEC-exposed IPEC-1 cells was too low to display any regulatory effect of Sc. We also analysed whether or not the inhibition of the pro-inflammatory responses might be due to a modulation of Akt and AMPK activity. Previous studies described that Akt and AMPK are activated during intestinal inflammation [51], [52]. Moreover, Sb has been shown to decrease Akt activation in human HT29 colonocytes [53]. Neither Sc nor ETEC modulated Akt and AMPK signaling pathways in our model. This result could be explained by their activation at a different time point. In addition, Sc and ETEC could stimulate pathogen recognition receptors which did not activate these protein kinases in IPEC-1 cells. Taken together, these results show that Sc (strain CNCM I-3856) interfer with MAPK (ERK1/2, p38) signaling pathways and decrease pro-inflammatory responses in porcine intestinal epithelial IPEC-1 cells exposed with ETEC. These anti-inflammatory properties are in accordance with those described with the probiotic yeast Sb [16], [18], [20] and indicate that both Sc and Sb can inhibit cell signaling pathways despite their genetic, metabolic and physiologic differences [27], [28]. Several reports displayed also that Sb decrease NF-κB activation in IEC infected with enteric pathogens such as enterohemorrhagic E. coli, S. flexneri and S. enterica [16], [18], [22]. However, Sc effect on NF-κB activation was not assessed in this study and thus requires further investigations.
In addition, we showed that Sc up-regulated significantly the mRNA levels of the pro-Th1 cytokine IL-12p35, the chemokine CCL25 and the nuclear receptor PPAR-γ. CCL25 is known to be involved in T and B cells recruitment into the small intestine [54], [55]. Thus, Sc might stimulate the mucosal immune response and IgA plasma cell recruitment through enhancement of CCL25 expression. Previous in vivo studies displayed that oral administration of Sb to mice increases the concentration of intestinal IgA [17], [56]. The up-regulation of CCL25 expression could potentially explain, at the molecular level, one mechanism used by probiotic yeasts to stimulate the mucosal immune response. Because IPEC-1 cells express CCL25, we also assessed the expression of Cdx-1 and -2 transcription factors which has been shown to regulate CCL25 transcription [57]. Our study did not show any Cdx-1 and -2 transcript expressions by IPEC-1 cells. This result is in agreement with previous studies reporting that Cdx-1 and -2 were not expressed by immortalized epithelial cell lines [57], [58] suggesting either a different mechanism for CCL25 regulation or an activation of Cdx-1 and -2 expression at a different time point. Sc increased the transcript expression of IL-12p35 whereas ETEC did not. This result is in accordance with previous studies showing that Sc stimulates the secretion of IL-12p70 in activated human neutrophil-like 60 cells and monocyte-derived dendritic cells [59], [60]. In contrast, E. coli pathovars such as ETEC failed to stimulate IL-12p40 expression in bovine primary colonocytes [61]. These results may suggest that Sc and ETEC could influence differently the immune response in part through modulation of IL-12 expression level. PPAR-γ is a nuclear receptor expressed by several cell types including IEC, dendritic cells, T and B cells and acts as a regulator of the inflammation [62], [63], [64]. In our study we shown an up-regulation of PPAR-γ transcripts and thus Sc could also mediate anti-inflammatory effects through activation of PPAR-γ. This result could be correlated with previous studies showing that Sb up-regulated PPAR-γ expression in human colonocytes [65] and that PPAR-γ decreased pro-inflammatory cytokine levels in IEC [66]. Moreover, previous studies showed that activation of ERK1/2 regulated negatively the expression of PPAR-γ [67], [68] suggesting that both Sc stimulatory effects on PPAR-γ expression and inhibitory effects on ERK1/2 activation could be linked [69].
In this study, we also showed that the number of cell-associated ETEC is increased in presence of Sc while the number of non cell-associated ETEC is decreased. Consequently, we hypothesized that adherent yeasts on IPEC-1 cells could agglutinate ETEC thus forming agglutinates on the surface monolayer as shown previously with S. enterica serovar Typhimurium on the surface of T84 cells [18]. An agglutination of ETEC by Sc was observed by phase-contrast microscopy in IPEC-1 cell culture supernatant. This result suggests that the higher number of cell-associated bacteria observed in presence of Sc might be due to yeast-bacteria agglutinates on the surface monolayer despite the lack of confirmation by scanning electron microscopy. Furthermore, the agglutination of ETEC by yeasts may explain partially the inhibition of pro-inflammatory gene expressions in IPEC-1 cells which could be less stimulated by bacterial components such as LPS or flagellin. We also determined that ETEC decreased significantly the TER of IPEC-1 cells exposed to ETEC. This result is in agreement with others reports showing that ETEC decreased the TER in porcine IPEC-1 and IPEC-J2 cells [70], [71] and altered tight-junction structure in IPEC-1 cells [35]. TER measurement displayed that Sc failed to preserve the barrier function of infected IPEC-1 cells despite its anti-inflammatory activity. This result differed from a previous report showing that yeast extracts prevent both TER and membrane permeability decreases induced by ETEC [71]. However, previous studies suggested that TER decrease may reflect alterations in ion channel function [72], [73]. Consequently, this result suggests that Sc does not inhibit ETEC enterotoxin activity leading to the disruption of the transepithelial resistance.
