Abstract
Transcriptional regulation at the level of elongation is vital for the control of gene expression and metazoan development. The mixed lineage leukemia (MLL) protein and its Drosophila homolog, Trithorax, which exist within COMPASS (complex of proteins associated with Set1)-like complexes, are master regulators of development. They are required for proper homeotic gene expression, in part through methylation of histone H3 on Lys 4. In humans, the MLL gene is involved in a large number of chromosomal translocations that create chimeric proteins, fusing the N terminus of MLL to several proteins that share little sequence similarity. Several frequent translocation partners of MLL were found recently to coexist in a super elongation complex (SEC) that includes known transcription elongation factors such as eleven-nineteen lysine-rich leukemia (ELL) and P-TEFb. Importantly, the SEC is required for HOX gene expression in leukemic cells, suggesting that chromosomal translocations involving MLL could lead to the overexpression of HOX and other genes through the involvement of the SEC. Here, we review the normal developmental roles of MLL and the SEC, and how MLL fusion proteins can mediate leukemogenesis.
Keywords: transcription elongation, H3K4 methylation, P-TEFb, HOX genes, leukemia
The program of developmental gene expression has long focused on the recruitment of RNA polymerase II (Pol II) by sequence-specific binding factors and their cofactors. More recently, an additional level of gene expression, regulating the release of paused Pol II, was found to be widely used during development in metazoans. Genome-wide binding studies have found paused Pol II at genes that are expressed later in development (Guenther et al. 2007). A similar finding was found in Drosophila melanogaster embryos, where it was demonstrated that 10% of the genes have paused Pol II, and that paused Pol II were overwhelmingly enriched for developmentally regulated genes (Muse et al. 2007; Zeitlinger et al. 2007). Paused RNA Pol II has been best characterized as a regulatory point at heat-shock loci in Drosophila, where RNA Pol II sits in an engaged state at the promoter, nonproductively transcribing ∼25 nucleotides (nt) in the nonstressed state. Upon heat shock, Pol II is released for the production of full-length transcripts (Gilmour and Lis 1986; Rougvie and Lis 1988; Gerber et al. 2001, 2005; Smith et al. 2008b). Other genes that have been long known to be regulated at the level of transcription elongation encode the proto-oncogenes FOS, JUN, and MYC (Saunders et al. 2006). Retroviruses have also been known to use transcriptional elongation as a regulatory mechanism, most notably by the human immunodeficiency virus (HIV) in the control of the production of full-length proviral transcripts (Kao et al. 1987; Laspia et al. 1989).
In order to find factors involved in the control of transcription elongation, several groups have taken a biochemical approach, fractionating nuclear extracts to search for activities that can stimulate the production of full-length transcripts in vitro. One factor that was identified in this manner was ELL (Shilatifard et al. 1996). Originally isolated from rat liver nuclear extracts as a factor that could increase the Vmax of the transcription rate by RNA Pol II, this protein was found to be related to the human eleven-nineteen lysine-rich leukemia (ELL) protein that is fused to MLL (mixed lineage leukemia) (MLL1, KMT2A) in a subset of MLL-rearranged leukemias (Thirman et al. 1994; Shilatifard et al. 1996). MLL is involved in a large number of chromosomal rearrangements that lead to myeloid and lymphoblastic leukemia. The Drosophila homolog of MLL is the Trithorax protein, which shares a conserved function with MLL as a master regulator of development, as it is required for transcription of Hox loci (Eissenberg and Shilatifard 2010). MLL is homologous to yeast Set1, an H3K4 methyltransferase that forms a large macromolecular complex, COMPASS (complex of proteins associated with Set1) (Miller et al. 2001). Similar to yeast Set1, MLL exists in a COMPASS-like complex that methylates H3K4, a modification associated with transcriptionally active genes (Hughes et al. 2004; Wu et al. 2008). MLL is responsible for H3K4 trimethylation at a subset of genes that includes Hox loci as well as many other genes that encode master regulators of development (P Wang et al. 2009). The recent finding that several translocation partners of MLL associate with the known transcription elongation factors ELL and P-TEFb in the super elongation complex (SEC) (Lin et al. 2010; Mohan et al. 2010) suggests that a major mechanism of leukemogenesis could be the misregulation of the elongation stage of transcription—as it was originally proposed >15 years ago (Shilatifard et al. 1996).
In this review, we discuss the normal developmental roles of MLL and SEC, and how the intricate control of developmental genes at the level of transcription elongation can be misregulated in leukemia.
Regulation of transcription at the level of elongation
Numerous factors that stimulate elongation by RNA Pol II have been identified from in vitro transcription assays. These factors can be categorized according to the stage of transcription elongation that they regulate: promoter clearance, promoter-proximal pausing, and productive elongation (Sims et al. 2004; Saunders et al. 2006). TFIIF and TFIIH are required for early promoter clearance. TFIIH has both helicase and kinase activities. The helicase activity helps melt the template DNA to form the transcription bubble, while the kinase activity phosphorylates RNA Pol II at Ser 5 within the C-terminal repeat domain (CTD) (Akoulitchev et al. 1995; Holstege et al. 1996; Dvir et al. 1997). Phosphorylation of the RNA Pol II CTD on Ser 5 is necessary for recruitment of the mRNA capping machinery. TFIIF is one of several factors that helps recruit RNA Pol II to the promoter, but it is also required for the synthesis of the first several bases of the transcript (Yan et al. 1999). After promoter clearance, the polymerase continues to exhibit some backtracking of up to +30 nt from the promoter. Productive elongation beyond this point is associated with phosphorylation of the CTD on Ser 2, mediated by P-TEFb. Many of the genes in metazoans are regulated at this step, characterized by Pol II stalling at 25–40 nt from the promoter (Core et al. 2008; Nechaev et al. 2010). The mechanism underlying the generation of these short transcripts could involve either polymerases that terminate prematurely or polymerases that are held in place until an appropriate signal is received (Krumm et al. 1992; Marshall and Price 1992; Aida et al. 2006; Ni et al. 2008). Stalling Pol II is phosphorylated on Ser 5 of the CTD and associates with the negative elongation factor (NELF) and the DRB sensitivity-inducible factor (DSIF). DSIF is composed of Spt4 and Spt5, which were originally identified genetically in yeast as transcription factors and were later identified from biochemical assays as transcription elongation factors (Winston et al. 1984; Wada et al. 1998). DSIF acts as both a positive and negative elongation factor. DSIF interacts with the multisubunit NELF complex at genes with paused Pol II, but, upon phosphorylation of NELF and DSIF by P-TEFb, DSIF is released from NELF and travels with Pol II during productive transcription elongation (Chen et al. 2009).
Other factors that facilitate transcription elongation include FACT, a histone chaperone complex that facilitates passage of the polymerase through nucleosomes (Orphanides et al. 1998), and SII, which prevents arrest of RNA Pol II during backtracking by stimulating cleavage of the nascent transcript to generate a new 3′ end that is properly aligned with the template (Reines et al. 1996). Elongin and ELL, like TFIIF, were identified as factors that stimulate the rate of Pol II transcription on naked DNA templates (Aso et al. 1995; Shilatifard et al. 1996; Shilatifard et al. 2003). Elongin is composed of three subunits: Elongin A, Elongin B, and Elongin C. Elongin A interacts directly with RNA Pol II, while Elongin B and Elongin C associate with an E3 ubiquitin ligase complex that can mediate degradation of RNA Pol II after DNA damage (Yasukawa et al. 2008). ELL was identified as a single polypeptide from rat liver extracts that could stimulate transcription elongation in vitro, and was related to the human gene that was a frequent translocation partner of MLL in acute myeloid leukemia (Shilatifard et al. 1996). MLL was known to be involved in translocations with many partners, but the elongation stimulatory activity of ELL provided the first biological function attributable to a translocation partner of MLL. More recently, biochemical purification of several of the commonly observed MLL chimeras identified the SEC that contains several MLL translocation partners, including ELL, as well as the CTD kinase P-TEFb (Lin et al. 2010).
The association of both P-TEFb and ELL—factors that were identified independently as stimulating transcription elongation in vitro—in a complex with other MLL chimeras supports the notion that transcription elongation is misregulated in these leukemias (Shilatifard et al. 1996; Lin et al. 2010). Combined with the recent demonstrations that many developmental genes are regulated at the level of paused Pol II, these findings suggest that the release of paused Pol II by the SEC could mediate the enhanced transcription of HOX and other loci in MLL-rearranged leukemias (Lin et al. 2010; Mohan et al. 2010).