In conclusion, results demonstrate that Sc (strain CNCM I-3856) inhibits pro-inflammatory gene expressions. This inhibition is associated to the modulation of both ERK1/2 and p38 signaling pathways, the increase of PPAR-γ transcript expression and the ETEC agglutination by yeasts. Sc stimulates also the transcript expression of CCL25 and IL-12p35. These results indicate that Sc exerts immuno-modulatory effects at the molecular level in IPEC-1 cells encouraging the assessment of Sc in vivo modulatory effects in the immune and inflammatory responses.
Materials and Methods
Epithelial cell line culture
The non-transformed porcine intestinal epithelial cell line IPEC-1 was derived from the small intestine of a newborn unsuckled piglet [74]. Cells were cultured in DMEM/F-12 medium (Invitrogen, Cergy Pontoise, France) supplemented with 5% foetal calf serum (Sigma–Aldrich, Saint-Quentin, France), 2 mM L-glutamine (Invitrogen), 1% insulin-transferrin-selenium (Sigma-Aldrich, Saint-Quentin, France), 10 ng/ml epidermal growth factor (Invitrogen), 50 U/ml penicillin and 50 µg/ml streptomycin (Invitrogen). For co-culture experiments, IPEC-1 cells were seeded onto 4.2 cm2 cell culture inserts (pore size 0.4 µm, Becton Dickinson Labware, Le Pont De Claix, France) at 3.5×105 cells per inserts. Inserts were not collagen-coated. IPEC-1 cells were grown for 2 days until 100% of confluence and then differentiated for 10 days. For differentiation culture, the medium described above was modified with the omission of foetal calf serum and the addition of 10-7 M dexamethasone (Sigma–Aldrich). Differentiation culture medium was changed every 2 days. Once differentiated, IPEC-1 cells had an average cell density of 1×106 cells/well and the transepithelial electrical resistance measured with a Millicell-ERS volt-ohm meter (Millipore, Molsheim, France) was typically around 8000 ohm.cm2. Differentiated IPEC-1 cells exhibit apical and basolateral surfaces, form apical microvilli and express tight junction proteins [35], [74].
Microorganisms' growth
The ETEC strain GIS26 (O149:K91, F4ac+, LT+ STa+ STb+ : H19) was grown in Luria-Bertani (LB) medium containing 1% tryptone, 0.5% yeast extract, 1% NaCl, pH 7. After overnight incubation at 37°C with vigorous shaking, bacteria were diluted at 1∶400 in fresh LB and grown until midlog phase (∼4 h) for all experiments.
Saccharomyces cerevisiae (strain CNCM I-3856, Lesaffre yeast collection, CNCM: French National deposit Collection of Microorganism Cultures, Institut Pasteur, Paris, France) were provided by Lesaffre (Société industrielle Lesaffre, Marcq-en-Baroeul, France) as a dry form at a concentration of 1×1010 yeasts/g. Sc was rehydrated in free-DMEM/F-12 medium (Invitrogen, Cergy Pontoise, France) for 45 min at 30°C. For the different experiments, concentration of viable Sc was established by methylene blue exclusion (0.3 g/L methylene blue, 20 g/L sodium citrate). To evaluate the effects of killed Sc, yeasts were rehydrated in DMEM/F-12 medium and then frozen in liquid nitrogen. After ten cycles of freezing/thawing, mortality resulted in 100% of killed yeasts.
Antibodies
Immunoblots were performed using the phosphorylated antibodies: (p)-ERK1/2 (4377S, Ozyme, Saint-Quentin-en-Yvelines, France), p-p38 (9211S, Ozyme), p-JNK 1/2/3 (9251, Ozyme), p-AMPK (2535L, Ozyme), p-Akt (sc-7985-R, Santa Cruz Biotechnology, Heidelberg, Germany), and the total antibodies: ERK2 (sc-154, Santa Cruz Biotechnology, cross-reacts with ERK1), p38 (sc-535, Santa Cruz Biotechnology), JNK (9252, Ozyme), AMPK (2532L, Ozyme), Akt (9272, Ozyme). According to the manufacturers, all antibodies react with human whereas only antibodies specific for p-ERK1/2, p-p38 and Akt cross-react with pig.
IPEC-1 exposure to ETEC
Prior to exposure, IPEC-1 cells (∼1×106 cells/well) were washed three times in medium without serum and antibiotics. The ETEC strain GIS26 was grown for 4 h in LB medium, pelleted by centrifugation at 2000 g for 20 min, resuspended in sterile phosphate-buffered saline and added to IPEC-1 cells at 3×107 CFU/well (30 bacteria/cell) for 3 h. When experiments were performed in presence of Sc, IPEC-1 cells were pre-incubated overnight with 3×106 yeasts/well (3 yeasts/cell) and then exposed to ETEC for 3 h. Overnight pre-incubation of IPEC-1 cells with Sc was chosen in order to assess the yeast preventive effects as described previously [22], [23]. Both ETEC and Sc were added to the apical compartment.
Analysis of relative mRNA expression using quantitative real-time PCR
IPEC-1 cells (∼1×106 cells/well) were incubated overnight with viable or killed Sc (3×106 yeasts/well) and then exposed to ETEC (3×107 CFU/well) for 3 h. IPEC-1 cells were lyzed with Trizol reagent (Invitrogen, Cergy-Pontoise, France) and total RNA was isolated using RNeasy Mini Kit (Qiagen, Courtaboeuf, France). Quantitative real-time PCR (qPCR) was performed using cDNA synthesized as previously described [58]. Diluted cDNA (10X) was combined with primer/probe sets and MESA GREEN qPCR MasterMix (Eurogentec, Liège, France) according to the manufacturer's recommendations. The qPCR conditions were 95°C for 30 s, followed by 37 cycles with denaturation at 95°C for 15 s and annealing/elongation for 45 s. To minimize sample variation, we used high quality RNA. Samples were normalized internally using simultaneously the average cycle quantification (Cq) of Hypoxanthine PhosphoRibosyl-Transferase 1 (HPRT-1), Ribosomal Protein L 19 (RPL-19) and Hydroxymethylbilane synthase 2 (HMBS2) [75] as references in each sample to avoid any artefact of variation in the target gene. These reference genes were selected for their stable expression in IPEC-1 cell line as described previously [76]. The primer sequences and the annealing temperatures are described in Table 1. Real time assays were run on a Bio-Rad Chromo4 (Bio-Rad, Hercules, CA, USA). Expression data are expressed as relative values after Genex macro analysis (Bio-Rad, Hercules, CA, USA) [77]. The design of qPCR experiments and the analysis of results were performed following the MIQE guidelines [78].
Measurement of apical IL-6 and IL-8 production
IPEC-1 cells (∼1×106 cells/well) were incubated overnight with viable or killed Sc (3×106 yeasts/well) and then exposed to ETEC (3×107 CFU/well) for 30 min. The time of ETEC exposure was chosen to avoid a massive alteration of the monolayer after the subsequent washes. After 3 washes, differentiation culture medium containing 50 µg/mL of gentamycin (Invitrogen) was added on IPEC-1 cells for 24 h. Apical and basolateral supernatants were then removed and cytokine production was measured by ELISA using commercial kits (R&D for IL-6 assay and Invitrogen for IL-8 assay).
Western Blotting
IPEC-1 cells (∼1×106 cells/well) were incubated overnight with Sc (3×106 yeasts/well) and then exposed to ETEC (3×107 CFU/well) for 30 and 60 min. Cells were then lysed in TNET lysis buffer (20 mM Tris pH 7.8, 50 mM NaCl, 5 mM EGTA, 1% (v/v) Triton X-100, 1 mM phenylmethylsulfonyl fluoride, 4 mM Na3VO4, 5 µg/ml leupeptin, 5 µg/ml pepstatin, and 5 µg/ml aprotinin). Equal amounts of proteins were separated on SDS-PAGE and transferred onto a nitrocellulose membrane. Membranes were incubated for 1 h at room temperature with Tris-buffered saline (TBS, 2 mM Tris–HCl, pH 8.0, 15 mM NaCl, pH 7.6), containing 5% non fat dry milk powder (NFDMP) and 0.1% Tween-20 to saturate non specific sites. Then, membranes were incubated overnight at 4°C with appropriate primary antibodies (final dilution 1∶1000) in TBS containing 0.1% Tween-20 and 5% NFDMP. After washing in TBS–0.1% Tween-20, the membranes were incubated for 2 h at room temperature with a HRP-conjugated goat anti-rabbit secondary antibody (final dilution 1∶10000; Diagnostic Pasteur, Marnes-la-Coquette, France) in TBS–0.1% Tween-20. After washing in TBS–0.1% Tween-20, the signal was detected by ECL (ECL, Amersham Pharmacia Biotech, Orsay, France). The films were analysed and signals quantified with the software Scion Image (4.0.3.2 version; Scion Corporation, Frederick, MD, USA).
Saccharomyces cerevisiae effects on ETEC adhesion to IPEC-1 cells
The number of cell-associated ETEC was determined by agar plating. IPEC-1 cells (∼1×106 cells/well) were infected with ETEC (3×107 CFU/well) for 30 min in the presence or absence of Sc (3×106 yeasts/well). The time of ETEC exposure was decreased from 3 h to 30 min to avoid a massive alteration of the monolayer after the subsequent washes. After 6 washes, cells were lysed with ultra pure water supplemented with 1%-Triton-X-100. Cell lysates containing the cell-associated ETEC were diluted and plated on 5% sheep blood agar. After 20 h of growth at 37°C, the number of ETEC colony was determined.
The number of non cell-associated ETEC was determined by agar plating. IPEC-1 cells (∼1×106 cells/well) were infected with ETEC (3×107 CFU/well) for 3 h in the presence or absence of Sc (3×106 yeasts/well). Then, 100 µl of culture supernatant were harvested from the apical compartment, diluted and plated on 5% sheep blood agar. After 20 h of growth at 37°C, the number of non cell-associated ETEC was quantified.
Scanning electron microscopy and phase contrast microscopy
For scanning electron microscopy analysis, IPEC-1 cells (∼1×106 cells/well) were incubated overnight with Sc (3×106 yeasts/well) and then exposed to ETEC (3×107 CFU/well) for 45 min. This time was chosen to avoid a massive alteration of the monolayer induced by the subsequent washes. Then, IPEC-1 cells were washed 3 times in PBS and then fixed in a mixture of 4% paraformaldehyde and 1% glutaraldehyde in phosphate buffer (0.3 M; pH 7.4) for 1 h. IPEC-1 cells were washed with phosphate buffer pH 7.4 with 0.4% NaCl (w/v) and dehydrated through a graded series of 50%, 70%, 90% and 100% alcohol solution and HMDS (hexa-methyl-disilazan). Then, they were let dried and coated with a thin layer of platinium on a PECS Gatan coater. Samples were observed on Zeiss Ultra+FEGSEM scanning electron micrograph (Carl Zeiss S.A.S, Le Pecq, France).