Regulation of development by MLL
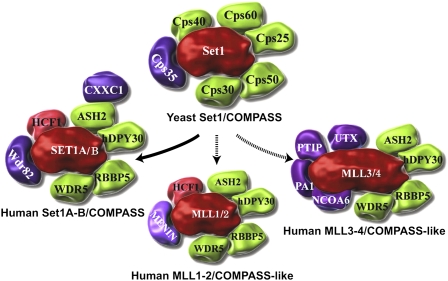
In order to gain a better insight into the molecular function of MLL, several years ago, we purified its closest yeast homolog, Set1 (Miller et al. 2001). Yeast Set1 was found to exist as part of an ∼1-MDa complex named COMPASS with six other proteins named for their apparent molecular weight by SDS-PAGE: Cps25, Cps30, Cps35, Cps40, Cps50, and Cps60 (Fig. 1). In mammalian cells, homologs of Cps25 (DPY30), Cps30 (WDR5), Cps50 (RBBP5), and Cps60 (ASH2) were found in a COMPASS-like complex with MLL (Fig. 1; Hughes et al. 2004; Yokoyama et al. 2004; Dou et al. 2006; Steward et al. 2006; Wu et al. 2008). COMPASS and COMPASS-like complexes can mono-, di-, or trimethylate H3K4 through a conserved SET domain (Shilatifard 2006; Wu et al. 2008). The Cps35 subunit of COMPASS, which is required for proper H3K4 trimethylation via H2B monoubiquitination cross-talk (JS Lee et al. 2007; Zheng et al. 2010), is conserved in humans as the WDR82 protein (Fig. 1; Wu et al. 2008). However, WDR82 is only found to be associated with human Set1/COMPASS and not the MLL/COMPASS-like complexes (Fig. 1; Wu et al. 2008; P Wang et al. 2009).
Figure 1.
COMPASS and COMPASS-like complexes from yeast to human. COMPASS was identified in yeast as a complex of proteins associated with Set1 that can methylate H3 on Lys 4. Subsequently, six COMPASS-like complexes were identified in humans. All complexes share the core components Cps30 (WDR5), Cps50 (RBBP5), Cps25 (DPY30), and Cps60 (ASH2). COMPASS in humans also has CXXC and WDR82, which are homologous to Cps40 and Cps35 in yeast and regulate H3K4 trimethylation by COMPASS in vivo. Cps35 and WDR82 mediate the stimulation of H3K4 trimethylation upon H2B monoubiquitination, a process that occurs post-Pol II recruitment via the PAF complex. The MLL1/2 complexes contain the tumor suppressor Menin, which helps recruit the MLL1/2 complexes to HOX and other loci. The MLL3/4 complexes contain NCOA6, PTIP, and PA-1, which help target the MLL3/4 complexes to a distinct set of genes. The MLL3/4 complexes also contain the H3K27 demethylase UTX. The MLL1–4 COMPASS-like complexes function as coactivators of gene transcription in contrast to the canonical COMPASS complexes in yeast and humans. Set1s/MLLs are colored red, core components are colored green, and subunits with complex-specific functions are colored purple.
Clues to the developmental function of MLL came with its cloning, since it encoded a protein highly similar to Drosophila Trithorax (Gu et al. 1992; Tkachuk et al. 1992). Trithorax was identified as a homeotic mutation in flies. trx1 mutants had homeotic transformations of abdominal and thoracic segments to more anterior segmental identities, similar to mutations in the Hox genes from the Bithorax cluster (Ingham and Whittle 1980; Eissenberg and Shilatifard 2010). A role for H3K4 methylation in Hox gene regulation was demonstrated by a homeotic mutant allele (trxz11) that was a point mutation within the SET domain, which prevents histone binding and methylation by Trithorax (Katsani et al. 2001).
MLL, like its Drosophila ortholog, Trithorax, is a large, ∼4000-amino-acid protein that is essential for embryonic development and proper Hox gene expression (Yu et al. 1995). MLL is found in a COMPASS-like complex and, like yeast COMPASS, is capable of methylating histone H3K4 (Hughes et al. 2004; Shilatifard 2008). Despite being a histone methyltransferase, MLL can also have methylation-independent functions. Mll1ΔSET domain mice are viable, but there are defects in Hox expression (Terranova et al. 2006). The myriad of potential protein and DNA interaction motifs within the MLL N terminus may play important roles in leukemogenesis or MLL chimeras (Fig. 2). However, cells with a MLL translocation also contain a wild-type allele of MLL. Using a mouse model system, the wild-type copy of MLL was shown to be required for leukemogenesis (Thiel et al. 2010). One model to explain the requirement for both wild-type and translocated MLL alleles is that the wild-type copy of MLL may be required to initially recruit or stabilize the basal transcriptional machinery, while the translocated allele binding to the same genes via the MLL N-terminal domain interacts with transcription elongation factors through the translocation partner (Mohan et al. 2010).
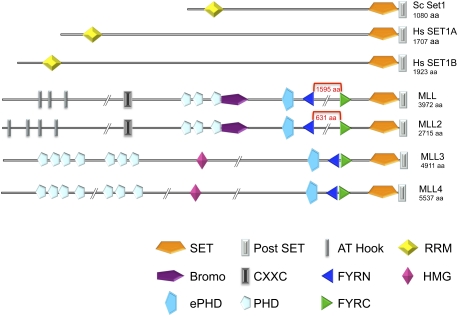
Figure 2.
Domain organization of Set1 and MLL-related proteins. Both Set1 and MLL-related proteins have highly conserved SET and post-SET domains. The Set1 subfamily also has N-terminal RRM motifs. MLL-related proteins have FYRN and FYRC regions. The MLL1–4 complexes have several PHD fingers and other domains associated with chromatin-associated proteins that contribute to targeting and activating properties of these complexes. The FYRN and FYRC regions of MLL and MLL2 are separated by hundreds of amino acids, while being juxtaposed in MLL3 and MLL4.
Differences between COMPASS and COMPASS-like complexes
While there is only one COMPASS in yeast, mammals have six COMPASS-like complexes, each capable of methylating H3K4 with essential and nonredundant functions (Fig. 1). All members of the Set1/MLL family have a highly conserved C-terminal SET and post-SET domain (Fig. 2; Dillon et al. 2005). Consistent with the relatively invariant C terminus of the Set1 family members, epitope tagging of yeast Set1 at the C terminus will kill its enzymatic activity (Krogan et al. 2002). We also demonstrated that tagging the C terminus of mammalian Set1 and MLL1 will also render the enzyme nonfunctional (M Wu and A Shilatifard, unpubl.). Outside of the SET domain and surrounding regions, the Set1/MLL proteins have substantial differences in their domain structure. Set1 proteins have one or two RNA recognition motifs (RRM), which are found in many RNA-binding proteins (Fig. 2; Tresaugues et al. 2006; Avramova 2009). The MLL1–4 proteins have multiple PHD fingers and a FYRN/FYRC (phenylalanine and tyrosine-rich N-terminal and C-terminal) domain. FYRN and FYRC domains are found in several different chromatin-associated factors. MLL1/2 and Trithorax proteins can be easily distinguished from MLL3/4 due to a large gap (1600 amino acids in the longest isoforms of human MLL) that splits FYRN and FYRC (Figs. 2, 3). Within this large gap are the Taspase cleavage sites where MLL1/2 and Trithorax are proteolytically cleaved in cells (Yokoyama et al. 2002; Hsieh et al. 2003; Takeda et al. 2006). The N-terminal and C-terminal cleavage products both participate in the same COMPASS-like complex (Hsieh et al. 2003; Hughes et al. 2004; Yokoyama et al. 2004; Wu et al. 2008). Based on a crystal structure of transforming growth factor β regulator 1 (TBRG1), we now know that the FYRN and FYRC fold together into a single domain (Fig. 3; Garcia-Alai et al. 2010), which may help explain why the N-terminal and C-terminal halves of MLL stay together in the cell after cleavage by Taspase. The MLL3/4 proteins are identified by having a FYRN/ FYRC and an ePHD finger just before the SET domains (Fig. 2; Avramova 2009).