For phase contrast microscopy, IPEC-1 cells (∼1×106 cells/well) were incubated overnight with Sc (3×106 yeasts/well) and then exposed to ETEC (3×107 CFU/well) for 3 h. The apical cell culture supernatant was then removed and the interaction between Sc and ETEC was observed by phase contrast microscopy (Olympus BX51, Olympus S.A.S, Rungis, France).
Transepithelial electrical resistance measurements
Transepithelial electrical resistance (TER) was measured to assess the integrity of epithelial monolayers using a Millicell-ERS volt-ohm meter (Millipore, Molsheim, France). IPEC-1 cells cultured onto 4.2 cm2 cell culture inserts were pre-incubated overnight with Sc (3×106 yeasts/well) and then exposed to ETEC (3×107 CFU/well) for 3 h. TER was measured each hour after ETEC addition.
Statistical analysis
The comparison of the differences in mRNA relative expression and cytokine production were evaluated by one-way ANOVA and differences tested by non-parametric Dunnett's test. (using GraphPad Prism software version 4.00, GraphPad Software Inc., San Diego, CA, USA). Differences were considered significant when P<0.05.
Acknowledgments
We gratefully acknowledge Dr. Eric Auclair, Dr. Pascal Vandekerckove, Dr. Georges Pignède (Lesaffre) and Dr. Pascale Crépieux (Institut National de la Recherche Agronomique, Tours, France) for their assistance in this work. We gratefully acknowledge Dr. E. Cox (Gent University, Belgium) for kindly providing the F4+ ETEC (strain GIS26) and Dr. I. Oswald (Institut National de la Recherche Agronomique, Toulouse, France) for kindly providing the IPEC-1 cell line. We also acknowledge D. Bourry (Université François-Rabelais, Tours, France) for the improvement of figure design.
Footnotes
Competing Interests: GZ and RDI are employees of Lesaffre. Lesaffre has a patent application related to strain CNCM I-3856, which is a commercial strain. This does not alter the authors' adherence to all the PLoS ONE policies on sharing data and materials, as detailed online in the guide for authors.
Funding: The study has been funded mostly by Lesaffre company and partially by INRA (<10%). GZ received a CIFRE PhD grant from Lesaffre company and the French government (50/50). GZ performed all the study at INRA under the supervision of HS and FM (INRA employees). The funders had no role in study design, data collection and analysis, or preparation of the manuscript.
References
- 1.Fairbrother JM, Nadeau E, Gyles CL. Escherichia coli in postweaning diarrhea in pigs: an update on bacterial types, pathogenesis, and prevention strategies. Anim Health Res Rev. 2005;6:17–39. doi: 10.1079/ahr2005105. [DOI] [PubMed] [Google Scholar]
- 2.Fang L, Gan Z, Marquardt RR. Isolation, affinity purification, and identification of piglet small intestine mucosa receptor for enterotoxigenic Escherichia coli k88ac+ fimbriae. Infect Immun. 2000;68:564–569. doi: 10.1128/iai.68.2.564-569.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jin LZ, Zhao X. Intestinal receptors for adhesive fimbriae of enterotoxigenic Escherichia coli (ETEC) K88 in swine–a review. Appl Microbiol Biotechnol. 2000;54:311–318. doi: 10.1007/s002530000404. [DOI] [PubMed] [Google Scholar]
- 4.Levine MM. Adhesion of enterotoxigenic Escherichia coli in humans and animals. Ciba Found Symp. 1981;80:142–160. doi: 10.1002/9780470720639.ch10. [DOI] [PubMed] [Google Scholar]
- 5.Nagy B, Fekete PZ. Enterotoxigenic Escherichia coli in veterinary medicine. Int J Med Microbiol. 2005;295:443–454. doi: 10.1016/j.ijmm.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 6.Sanchez J, Holmgren J. Virulence factors, pathogenesis and vaccine protection in cholera and ETEC diarrhea. Curr Opin Immunol. 2005;17:388–398. doi: 10.1016/j.coi.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 7.Van den Broeck W, Cox E, Goddeeris BM. Receptor-dependent immune responses in pigs after oral immunization with F4 fimbriae. Infect Immun. 1999;67:520–526. doi: 10.1128/iai.67.2.520-526.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frydendahl K. Prevalence of serogroups and virulence genes in Escherichia coli associated with postweaning diarrhoea and edema disease in pigs and a comparison of diagnostic approaches. Vet Microbiol. 2002;85:169–182. doi: 10.1016/s0378-1135(01)00504-1. [DOI] [PubMed] [Google Scholar]
- 9.Docic M, Bilkei G. Differences in antibiotic resistance in Escherichia coli, isolated from East-European swine herds with or without prophylactic use of antibiotics. J Vet Med B Infect Dis Vet Public Health. 2003;50:27–30. doi: 10.1046/j.1439-0450.2003.00609.x. [DOI] [PubMed] [Google Scholar]
- 10.Lanz R, Kuhnert P, Boerlin P. Antimicrobial resistance and resistance gene determinants in clinical Escherichia coli from different animal species in Switzerland. Vet Microbiol. 2003;91:73–84. doi: 10.1016/s0378-1135(02)00263-8. [DOI] [PubMed] [Google Scholar]
- 11.Maynard C, Fairbrother JM, Bekal S, Sanschagrin F, Levesque RC, et al. Antimicrobial resistance genes in enterotoxigenic Escherichia coli O149:K91 isolates obtained over a 23-year period from pigs. Antimicrob Agents Chemother. 2003;47:3214–3221. doi: 10.1128/AAC.47.10.3214-3221.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zanello G, Meurens F, Berri M, Salmon H. Saccharomyces boulardii effects on gastrointestinal diseases. Curr Issues Mol Biol. 2009;11:47–58. [PubMed] [Google Scholar]
- 13.Wu X, Vallance BA, Boyer L, Bergstrom KS, Walker J, et al. Saccharomyces boulardii ameliorates Citrobacter rodentium-induced colitis through actions on bacterial virulence factors. Am J Physiol Gastrointest Liver Physiol. 2008;294:G295–306. doi: 10.1152/ajpgi.00173.2007. [DOI] [PubMed] [Google Scholar]
- 14.Castagliuolo I, Riegler MF, Valenick L, LaMont JT, Pothoulakis C. Saccharomyces boulardii protease inhibits the effects of Clostridium difficile toxins A and B in human colonic mucosa. Infect Immun. 1999;67:302–307. doi: 10.1128/iai.67.1.302-307.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Czerucka D, Dahan S, Mograbi B, Rossi B, Rampal P. Saccharomyces boulardii preserves the barrier function and modulates the signal transduction pathway induced in enteropathogenic Escherichia coli-infected T84 cells. Infect Immun. 2000;68:5998–6004. doi: 10.1128/iai.68.10.5998-6004.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mumy KL, Chen X, Kelly CP, McCormick BA. Am J Physiol Gastrointest Liver Physiol; 2007. Saccharomyces boulardii interferes with Shigella pathogenesis by post-invasion signaling events. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rodrigues AC, Cara DC, Fretez SH, Cunha FQ, Vieira EC, et al. Saccharomyces boulardii stimulates sIgA production and the phagocytic system of gnotobiotic mice. J Appl Microbiol. 2000;89:404–414. doi: 10.1046/j.1365-2672.2000.01128.x. [DOI] [PubMed] [Google Scholar]
- 18.Martins FS, Dalmasso G, Arantes RM, Doye A, Lemichez E, et al. Interaction of Saccharomyces boulardii with Salmonella enterica serovar Typhimurium protects mice and modifies T84 cell response to the infection. PLoS One. 2010;5:e8925. doi: 10.1371/journal.pone.0008925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oswald IP. Role of intestinal epithelial cells in the innate immune defence of the pig intestine. Vet Res. 2006;37:359–368. doi: 10.1051/vetres:2006006. [DOI] [PubMed] [Google Scholar]
- 20.Chen X, Kokkotou EG, Mustafa N, Bhaskar KR, Sougioultzis S, et al. Saccharomyces boulardii inhibits ERK1/2 mitogen-activated protein kinase activation both in vitro and in vivo and protects against Clostridium difficile toxin A-induced enteritis. J Biol Chem. 2006;281:24449–24454. doi: 10.1074/jbc.M605200200. [DOI] [PubMed] [Google Scholar]
- 21.Dahan S, Busuttil V, Imbert V, Peyron JF, Rampal P, et al. Enterohemorrhagic Escherichia coli infection induces interleukin-8 production via activation of mitogen-activated protein kinases and the transcription factors NF-kappaB and AP-1 in T84 cells. Infect Immun. 2002;70:2304–2310. doi: 10.1128/IAI.70.5.2304-2310.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dahan S, Dalmasso G, Imbert V, Peyron JF, Rampal P, et al. Saccharomyces boulardii interferes with enterohemorrhagic Escherichia coli-induced signaling pathways in T84 cells. Infect Immun. 2003;71:766–773. doi: 10.1128/IAI.71.2.766-773.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dalmasso G, Loubat A, Dahan S, Calle G, Rampal P, et al. Saccharomyces boulardii prevents TNF-alpha-induced apoptosis in EHEC-infected T84 cells. Res Microbiol. 2006;157:456–465. doi: 10.1016/j.resmic.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 24.Kagnoff MF, Eckmann L. Epithelial cells as sensors for microbial infection. J Clin Invest. 1997;100:6–10. doi: 10.1172/JCI119522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Berkes J, Viswanathan VK, Savkovic SD, Hecht G. Intestinal epithelial responses to enteric pathogens: effects on the tight junction barrier, ion transport, and inflammation. Gut. 2003;52:439–451. doi: 10.1136/gut.52.3.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Edwards-Ingram L, Gitsham P, Burton N, Warhurst G, Clarke I, et al. Genotypic and physiological characterization of Saccharomyces boulardii, the probiotic strain of Saccharomyces cerevisiae. Appl Environ Microbiol. 2007;73:2458–2467. doi: 10.1128/AEM.02201-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fietto JL, Araujo RS, Valadao FN, Fietto LG, Brandao RL, et al. Molecular and physiological comparisons between Saccharomyces cerevisiae and Saccharomyces boulardii. Can J Microbiol. 2004;50:615–621. doi: 10.1139/w04-050. [DOI] [PubMed] [Google Scholar]