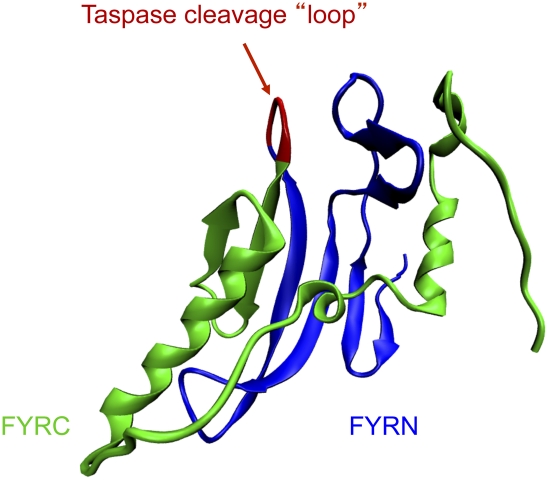
Figure 3.
Structure of the FYR domain. FYRN and FYRC motifs have been found in several chromatin-associated transcription factors in addition to the MLL family members. A crystal structure of the FYRN and FYRC regions of TGFBR was reported recently. It was found that FYRN and FYRC constitute a single domain, termed FYR. The FYR domain consists of N-terminal β sheets and C-terminal α helices. The N and C regions are separated by a small loop of approximately five amino acids in TGFBR, MLL3, and MLL4. For MLL, MLL2, and their Drosophila ortholog, Trithorax, this region is several hundred amino acids. MLL, MLL2, and Trithorax are proteolytically cleaved within this region by Taspase, but they are still found to self-associate within the same COMPASS-like complex. Recent genome-wide profiling also suggests that the N-terminal half of Trithorax has more binding sites in addition to those bound by the COMPASS-like complex.
Evidence for the importance of the cleavage of Trithorax or the MLL1/2 family proteins comes from mammalian studies, where it was shown that Taspase-deficient mice have homeotic transformations, suggesting that cleavage of these proteins may be necessary for their developmental functions (Takeda et al. 2006), although Taspase has been shown to cleave other substrates as well (Zhou et al. 2006; Capotosti et al. 2007). Furthermore, genome-wide profiling in Drosophila of N-terminal and C-terminal Trithorax reveals that, while both halves of Trithorax colocalize at many sites, they show major differences in their distributions (Schuettengruber et al. 2009; Schwartz et al. 2010). While both N-terminal and C-terminal halves of Trithorax are found to colocalize at Polycomb response elements (PREs) and transcription start sites of a subset of active genes, the N-terminal half of Trithorax is found over broad domains encompassing transcriptionally active PcG target genes (Schwartz et al. 2010). Separation of FYRN and FYRC and cleavage by Taspase may also have implications for the etiology of some MLL-rearranged leukemias, where FYRN and FYRC can be found in the reciprocal translocation that fuses the N terminus of the fusion partner to the C terminus of MLL. In the case of the most common translocation, MLL fuses to AFF1 (also known as AF4), Taspase cleaves the reciprocal AFF1-MLL fusion protein, and inhibition of this cleavage can lead to instability of this chimera. However, there are differing opinions on the contribution of the reciprocal AFF1-MLL fusion compared with MLL-AFF1 in the pathogenesis of leukemia (Bursen et al. 2010; Kumar et al. 2010).
COMPASS and COMPASS-like complexes can be characterized by their unique subunit compositions, whose identities provide insight into the different biological functions of these complexes. The SetD1A and SetD1B (KMT2F–G) complexes are the closest in composition to yeast COMPASS, and are collectively referred to as mammalian COMPASS (Fig. 1). One unique subunit of mammalian COMPASS is WDR82, which is related to yeast Cps35 (JH Lee et al. 2007; Lee and Skalnik 2008; Wu et al. 2008). Cps35 and its homolog, WDR82, are required for the observed cross-talk between histone H2B ubiquitination and H3K4 methylation (JS Lee et al. 2007; Wu et al. 2008; Zheng et al. 2010). As recruitment of the H2B monoubiquitination machinery occurs post-Pol II recruitment, H3K4 trimethylation by COMPASS occurs after the establishment of the basal transcription machinery, and therefore does not act as a major activator of transcription. This is different from the H3K4 methylation that is implemented by the MLL/COMPASS-like complexes.
MLL1 and MLL2 (KMT2A–B) both have an N-terminal domain that interacts with the tumor suppressor Menin. Menin helps target MLL1/2 to the Hox loci, and, in the absence of Menin, H3K4 trimethylation throughout the entire Hox cluster is abolished (Hughes et al. 2004; P Wang et al. 2009). Although MLL1 and MLL2 are found in identical macromolecular complexes, they appear to have both redundant and nonredundant functions at Hox loci (P Wang et al. 2009). Mll1 and Mll2 mouse knockouts also demonstrate that these two closely related genes are not redundant (Yu et al. 1995; Lubitz et al. 2007). Further evidence of nonredundancy between these two proteins is that Mll2 cannot substitute for Mll1 in mouse models of leukemogenesis (Bach et al. 2009).
MLL3 and MLL4 (KMT2C–D), which are orthologous to Trithorax-related in Drosophila, contain subunits important for targeting by nuclear receptors (Cho et al. 2007). One key subunit of these complexes may be UTX (KDM6A), an enzyme that can demethylate H3K27 (Hong et al. 2007; Lan et al. 2007; MG Lee et al. 2007; Smith et al. 2008a). H3K4 and H3K27 methylations are thought to be antagonistic. Indeed, Drosophila genetic studies originally identified Trithorax and Polycomb group (PcG) proteins as having opposing roles in the regulation of Hox genes, long before their roles in implementing H3K4 and H3K27 methylations were known (Eissenberg and Shilatifard 2010). Although MLL1/2 and Trithorax proteins are thought to be primary regulators of the Hox genes, MLL3/4 and Trithorax-related proteins, or their complexes, may also have a role in regulating Hox loci (Agger et al. 2007; De Santa et al. 2007; Lan et al. 2007; MG Lee et al. 2007). However, loss of Menin, which is a component of the MLL1/2 complexes and not the MLL3/4 complexes (Fig. 1), leaves little H3K4 methylation at Hox loci and almost no transcription of Hox genes (P Wang et al. 2009). Furthermore, mouse embryonic fibroblasts (MEFs) lacking Ptip, a component of the MLL3/4 complexes, show little change in H3K4 methylation or Hox transcription (P Wang et al. 2009).
In Drosophila, a presumptive null allele of trithorax-related (MLL3/4 ortholog) showed no homeotic mutations, nor did this allele act as a dominant modifier of Polycomb and trithorax alleles, further supporting that Trithorax-related is not a major regulator of Hox loci in Drosophila (Eissenberg and Shilatifard 2010). Interestingly, the nematode Caenorhabditis elegans has only one MLL-related protein, which is most similar to MLL3/4 in mammals and Trithorax-related in Drosophila (Fig. 4). C. elegans also has a markedly reduced Hox gene cluster, and its development is determined more by cell lineage than position within the embryo, which is regulated by the Hox genes and determines development in other animals such as flies and vertebrates (Aboobaker and Blaxter 2003).
Figure 4.
Ciliates contain a single Set1-related protein that most closely resembles the MLL1–4 proteins. (A) Dendogram of Set1/MLL family members from the ciliated protozoans T. thermophila and Paramecium tetraurelia, the nematodes C. elegans and Brugia malayi, and the fruit fly D. melanogaster, as well as humans. For MLL family members, the region from the ePHD through the post-Set domain was used for alignment with the full-length Set1 proteins from worms, flies, and humans. ClustalW and QuickTree (http://www.ngbw.org) were used to generate the phylogenetic tree (which was further processed at http://iTOL.embl.de). Branch distances were ignored for clarity of presentation. (B) Domain organization of MLL-related proteins in C. elegans and T. thermophila. Protein domain representations are the same as in Figure 2. Domains were identified by SMART (http://smart.embl-heidelberg.de), except for the FYR domain in Tetrahymena, which we predicted based on similarities with the FYR domain of Drosophila Trithorax-related and C. elegans set-16, as detected by HHpred (http://toolkit.tuebingen.mpg.de/hhpred).