- 28.Hennequin C, Thierry A, Richard GF, Lecointre G, Nguyen HV, et al. Microsatellite typing as a new tool for identification of Saccharomyces cerevisiae strains. J Clin Microbiol. 2001;39:551–559. doi: 10.1128/JCM.39.2.551-559.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jurgens MH, Rikabi RA, Zimmerman DR. The effect of dietary active dry yeast supplement on performance of sows during gestation-lactation and their pigs. J Anim Sci. 1997;75:593–597. doi: 10.2527/1997.753593x. [DOI] [PubMed] [Google Scholar]
- 30.Perez-Sotelo LS, Talavera-Rojas M, Monroy-Salazar HG, Lagunas-Bernabe S, Cuaron-Ibarguengoytia JA, et al. In vitro evaluation of the binding capacity of Saccharomyces cerevisiae Sc47 to adhere to the wall of Salmonella spp. Rev Latinoam Microbiol. 2005;47:70–75. [PubMed] [Google Scholar]
- 31.Foligne B, Dewulf J, Vandekerckove P, Pignede G, Pot B. Probiotic yeasts: anti-inflammatory potential of various non-pathogenic strains in experimental colitis in mice. World J Gastroenterol. 2010;16:2134–2145. doi: 10.3748/wjg.v16.i17.2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Desreumaux P, Neut C, Cazaubiel M, Pichelin M, Pelerin F, Justen P. Barcelona, Spain: 18th United European Gastroenterology Week; 2010. Saccharomyces cerevisiae CNCM I-3856 reduces digestive discomfort and abdominal pain in subjects with irritable bowel syndrome: a randomized double-blinded placebo-controlled clinical trial. [Google Scholar]
- 33.Etienne-Mesmin L, Livrelli V, Privat M, Denis S, Cardot JM, et al. Appl Environ Microbiol; 2010. Effect of a new probiotic Saccharomyces cerevisiae strain on the survival of Escherichia coli O157:H7 in a dynamic gastrointestinal model. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koh SY, George S, Brozel V, Moxley R, Francis D, et al. Porcine intestinal epithelial cell lines as a new in vitro model for studying adherence and pathogenesis of enterotoxigenic Escherichia coli. Vet Microbiol. 2008;130:191–197. doi: 10.1016/j.vetmic.2007.12.018. [DOI] [PubMed] [Google Scholar]
- 35.Roselli M, Finamore A, Britti MS, Konstantinov SR, Smidt H, et al. The novel porcine Lactobacillus sobrius strain protects intestinal cells from enterotoxigenic Escherichia coli K88 infection and prevents membrane barrier damage. J Nutr. 2007;137:2709–2716. doi: 10.1093/jn/137.12.2709. [DOI] [PubMed] [Google Scholar]
- 36.van der Aa Kuhle A, Skovgaard K, Jespersen L. In vitro screening of probiotic properties of Saccharomyces cerevisiae var. boulardii and food-borne Saccharomyces cerevisiae strains. Int J Food Microbiol. 2005;101:29–39. doi: 10.1016/j.ijfoodmicro.2004.10.039. [DOI] [PubMed] [Google Scholar]
- 37.Devriendt B, Stuyven E, Verdonck F, Goddeeris BM, Cox E. Enterotoxigenic Escherichia coli (K88) induce proinflammatory responses in porcine intestinal epithelial cells. Dev Comp Immunol. 2010;34:1175–1182. doi: 10.1016/j.dci.2010.06.009. [DOI] [PubMed] [Google Scholar]
- 38.Konstantinov SR, Smidt H, Akkermans AD, Casini L, Trevisi P, et al. Feeding of Lactobacillus sobrius reduces Escherichia coli F4 levels in the gut and promotes growth of infected piglets. FEMS Microbiol Ecol. 2008;66:599–607. doi: 10.1111/j.1574-6941.2008.00517.x. [DOI] [PubMed] [Google Scholar]
- 39.Lessard M, Dupuis M, Gagnon N, Nadeau E, Matte JJ, et al. Administration of Pediococcus acidilactici or Saccharomyces cerevisiae boulardii modulates development of porcine mucosal immunity and reduces intestinal bacterial translocation after Escherichia coli challenge. J Anim Sci. 2009;87:922–934. doi: 10.2527/jas.2008-0919. [DOI] [PubMed] [Google Scholar]
- 40.Cario E, Rosenberg IM, Brandwein SL, Beck PL, Reinecker HC, et al. Lipopolysaccharide activates distinct signaling pathways in intestinal epithelial cell lines expressing Toll-like receptors. J Immunol. 2000;164:966–972. doi: 10.4049/jimmunol.164.2.966. [DOI] [PubMed] [Google Scholar]
- 41.Tallant T, Deb A, Kar N, Lupica J, de Veer MJ, et al. Flagellin acting via TLR5 is the major activator of key signaling pathways leading to NF-kappa B and proinflammatory gene program activation in intestinal epithelial cells. BMC Microbiol. 2004;4:33. doi: 10.1186/1471-2180-4-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chow AW, Liang JF, Wong JS, Fu Y, Tang NL, et al. Polarized secretion of interleukin (IL)-6 and IL-8 by human airway epithelia 16HBE14o- cells in response to cationic polypeptide challenge. PLoS One. 2010;5:e12091. doi: 10.1371/journal.pone.0012091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fahey JV, Schaefer TM, Channon JY, Wira CR. Secretion of cytokines and chemokines by polarized human epithelial cells from the female reproductive tract. Hum Reprod. 2005;20:1439–1446. doi: 10.1093/humrep/deh806. [DOI] [PubMed] [Google Scholar]