One way in which H3K4 methylation by MLL/COMPASS-like complexes could help activate genes is through recruitment of trimethylated H3K4 (H3K4me3)-binding proteins (Yap and Zhou 2010). For example, H3K4me3 can directly recruit the basal transcription factors via the TAF3 subunit's PHD finger (Vermeulen et al. 2007). This enhanced affinity for TFIID at the promoters could be particularly important for genes with TATA-less promoters, a class of genes that includes many of the Hox loci in flies and mammals (Juven-Gershon and Kadonaga 2010). Thus, MLL1–4 recruitment to chromatin and methylation of H3K4 are likely to be independent of H2B monoubiquitination and generally precede transcription activation, in contrast to Set1/COMPASS. Sequence comparisons suggest that the ciliate Tetrahymena thermophila may have only a MLL-like protein, and not a Set1-like H3K4 methylase enzyme (Fig. 4). Interestingly, it was reported recently that H3K4me3 levels were not affected by loss of H2B monoubiquitination in Tetrahymena (Z Wang et al. 2009a). In support of Tetrahymena having only a MLL-type H3K4 methylase, loss of the ability to methylate H3K4 results in the loss of transcription of several hundred genes in Tetrahymena (Z Wang et al. 2009a). This differs from the situation in budding and fission yeast, where loss of H3K4 methylation by Set1/COMPASS has little effect on gene expression (Miller et al. 2001; Tanny et al. 2007). Tetrahymena, unlike Saccharomyces cerevisiae, has silencing programs associated with H3K27 methylation and Polycomb group proteins (Liu et al. 2007). Thus, Trithorax and Polycomb antagonism/regulation appears to be an ancient process.
Several subunits of COMPASS are shared components of other macromolecular complexes
Some components of Set1/COMPASS and MLL/COMPASS-like complexes have also been found in additional complexes. For example, Cps35/WDR82, a unique component of Set1/COMPASS, also participates in a complex with protein phosphatase 1 (PP1) in yeast and mammals (Nedea et al. 2008; JH Lee et al. 2010). LEDGF, which together with Menin is required for proper targeting of the MLL1/2 complexes, also participates in the HIV integrase complex (Yokoyama and Cleary 2008). PTIP and PA-1, components of the MLL3/4 complexes, also exist in a complex implicated in DNA repair (Gong et al. 2009).
One of the shared components of all COMPASS and COMPASS-like complexes is Cps30/WDR5. In yeast and mammals, Cps30/WDR5 is critical for assembly of COMPASS and COMPASS-like complexes (Krogan et al. 2002; Steward et al. 2006). Based on this information, WDR5 is sometimes tagged to isolate H3K4 methyltransferase complexes or targeted to eliminate H3K4 methylation in cells (Dou et al. 2005; Z Wang et al. 2009b). However, WDR5 is also a component of at least two different histone acetyltransferase complexes: the NSL/MSL1v complex, which acetylates histone H4 and p53 (Li et al. 2009; Cai et al. 2010), and the ATAC complex, which contains the H3K9 and K14 acetyltransferase GCN5 (KAT2) (Suganuma et al. 2008; Wang et al. 2008). Therefore, tagging Wdr5 will pull down both the H3K4 methylase complexes and the histone acetyltransferase complexes. Furthermore, the association of WDR5 with so many transcription-associated complexes makes it a poor choice for targeting by RNAi for the purpose of looking for effects due to only H3K4 methylation (JS Lee et al. 2010).
Is there a common function for WDR5 in these distinct chromatin-modifying complexes? One proposal has been that WDR5 helps target complexes to chromatin through the interaction with H3 tails. This is based on the observation that a dimethylated H3 tail peptide could pull WDR5 out of cell extracts (Wysocka et al. 2005). It was later shown that WDR5 could interact with the H3 tails independently of the methylation status of the lysine (Couture et al. 2006; Ruthenburg et al. 2006). One of the major determinants of WDR5 binding to the H3 tail is Arg 2 (Trievel and Shilatifard 2009). Another arginine-bearing sequence was found in Set1 and MLL-related proteins; interaction of WDR5 with this motif (Win) is required for the assembly and activity of COMPASS and COMPASS-like complexes (Patel et al. 2008; Song and Kingston 2008). Since WDR5 cannot interact simultaneously with COMPASS and the H3 tail, this rules out a role for WDR5 in presenting the H3 tail for COMPASS to methylate (Trievel and Shilatifard 2009). The presence of WDR5 in multiple complexes (many of which do not use H3K4 as a substrate) could reflect a common role as a scaffold in complex assembly, rather than in substrate recognition. Clearly, WDR5 is not the component of the MLL complexes required for gene target specificity, but rather functions in the assembly of both the Set1/COMPASS and MLL/COMPASS-like complexes. However, the MLL target genes play an essential role in MLL translocation-based leukemic pathogenesis. Therefore, identification of such target genes is central for a better molecular understanding and the treatment of MLL translocation-based leukemia.
Misregulation of MLL target genes via the SEC
MLL has >60 different molecularly characterized translocation partners, which share no single common feature or cellular role (Meyer et al. 2009). However, the most common translocation partners are nuclear. In order to determine how MLL translocations lead to leukemia, we purified several MLL fusion proteins: MLL-AFF1, MLL-ENL, MLL-AF9, and MLL-ELL (Lin et al. 2010). Our purification of these MLL chimeras led to the identification of SEC that contains ELL1–3, ELL-associated factor (EAF1/2), several of the MLL translocation partners, and the Pol II CTD kinase P-TEFb (Lin et al. 2010).
The ELL gene on chromosome 19p13.1 was identified as one of the translocation partners of MLL found in hematological malignancies (Thirman et al. 1994; Rowley 1998). Human ELL was demonstrated to be a Pol II elongation factor capable of increasing the catalytic rate of transcription elongation by reducing transient pausing (Shilatifard et al. 1996; Shilatifard 1998). To date, ELL is the best functionally characterized MLL partner (Shilatifard 1998). In humans, there are three family members of ELL; namely, ELL (or ELL1), ELL2, and ELL3 (Miller et al. 2000). In Drosophila, there is only one ELL family member (dELL) also capable of functioning as a Pol II elongation factor both in vitro and in vivo (Gerber et al. 2001; Eissenberg et al. 2002; Smith et al. 2008b).
P-TEFb, like ELL, was identified through in vitro transcription assays (Marshall and Price 1995). P-TEFb is comprised of two subunits: the kinase Cdk9 and its regulatory subunit, Cyclin T1 or Cyclin T2. P-TEFb stimulates transcription elongation through the phosphorylation of the RNA Pol II CTD as well as through the phosphorylation of NELF and DSIF (Yamaguchi et al. 1999; Peterlin and Price 2006). DSIF/NELF plays a role in the pausing of Pol II at genes, and phosphorylation of DSIF/NELF by P-TEFb is thought to be a regulatory step in the release of Pol II.
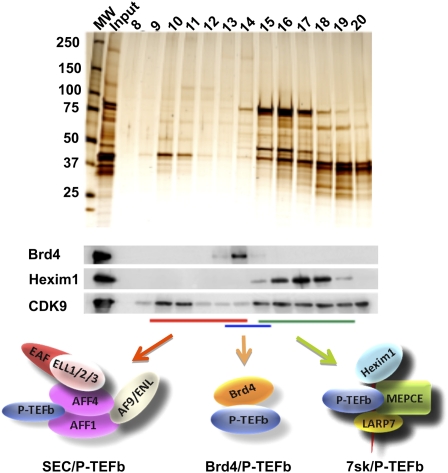
Size fractionation of nuclear extracts demonstrated that only a small fraction of P-TEFb is in the SEC (Fig. 5), but this fraction is highly active as a CTD kinase (Lin et al. 2010). P-TEFb had been isolated previously in two different complexes. The HEXIM1/7SK complex is a small nuclear RNA protein complex that inhibits P-TEFb activity (Zhou and Yik 2006). The double bromodomain protein BRD4 has also been found to associate with P-TEFb (Jang et al. 2005; Yang et al. 2005). BRD4/P-TEFb, unlike the HEXIM1 complex, is active as a CTD kinase, although it was shown not to be required for P-TEFb-dependent enhancement of HIV transcription (Jang et al. 2005; Yang et al. 2005).
Figure 5.
Multiple P-TEFb complexes. P-TEFb, consisting of CDK9 and Cyclin T, is found in multiple complexes, which can be separated by gel filtration chromatography. CDK9-containing complexes from 293 cells were isolated by Flag affinity purification and subjected to size exclusion analysis. The resulting fractions were analyzed by silver staining and Western blotting. The inactive HEXIM1-containing P-TEFb complex, represented by HEXIM1, was enriched in fractions 15–19. The BRD4/P-TEFb complex peaked at fractions 13–15. SEC complexes are the largest P-TEFb complexes and are found in fractions 10–14, as described previously (Lin et al. 2010).
AFF4 is required for SEC integrity, and its knockdown in MLL translocation-carrying leukemic cells led to the loss of HOXA9 expression, a key target of MLL fusion proteins (Lin et al. 2010). The fact that so many MLL fusion proteins associate with the SEC suggests that regulation of transcription elongation is disrupted in the MLL-rearranged leukemias. These translocations, which lead to the fusion of MLL's N terminus with a C-terminal portion of another protein, can result in the recruitment of these chimera complexes to MLL's normal target genes, leading to premature activation of transcription elongation. It is also possible that genes that normally recruit the SEC could gain altered activity by corecruiting MLL's N-terminal domain, the associated Menin, and other proteins to give aberrant transcription. Gene expression studies have identified genes within the HOX clusters as the most frequently up-regulated in MLL-rearranged leukemias, and these are thought to be among the most important MLL target genes because their misexpression is required for leukemogenesis (Yu et al. 1995; Ferrando et al. 2003). Since the SEC forms an active P-TEFb complex, and since some HOX genes in undifferentiated cells have paused Pol II at their promoters, HOX genes may also be regulated by the SEC during normal development. The misexpression of the HOX genes in the MLL-rearranged leukemias could result from the stabilized recruitment of SEC and the concomitant release of Pol II in an unregulated manner (Mohan et al. 2010).
The SEC is required for HIV pathogenesis
The SEC was also found to play a critical role in HIV replication (He et al. 2010; Sobhian et al. 2010). Transcription of the provirus has long been known to be regulated at one of the steps of transcription elongation (Peterlin and Price 2006). The HIV-encoded Tat protein is an RNA-binding protein that binds a stem–loop structure Tat response element (TAR) early in transcription from the long terminal repeat (LTR). At early stages of HIV transcription, not enough Tat protein has accumulated and full-length HIV transcripts are not made. When enough Tat is present, Tat interacts with TAR and stimulates Pol II to transcribe through the recruitment of P-TEFb. Purifications of Tat and CDK9 identified the same set of components found in the SEC (He et al. 2010; Sobhian et al. 2010). Unlike the BRD4-containing P-TEFb complex, the SEC is required for Tat-mediated transactivation (He et al. 2010; Sobhian et al. 2010).
Tat competes with BRD4 for binding to P-TEFb (Yang et al. 2005). Indeed, a peptide of BRD4 that corresponds to its P-TEFb interaction domain can inhibit HIV transcription (Bisgrove et al. 2007). By disrupting Tat's interaction with P-TEFb, this peptide would also prevent recruitment of other SEC components to the HIV LTR. It will be interesting to determine if this peptide disrupts SEC components such as AFF1 and AFF4 from interacting with P-TEFb. The ability of Tat to interact with the SEC, but not with the BRD4/P-TEFb complexes, suggests that the BRD4 peptide might not interfere with normal SEC function, making it an attractive therapeutic strategy, since inhibiting normal SEC function might interfere with essential cellular functions.
Are there different versions of the SEC in cells on chromatin?
AFF1 is the most frequent translocation partner of MLL. AFF4, a related protein, is a less frequent partner, and AFF3 has been found in one translocation with MLL. AFF2 is silenced by triplet repeat expansion and transcriptional silencing in FRAXE mental retardation (Gecz et al. 1996; Gu et al. 1996). Thus, all members of this family are linked to human disease. Altogether, four AFF proteins and three ELL proteins, together with the ENL/AF9 paralogs, give enormous regulatory potential for SEC complexes in mammals. In contrast, Drosophila has one ELL, one ENL/AF9, and a single AFF protein. The Drosophila homolog of the AFF1–4 proteins is encoded by the lilliputian (lilli) gene. Lilli is essential for development, with loss-of-function alleles being embryonic-lethal (Su et al. 2001; Tang et al. 2001; Wittwer et al. 2001). Lilli is required for some of the earliest transcription in Drosophila embryos, regulating ftz and Sxl genes (Vanderzwan-Butler et al. 2007). Lilli has been identified genetically as a dosage-sensitive modulator of many signaling pathways (Su et al. 2001; Tang et al. 2001; Wittwer et al. 2001). Importantly, alleles of dELL and lilli were frequently identified in genetic screens looking for modifiers of Ras signaling in the Drosophila eye (Neufeld et al. 1998; Rebay et al. 2000; Su et al. 2001; Tang et al. 2001; Wittwer et al. 2001; Eissenberg et al. 2002). Drosophila ELL, like Lilli, is essential for development, with loss-of-function alleles leading to embryonic lethality with defects in patterning of the embryo (Eissenberg et al. 2002). Consistent with Drosophila ELL interacting in a fly version of the SEC, the knockdown of ELL protein levels in larvae leads to reductions in Ser 2 phosphorylation (Smith et al. 2008b). Our biochemical analysis indicates that a Drosophila SEC consists of ELL, Lilli, Cdk9, Cyclin T, and Ear, the Drosophila homolog of ENL and AF9 (E Smith and A Shilatifard, unpubl.). Drosophila SEC components localize to Hsp70 upon heat shock, a well-characterized model for the regulation of transcription by the controlled release of Pol II (Lin et al. 2010). This raises the question of whether, and to what extent, the Drosophila SEC regulates the large number of developmental genes with poised Pol II identified in Drosophila embryos (Muse et al. 2007; Zeitlinger et al. 2007; Boettiger and Levine 2009). Since Drosophila has only one ELL, one AFF4-related protein, and one ENL, it is an attractive model system for studying the role of these factors during development. However, due to their role in human diseases, determining the specificity of function of the four AFF proteins in mammals will also be required (Fig. 6).
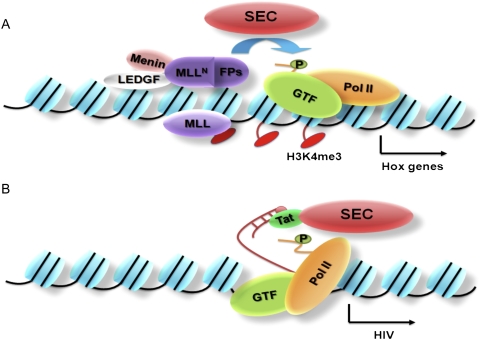
Figure 6.
The SEC regulates transcription elongation in disease pathogenesis. (A) Purification of MLL fusion proteins led to the identification of the SEC, which contains the transcription elongation factor ELL and the RNA Pol II CTD kinase P-TEFb. The SEC contains several proteins frequently fused to the MLL N-terminal region in MLL, including ELL, AFF1, AFF4, ENL, and AF9. The N-terminal portion of MLL, which lacks the SET domain, can still interact with chromatin with the help of Menin and LEDGF at genes with poised Pol II, such as the Hox loci. When fused to SEC components, MLLN can direct improper stimulation of transcription elongation through phosphorylation of the RNA Pol II CTD at Ser 2. (B) The SEC was later identified in a complex with the HIV Tat protein. Tat associates with the TAR region of nascent HIV transcripts, and subsequently stimulates Pol II elongation in a step that requires Pol II CTD phosphorylation at Ser 2. We now know that this regulatory step requires SEC components ELL2 and AFF4.
Summary and future directions
Despite years of studying the activities that stimulate transcription elongation in vitro, an understanding of the in vivo role of these factors has been very limited. Recent work, however, has begun to provide new insight into some of these factors. For example, NELF, identified as an inhibitor of transcription elongation in vitro, is now known to be required for the proper expression, and not just the repression, of many genes (Gilchrist et al. 2008). NELF may help set up the transcriptionally poised state until it is phosphorylated by P-TEFb (Gilchrist et al. 2008). The purification of common translocation partners of MLL led to the identification of the SEC, containing ELL and P-TEFb, each previously identified independently by in vitro studies (Lin et al. 2010). We now know that the SEC is required for the overexpression of HOX genes by MLL chimeras (Lin et al. 2010). Determining the way in which the SEC, NELF, and other transcription elongation factors interact with each other to regulate specific target genes is an exciting avenue for future investigation. The growing evidence that transcriptional elongation is a major regulatory step during early development, along with the interactions between MLL and the SEC in the expression of the HOX genes in leukemia, demonstrates that transcription elongation and histone methylation are just two parts of a complex chromatin signaling pathway in development. Understanding more of these links will provide additional potential therapeutic targets for MLL-rearranged leukemias as well as other diseases.