- 44.Jurczyluk J, Brown D, Stanley KK. Polarised secretion of cytokines in primary human microvascular endothelial cells is not dependent on N-linked glycosylation. Cell Biol Int. 2003;27:997–1003. doi: 10.1016/j.cellbi.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 45.Sougioultzis S, Simeonidis S, Bhaskar KR, Chen X, Anton PM, et al. Saccharomyces boulardii produces a soluble anti-inflammatory factor that inhibits NF-kappaB-mediated IL-8 gene expression. Biochem Biophys Res Commun. 2006;343:69–76. doi: 10.1016/j.bbrc.2006.02.080. [DOI] [PubMed] [Google Scholar]
- 46.Zanello G, Meurens F, Berri M, Chevaleyre C, Melo S, et al. Vet Immunol Immunopathol; Saccharomyces cerevisiae decreases inflammatory responses induced by F4+ enterotoxigenic Escherichia coli in porcine intestinal epithelial cells. (In press) [DOI] [PubMed] [Google Scholar]
- 47.Cekic C, Casella CR, Eaves CA, Matsuzawa A, Ichijo H, et al. Selective activation of the p38 MAPK pathway by synthetic monophosphoryl lipid A. J Biol Chem. 2009;284:31982–31991. doi: 10.1074/jbc.M109.046383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dommisch H, Chung WO, Jepsen S, Hacker BM, Dale BA. Phospholipase C, p38/MAPK, and NF-kappaB-mediated induction of MIP-3alpha/CCL20 by Porphyromonas gingivalis. Innate Immun. 2010;16:226–234. doi: 10.1177/1753425909339237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huang FC. Upregulation of Salmonella-induced IL-6 production in Caco-2 cells by PJ-34, PARP-1 inhibitor: involvement of PI3K, p38 MAPK, ERK, JNK, and NF-kappaB. Mediators Inflamm. 2009;2009:103890. doi: 10.1155/2009/103890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Valledor AF, Sanchez-Tillo E, Arpa L, Park JM, Caelles C, et al. Selective roles of MAPKs during the macrophage response to IFN-gamma. J Immunol. 2008;180:4523–4529. doi: 10.4049/jimmunol.180.7.4523. [DOI] [PubMed] [Google Scholar]
- 51.Bandyopadhaya A, Bhowmick S, Chaudhuri K. Activation of proinflammatory response in human intestinal epithelial cells following Vibrio cholerae infection through PI3K/Akt pathway. Can J Microbiol. 2009;55:1310–1318. doi: 10.1139/w09-093. [DOI] [PubMed] [Google Scholar]
- 52.Scharl M, Paul G, Barrett KE, McCole DF. AMP-activated protein kinase mediates the interferon-gamma-induced decrease in intestinal epithelial barrier function. J Biol Chem. 2009;284:27952–27963. doi: 10.1074/jbc.M109.046292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen X, Fruehauf J, Goldsmith JD, Xu H, Katchar KK, et al. Saccharomyces boulardii inhibits EGF receptor signaling and intestinal tumor growth in Apc(min) mice. Gastroenterology. 2009;137:914–923. doi: 10.1053/j.gastro.2009.05.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kunkel EJ, Campbell JJ, Haraldsen G, Pan J, Boisvert J, et al. Lymphocyte CC chemokine receptor 9 and epithelial thymus-expressed chemokine (TECK) expression distinguish the small intestinal immune compartment: Epithelial expression of tissue-specific chemokines as an organizing principle in regional immunity. J Exp Med. 2000;192:761–768. doi: 10.1084/jem.192.5.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Meurens F, Berri M, Whale J, Dybvig T, Strom S, et al. Expression of TECK/CCL25 and MEC/CCL28 chemokines and their respective receptors CCR9 and CCR10 in porcine mucosal tissues. Vet Immunol Immunopathol. 2006;113:313–327. doi: 10.1016/j.vetimm.2006.05.014. [DOI] [PubMed] [Google Scholar]
- 56.Qamar A, Aboudola S, Warny M, Michetti P, Pothoulakis C, et al. Saccharomyces boulardii stimulates intestinal immunoglobulin A immune response to Clostridium difficile toxin A in mice. Infect Immun. 2001;69:2762–2765. doi: 10.1128/IAI.69.4.2762-2765.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ericsson A, Kotarsky K, Svensson M, Sigvardsson M, Agace W. Functional characterization of the CCL25 promoter in small intestinal epithelial cells suggests a regulatory role for caudal-related homeobox (Cdx) transcription factors. J Immunol. 2006;176:3642–3651. doi: 10.4049/jimmunol.176.6.3642. [DOI] [PubMed] [Google Scholar]
- 58.Meurens F, Berri M, Siggers RH, Willing BP, Salmon H, et al. Commensal bacteria and expression of two major intestinal chemokines, TECK/CCL25 and MEC/CCL28, and their receptors. PLoS One. 2007;2:e677. doi: 10.1371/journal.pone.0000677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rizzetto L, Kuka M, De Filippo C, Cambi A, Netea MG, et al. Differential IL-17 production and mannan recognition contribute to fungal pathogenicity and commensalism. J Immunol. 2010;184:4258–4268. doi: 10.4049/jimmunol.0902972. [DOI] [PubMed] [Google Scholar]