Acknowledgments
We are grateful to Laura Shilatifard for editorial assistance. C.L. is a PhD student registered with the Open University. The work in our laboratory is supported in part by generous funding from the National Institutes of Health (R01GM069905, R01CA150265, and R01CA89455).
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.2015411.
References
- Aboobaker AA, Blaxter ML 2003. Hox gene loss during dynamic evolution of the nematode cluster. Curr Biol 13: 37–40 [DOI] [PubMed] [Google Scholar]
- Agger K, Cloos PA, Christensen J, Pasini D, Rose S, Rappsilber J, Issaeva I, Canaani E, Salcini AE, Helin K 2007. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature 449: 731–734 [DOI] [PubMed] [Google Scholar]
- Aida M, Chen Y, Nakajima K, Yamaguchi Y, Wada T, Handa H 2006. Transcriptional pausing caused by NELF plays a dual role in regulating immediate-early expression of the junB gene. Mol Cell Biol 26: 6094–6104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akoulitchev S, Makela TP, Weinberg RA, Reinberg D 1995. Requirement for TFIIH kinase activity in transcription by RNA polymerase II. Nature 377: 557–560 [DOI] [PubMed] [Google Scholar]
- Aso T, Lane WS, Conaway JW, Conaway RC 1995. Elongin (SIII): a multisubunit regulator of elongation by RNA polymerase II. Science 269: 1439–1443 [DOI] [PubMed] [Google Scholar]
- Avramova Z 2009. Evolution and pleiotropy of TRITHORAX function in Arabidopsis. Int J Dev Biol 53: 371–381 [DOI] [PubMed] [Google Scholar]
- Bach C, Mueller D, Buhl S, Garcia-Cuellar MP, Slany RK 2009. Alterations of the CxxC domain preclude oncogenic activation of mixed-lineage leukemia 2. Oncogene 28: 815–823 [DOI] [PubMed] [Google Scholar]
- Bisgrove DA, Mahmoudi T, Henklein P, Verdin E 2007. Conserved P-TEFb-interacting domain of BRD4 inhibits HIV transcription. Proc Natl Acad Sci 104: 13690–13695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boettiger AN, Levine M 2009. Synchronous and stochastic patterns of gene activation in the Drosophila embryo. Science 325: 471–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bursen A, Schwabe K, Ruster B, Henschler R, Ruthardt M, Dingermann T, Marschalek R 2010. The AF4.MLL fusion protein is capable of inducing ALL in mice without requirement of MLL.AF4. Blood 115: 3570–3579 [DOI] [PubMed] [Google Scholar]
- Cai Y, Jin J, Swanson SK, Cole MD, Choi SH, Florens L, Washburn MP, Conaway JW, Conaway RC 2010. Subunit composition and substrate specificity of a MOF-containing histone acetyltransferase distinct from the male-specific lethal (MSL) complex. J Biol Chem 285: 4268–4272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capotosti F, Hsieh JJ, Herr W 2007. Species selectivity of mixed-lineage leukemia/trithorax and HCF proteolytic maturation pathways. Mol Cell Biol 27: 7063–7072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Yamaguchi Y, Tsugeno Y, Yamamoto J, Yamada T, Nakamura M, Hisatake K, Handa H 2009. DSIF, the Paf1 complex, and Tat-SF1 have nonredundant, cooperative roles in RNA polymerase II elongation. Genes Dev 23: 2765–2777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho YW, Hong T, Hong S, Guo H, Yu H, Kim D, Guszczynski T, Dressler GR, Copeland TD, Kalkum M et al. 2007. PTIP associates with MLL3- and MLL4-containing histone H3 lysine 4 methyltransferase complex. J Biol Chem 282: 20385–20406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Core LJ, Waterfall JJ, Lis JT 2008. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science 322: 1845–1848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couture JF, Collazo E, Trievel RC 2006. Molecular recognition of histone H3 by the WD40 protein WDR5. Nat Struct Mol Biol 13: 698–703 [DOI] [PubMed] [Google Scholar]
- De Santa F, Totaro MG, Prosperini E, Notarbartolo S, Testa G, Natoli G 2007. The histone H3 lysine-27 demethylase Jmjd3 links inflammation to inhibition of polycomb-mediated gene silencing. Cell 130: 1083–1094 [DOI] [PubMed] [Google Scholar]
- Dillon SC, Zhang X, Trievel RC, Cheng X 2005. The SET-domain protein superfamily: protein lysine methyltransferases. Genome Biol 6: 227 doi: 10.1186/gb-2005-6-8-227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou Y, Milne TA, Tackett AJ, Smith ER, Fukuda A, Wysocka J, Allis CD, Chait BT, Hess JL, Roeder RG 2005. Physical association and coordinate function of the H3 K4 methyltransferase MLL1 and the H4 K16 acetyltransferase MOF. Cell 121: 873–885 [DOI] [PubMed] [Google Scholar]
- Dou Y, Milne TA, Ruthenburg AJ, Lee S, Lee JW, Verdine GL, Allis CD, Roeder RG 2006. Regulation of MLL1 H3K4 methyltransferase activity by its core components. Nat Struct Mol Biol 13: 713–719 [DOI] [PubMed] [Google Scholar]
- Dvir A, Conaway RC, Conaway JW 1997. A role for TFIIH in controlling the activity of early RNA polymerase II elongation complexes. Proc Natl Acad Sci 94: 9006–9010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eissenberg JC, Shilatifard A 2010. Histone H3 lysine 4 (H3K4) methylation in development and differentiation. Dev Biol 339: 240–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eissenberg JC, Ma J, Gerber MA, Christensen A, Kennison JA, Shilatifard A 2002. dELL is an essential RNA polymerase II elongation factor with a general role in development. Proc Natl Acad Sci 99: 9894–9899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrando AA, Armstrong SA, Neuberg DS, Sallan SE, Silverman LB, Korsmeyer SJ, Look AT 2003. Gene expression signatures in MLL-rearranged T-lineage and B-precursor acute leukemias: dominance of HOX dysregulation. Blood 102: 262–268 [DOI] [PubMed] [Google Scholar]
- Garcia-Alai MM, Allen MD, Joerger AC, Bycroft M 2010. The structure of the FYR domain of transforming growth factor β regulator 1. Protein Sci 19: 1432–1438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gecz J, Gedeon AK, Sutherland GR, Mulley JC 1996. Identification of the gene FMR2, associated with FRAXE mental retardation. Nat Genet 13: 105–108 [DOI] [PubMed] [Google Scholar]
- Gerber M, Ma J, Dean K, Eissenberg JC, Shilatifard A 2001. Drosophila ELL is associated with actively elongating RNA polymerase II on transcriptionally active sites in vivo. EMBO J 20: 6104–6114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber M, Tenney K, Conaway JW, Conaway RC, Eissenberg JC, Shilatifard A 2005. Regulation of heat shock gene expression by RNA polymerase II elongation factor, Elongin A. J Biol Chem 280: 4017–4020 [DOI] [PubMed] [Google Scholar]
- Gilchrist DA, Nechaev S, Lee C, Ghosh SK, Collins JB, Li L, Gilmour DS, Adelman K 2008. NELF-mediated stalling of Pol II can enhance gene expression by blocking promoter–proximal nucleosome assembly. Genes Dev 22: 1921–1933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour DS, Lis JT 1986. RNA polymerase II interacts with the promoter region of the noninduced hsp70 gene in Drosophila melanogaster cells. Mol Cell Biol 6: 3984–3989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Z, Cho YW, Kim JE, Ge K, Chen J 2009. Accumulation of Pax2 transactivation domain interaction protein (PTIP) at sites of DNA breaks via RNF8-dependent pathway is required for cell survival after DNA damage. J Biol Chem 284: 7284–7293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Y, Nakamura T, Alder H, Prasad R, Canaani O, Cimino G, Croce CM, Canaani E 1992. The t(4;11) chromosome translocation of human acute leukemias fuses the ALL-1 gene, related to Drosophila trithorax, to the AF-4 gene. Cell 71: 701–708 [DOI] [PubMed] [Google Scholar]
- Gu Y, Shen Y, Gibbs RA, Nelson DL 1996. Identification of FMR2, a novel gene associated with the FRAXE CCG repeat and CpG island. Nat Genet 13: 109–113 [DOI] [PubMed] [Google Scholar]
- Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA 2007. A chromatin landmark and transcription initiation at most promoters in human cells. Cell 130: 77–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He N, Liu M, Hsu J, Xue Y, Chou S, Burlingame A, Krogan NJ, Alber T, Zhou Q 2010. HIV-1 Tat and host AFF4 recruit two transcription elongation factors into a bifunctional complex for coordinated activation of HIV-1 transcription. Mol Cell 38: 428–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holstege FC, van der Vliet PC, Timmers HT 1996. Opening of an RNA polymerase II promoter occurs in two distinct steps and requires the basal transcription factors IIE and IIH. EMBO J 15: 1666–1677 [PMC free article] [PubMed] [Google Scholar]
- Hong S, Cho YW, Yu LR, Yu H, Veenstra TD, Ge K 2007. Identification of JmjC domain-containing UTX and JMJD3 as histone H3 lysine 27 demethylases. Proc Natl Acad Sci 104: 18439–18444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh JJ, Ernst P, Erdjument-Bromage H, Tempst P, Korsmeyer SJ 2003. Proteolytic cleavage of MLL generates a complex of N- and C-terminal fragments that confers protein stability and subnuclear localization. Mol Cell Biol 23: 186–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes CM, Rozenblatt-Rosen O, Milne TA, Copeland TD, Levine SS, Lee JC, Hayes DN, Shanmugam KS, Bhattacharjee A, Biondi CA, et al. 2004. Menin associates with a trithorax family histone methyltransferase complex and with the hoxc8 locus. Mol Cell 13: 587–597 [DOI] [PubMed] [Google Scholar]
- Ingham PW, Whittle R 1980. Trithorax: a new homeotic mutation of Drosophila melanogaster causing transformations of abdominal and thoracic imaginal segments. Mol Gen Genet 179: 607–614 [Google Scholar]
- Jang MK, Mochizuki K, Zhou M, Jeong HS, Brady JN, Ozato K 2005. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol Cell 19: 523–534 [DOI] [PubMed] [Google Scholar]
- Juven-Gershon T, Kadonaga JT 2010. Regulation of gene expression via the core promoter and the basal transcriptional machinery. Dev Biol 339: 225–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao SY, Calman AF, Luciw PA, Peterlin BM 1987. Anti-termination of transcription within the long terminal repeat of HIV-1 by tat gene product. Nature 330: 489–493 [DOI] [PubMed] [Google Scholar]
- Katsani KR, Arredondo JJ, Kal AJ, Verrijzer CP 2001. A homeotic mutation in the trithorax SET domain impedes histone binding. Genes Dev 15: 2197–2202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan NJ, Dover J, Khorrami S, Greenblatt JF, Schneider J, Johnston M, Shilatifard A 2002. COMPASS, a histone H3 (Lysine 4) methyltransferase required for telomeric silencing of gene expression. J Biol Chem 277: 10753–10755 [DOI] [PubMed] [Google Scholar]
- Krumm A, Meulia T, Brunvand M, Groudine M 1992. The block to transcriptional elongation within the human c-myc gene is determined in the promoter-proximal region. Genes Dev 6: 2201–2213 [DOI] [PubMed] [Google Scholar]
- Kumar AR, Yao Q, Li Q, Sam TA, Kersey JH 2010. t(4;11) leukemias display addiction to MLL-AF4 but not to AF4-MLL. Leuk Res doi: 10.1016/j.leukres.2010.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan F, Bayliss PE, Rinn JL, Whetstine JR, Wang JK, Chen S, Iwase S, Alpatov R, Issaeva I, Canaani E, et al. 2007. A histone H3 lysine 27 demethylase regulates animal posterior development. Nature 449: 689–694 [DOI] [PubMed] [Google Scholar]
- Laspia MF, Rice AP, Mathews MB 1989. HIV-1 Tat protein increases transcriptional initiation and stabilizes elongation. Cell 59: 283–292 [DOI] [PubMed] [Google Scholar]
- Lee JH, Skalnik DG 2008. Wdr82 is a C-terminal domain-binding protein that recruits the Setd1A Histone H3-Lys4 methyltransferase complex to transcription start sites of transcribed human genes. Mol Cell Biol 28: 609–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Tate CM, You JS, Skalnik DG 2007. Identification and characterization of the human Set1B histone H3-Lys4 methyltransferase complex. J Biol Chem 282: 13419–13428 [DOI] [PubMed] [Google Scholar]
- Lee JS, Shukla A, Schneider J, Swanson SK, Washburn MP, Florens L, Bhaumik SR, Shilatifard A 2007. Histone Crosstalk between H2B Monoubiquitination and H3 Methylation Mediated by COMPASS. Cell 131: 1084–1096 [DOI] [PubMed] [Google Scholar]
- Lee MG, Villa R, Trojer P, Norman J, Yan KP, Reinberg D, Di Croce L, Shiekhattar R 2007. Demethylation of H3K27 regulates polycomb recruitment and H2A ubiquitination. Science 318: 447–450 [DOI] [PubMed] [Google Scholar]
- Lee JH, You J, Dobrota E, Skalnik DG 2010. Identification and characterization of a novel human PP1 phosphatase complex. J Biol Chem 285: 24466–24476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JS, Smith E, Shilatifard A 2010. The language of histone crosstalk. Cell 142: 682–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Wu L, Corsa CA, Kunkel S, Dou Y 2009. Two mammalian MOF complexes regulate transcription activation by distinct mechanisms. Mol Cell 36: 290–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C, Smith ER, Takahashi H, Lai KC, Martin-Brown S, Florens L, Washburn MP, Conaway JW, Conaway RC, Shilatifard A 2010. AFF4, a component of the ELL/P-TEFb elongation complex and a shared subunit of MLL chimeras, can link transcription elongation to leukemia. Mol Cell 37: 429–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Taverna SD, Muratore TL, Shabanowitz J, Hunt DF, Allis CD 2007. RNAi-dependent H3K27 methylation is required for heterochromatin formation and DNA elimination in Tetrahymena. Genes Dev 21: 1530–1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubitz S, Glaser S, Schaft J, Stewart AF, Anastassiadis K 2007. Increased apoptosis and skewed differentiation in mouse embryonic stem cells lacking the histone methyltransferase Mll2. Mol Biol Cell 18: 2356–2366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall NF, Price DH 1992. Control of formation of two distinct classes of RNA polymerase II elongation complexes. Mol Cell Biol 12: 2078–2090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall NF, Price DH 1995. Purification of P-TEFb, a transcription factor required for the transition into productive elongation. J Biol Chem 270: 12335–12338 [DOI] [PubMed] [Google Scholar]
- Meyer C, Kowarz E, Hofmann J, Renneville A, Zuna J, Trka J, Ben Abdelali R, Macintyre E, De Braekeleer E, De Braekeleer M, et al. 2009. New insights to the MLL recombinome of acute leukemias. Leukemia 23: 1490–1499 [DOI] [PubMed] [Google Scholar]
- Miller T, Williams K, Johnstone RW, Shilatifard A 2000. Identification, cloning, expression, and biochemical characterization of the testis-specific RNA polymerase II elongation factor ELL3. J Biol Chem 275: 32052–32056 [DOI] [PubMed] [Google Scholar]
- Miller T, Krogan NJ, Dover J, Erdjument-Bromage H, Tempst P, Johnston M, Greenblatt JF, Shilatifard A 2001. COMPASS: a complex of proteins associated with a trithorax-related SET domain protein. Proc Natl Acad Sci 98: 12902–12907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohan M, Lin C, Guest E, Shilatifard A 2010. Licensed to elongate: a molecular mechanism for MLL-based leukaemogenesis. Nat Rev Cancer 10: 721–728 [DOI] [PubMed] [Google Scholar]
- Muse GW, Gilchrist DA, Nechaev S, Shah R, Parker JS, Grissom SF, Zeitlinger J, Adelman K 2007. RNA polymerase is poised for activation across the genome. Nat Genet 39: 1507–1511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nechaev S, Fargo DC, dos Santos G, Liu L, Gao Y, Adelman K 2010. Global analysis of short RNAs reveals widespread promoter-proximal stalling and arrest of Pol II in Drosophila. Science 327: 335–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nedea E, Nalbant D, Xia D, Theoharis NT, Suter B, Richardson CJ, Tatchell K, Kislinger T, Greenblatt JF, Nagy PL 2008. The Glc7 phosphatase subunit of the cleavage and polyadenylation factor is essential for transcription termination on snoRNA genes. Mol Cell 29: 577–587 [DOI] [PubMed] [Google Scholar]