- 60.Saegusa S, Totsuka M, Kaminogawa S, Hosoi T. Saccharomyces cerevisiae and Candida albicans stimulate cytokine secretion from human neutrophil-like HL-60 cells differentiated with retinoic acid or dimethylsulfoxide. Biosci Biotechnol Biochem. 2009;73:2600–2608. doi: 10.1271/bbb.90410. [DOI] [PubMed] [Google Scholar]
- 61.Bridger PS, Mohr M, Stamm I, Frohlich J, Follmann W, et al. Primary bovine colonic cells: a model to study strain-specific responses to Escherichia coli. Vet Immunol Immunopathol. 2010;137:54–63. doi: 10.1016/j.vetimm.2010.04.010. [DOI] [PubMed] [Google Scholar]
- 62.Escher P, Braissant O, Basu-Modak S, Michalik L, Wahli W, et al. Rat PPARs: quantitative analysis in adult rat tissues and regulation in fasting and refeeding. Endocrinology. 2001;142:4195–4202. doi: 10.1210/endo.142.10.8458. [DOI] [PubMed] [Google Scholar]
- 63.Mansen A, Guardiola-Diaz H, Rafter J, Branting C, Gustafsson JA. Expression of the peroxisome proliferator-activated receptor (PPAR) in the mouse colonic mucosa. Biochem Biophys Res Commun. 1996;222:844–851. doi: 10.1006/bbrc.1996.0832. [DOI] [PubMed] [Google Scholar]
- 64.Su CG, Wen X, Bailey ST, Jiang W, Rangwala SM, et al. A novel therapy for colitis utilizing PPAR-gamma ligands to inhibit the epithelial inflammatory response. J Clin Invest. 1999;104:383–389. doi: 10.1172/JCI7145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lee SK, Kim HJ, Chi SG, Jang JY, Nam KD, et al. [Saccharomyces boulardii activates expression of peroxisome proliferator-activated receptor-gamma in HT-29 cells]. Korean J Gastroenterol. 2005;45:328–334. [PubMed] [Google Scholar]
- 66.Adachi M, Kurotani R, Morimura K, Shah Y, Sanford M, et al. Peroxisome proliferator activated receptor gamma in colonic epithelial cells protects against experimental inflammatory bowel disease. Gut. 2006;55:1104–1113. doi: 10.1136/gut.2005.081745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kaplan JM, Hake PW, Denenberg A, Nowell M, Piraino G, et al. Phosphorylation of Extracellular Signal-Regulated Kinase (ERK)-1/2 Is Associated with the Downregulation of Peroxisome Proliferator-Activated Receptor (PPAR)-gamma during Polymicrobial Sepsis. Mol Med. 2010;16:491–497. doi: 10.2119/molmed.2010.00063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhou Y, Jia X, Wang G, Wang X, Liu J. PI-3 K/AKT and ERK signaling pathways mediate leptin-induced inhibition of PPARgamma gene expression in primary rat hepatic stellate cells. Mol Cell Biochem. 2009;325:131–139. doi: 10.1007/s11010-009-0027-3. [DOI] [PubMed] [Google Scholar]
- 69.Wei-guo Z, Hui Y, Shan L, Yun Z, Wen-cheng N, et al. PPAR-gamma agonist inhibits Ang II-induced activation of dendritic cells via the MAPK and NF-kappaB pathways. Immunol Cell Biol. 2009;88:305–312. doi: 10.1038/icb.2009.100. [DOI] [PubMed] [Google Scholar]
- 70.Johnson AM, Kaushik RS, Hardwidge PR. Vet Microbiol; 2009. Disruption of transepithelial resistance by enterotoxigenic Escherichia coli. [DOI] [PubMed] [Google Scholar]
- 71.Roselli M, Britti MS, Le Huerou-Luron I, Marfaing H, Zhu WY, et al. Effect of different plant extracts and natural substances (PENS) against membrane damage induced by enterotoxigenic Escherichia coli K88 in pig intestinal cells. Toxicol In Vitro. 2007;21:224–229. doi: 10.1016/j.tiv.2006.09.012. [DOI] [PubMed] [Google Scholar]
- 72.Moeser AJ, Nighot PK, Roerig B, Ueno R, Blikslager AT. Comparison of the chloride channel activator lubiprostone and the oral laxative Polyethylene Glycol 3350 on mucosal barrier repair in ischemic-injured porcine intestine. World J Gastroenterol. 2008;14:6012–6017. doi: 10.3748/wjg.14.6012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nighot PK, Moeser AJ, Ryan KA, Ghashghaei T, Blikslager AT. ClC-2 is required for rapid restoration of epithelial tight junctions in ischemic-injured murine jejunum. Exp Cell Res. 2009;315:110–118. doi: 10.1016/j.yexcr.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 74.Gonzalez-Vallina R, Wang H, Zhan R, Berschneider HM, Lee RM, et al. Lipoprotein and apolipoprotein secretion by a newborn piglet intestinal cell line (IPEC-1). Am J Physiol. 1996;271:G249–259. doi: 10.1152/ajpgi.1996.271.2.G249. [DOI] [PubMed] [Google Scholar]
- 75.Nygard AB, Jorgensen CB, Cirera S, Fredholm M. Selection of reference genes for gene expression studies in pig tissues using SYBR green qPCR. BMC Mol Biol. 2007;8:67. doi: 10.1186/1471-2199-8-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bruel T, Guibon R, Melo S, Guillen N, Salmon H, et al. Epithelial induction of porcine suppressor of cytokine signaling 2 (SOCS2) gene expression in response to Entamoeba histolytica. Dev Comp Immunol. 2010;34:562–571. doi: 10.1016/j.dci.2009.12.017. [DOI] [PubMed] [Google Scholar]
- 77.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]