- Neufeld TP, Tang AH, Rubin GM 1998. A genetic screen to identify components of the sina signaling pathway in Drosophila eye development. Genetics 148: 277–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni Z, Saunders A, Fuda NJ, Yao J, Suarez JR, Webb WW, Lis JT 2008. P-TEFb is critical for the maturation of RNA polymerase II into productive elongation in vivo. Mol Cell Biol 28: 1161–1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orphanides G, LeRoy G, Chang CH, Luse DS, Reinberg D 1998. FACT, a factor that facilitates transcript elongation through nucleosomes. Cell 92: 105–116 [DOI] [PubMed] [Google Scholar]
- Patel A, Dharmarajan V, Cosgrove MS 2008. Structure of WDR5 bound to mixed lineage leukemia protein-1 peptide. J Biol Chem 283: 32158–32161 [DOI] [PubMed] [Google Scholar]
- Peterlin BM, Price DH 2006. Controlling the elongation phase of transcription with P-TEFb. Mol Cell 23: 297–305 [DOI] [PubMed] [Google Scholar]
- Rebay I, Chen F, Hsiao F, Kolodziej PA, Kuang BH, Laverty T, Suh C, Voas M, Williams A, Rubin GM 2000. A genetic screen for novel components of the Ras/Mitogen-activated protein kinase signaling pathway that interact with the yan gene of Drosophila identifies split ends, a new RNA recognition motif-containing protein. Genetics 154: 695–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reines D, Conaway JW, Conaway RC 1996. The RNA polymerase II general elongation factors. Trends Biochem Sci 21: 351–355 [PMC free article] [PubMed] [Google Scholar]
- Rougvie AE, Lis JT 1988. The RNA polymerase II molecule at the 5′ end of the uninduced hsp70 gene of D. melanogaster is transcriptionally engaged. Cell 54: 795–804 [DOI] [PubMed] [Google Scholar]
- Rowley JD 1998. The critical role of chromosome translocations in human leukemias. Annu Rev Genet 32: 495–519 [DOI] [PubMed] [Google Scholar]
- Ruthenburg AJ, Wang W, Graybosch DM, Li H, Allis CD, Patel DJ, Verdine GL 2006. Histone H3 recognition and presentation by the WDR5 module of the MLL1 complex. Nat Struct Mol Biol 13: 704–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders A, Core LJ, Lis JT 2006. Breaking barriers to transcription elongation. Nat Rev Mol Cell Biol 7: 557–567 [DOI] [PubMed] [Google Scholar]
- Schuettengruber B, Ganapathi M, Leblanc B, Portoso M, Jaschek R, Tolhuis B, van Lohuizen M, Tanay A, Cavalli G 2009. Functional anatomy of polycomb and trithorax chromatin landscapes in Drosophila embryos. PLoS Biol 7: e13 doi: 10.1371/journal.pbio.1000013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz YB, Kahn TG, Stenberg P, Ohno K, Bourgon R, Pirrotta V 2010. Alternative epigenetic chromatin states of polycomb target genes. PLoS Genet 6: e1000805 doi: 10.1371/journal.pgen.1000805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shilatifard A 1998. Factors regulating the transcriptional elongation activity of RNA polymerase II. FASEB J 12: 1437–1446 [DOI] [PubMed] [Google Scholar]
- Shilatifard A 2006. Chromatin modifications by methylation and ubiquitination: implications in the regulation of gene expression. Annu Rev Biochem 75: 243–269 [DOI] [PubMed] [Google Scholar]
- Shilatifard A 2008. Molecular implementation and physiological roles for histone H3 lysine 4 (H3K4) methylation. Curr Opin Cell Biol 20: 341–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shilatifard A, Lane WS, Jackson KW, Conaway RC, Conaway JW 1996. An RNA polymerase II elongation factor encoded by the human ELL gene. Science 271: 1873–1876 [DOI] [PubMed] [Google Scholar]
- Shilatifard A, Conaway RC, Conaway JW 2003. The RNA polymerase II elongation complex. Annu Rev Biochem 72: 693–715 [DOI] [PubMed] [Google Scholar]
- Sims RJ 3rd, Belotserkovskaya R, Reinberg D 2004. Elongation by RNA polymerase II: the short and long of it. Genes Dev 18: 2437–2468 [DOI] [PubMed] [Google Scholar]
- Smith ER, Lee MG, Winter B, Droz NM, Eissenberg JC, Shiekhattar R, Shilatifard A 2008a. Drosophila UTX is a histone H3 Lys27 demethylase that colocalizes with the elongating form of RNA polymerase II. Mol Cell Biol 28: 1041–1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ER, Winter B, Eissenberg JC, Shilatifard A 2008b. Regulation of the transcriptional activity of poised RNA polymerase II by the elongation factor ELL. Proc Natl Acad Sci 105: 8575–8579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobhian B, Laguette N, Yatim A, Nakamura M, Levy Y, Kiernan R, Benkirane M 2010. HIV-1 Tat assembles a multifunctional transcription elongation complex and stably associates with the 7SK snRNP. Mol Cell 38: 439–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song JJ, Kingston RE 2008. WDR5 interacts with mixed lineage leukemia (MLL) protein via the histone H3-binding pocket. J Biol Chem 283: 35258–35264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steward MM, Lee JS, O'Donovan A, Wyatt M, Bernstein BE, Shilatifard A 2006. Molecular regulation of H3K4 trimethylation by ASH2L, a shared subunit of MLL complexes. Nat Struct Mol Biol 13: 852–854 [DOI] [PubMed] [Google Scholar]
- Su MA, Wisotzkey RG, Newfeld SJ 2001. A screen for modifiers of decapentaplegic mutant phenotypes identifies lilliputian, the only member of the Fragile-X/Burkitt's Lymphoma family of transcription factors in Drosophila melanogaster. Genetics 157: 717–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suganuma T, Gutierrez JL, Li B, Florens L, Swanson SK, Washburn MP, Abmayr SM, Workman JL 2008. ATAC is a double histone acetyltransferase complex that stimulates nucleosome sliding. Nat Struct Mol Biol 15: 364–372 [DOI] [PubMed] [Google Scholar]
- Takeda S, Chen DY, Westergard TD, Fisher JK, Rubens JA, Sasagawa S, Kan JT, Korsmeyer SJ, Cheng EH, Hsieh JJ 2006. Proteolysis of MLL family proteins is essential for taspase1-orchestrated cell cycle progression. Genes Dev 20: 2397–2409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang AH, Neufeld TP, Rubin GM, Muller HA 2001. Transcriptional regulation of cytoskeletal functions and segmentation by a novel maternal pair-rule gene, lilliputian. Development 128: 801–813 [DOI] [PubMed] [Google Scholar]
- Tanny JC, Erdjument-Bromage H, Tempst P, Allis CD 2007. Ubiquitylation of histone H2B controls RNA polymerase II transcription elongation independently of histone H3 methylation. Genes Dev 21: 835–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terranova R, Agherbi H, Boned A, Meresse S, Djabali M 2006. Histone and DNA methylation defects at Hox genes in mice expressing a SET domain-truncated form of Mll. Proc Natl Acad Sci 103: 6629–6634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel AT, Blessington P, Zou T, Feather D, Wu X, Yan J, Zhang H, Liu Z, Ernst P, Koretzky GA, et al. 2010. MLL-AF9-induced leukemogenesis requires coexpression of the wild-type Mll allele. Cancer Cell 17: 148–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thirman MJ, Levitan DA, Kobayashi H, Simon MC, Rowley JD 1994. Cloning of ELL, a gene that fuses to MLL in a t(11;19)(q23;p13.1) in acute myeloid leukemia. Proc Natl Acad Sci 91: 12110–12114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tkachuk DC, Kohler S, Cleary ML 1992. Involvement of a homolog of Drosophila trithorax by 11q23 chromosomal translocations in acute leukemias. Cell 71: 691–700 [DOI] [PubMed] [Google Scholar]
- Tresaugues L, Dehe PM, Guerois R, Rodriguez-Gil A, Varlet I, Salah P, Pamblanco M, Luciano P, Quevillon-Cheruel S, Sollier J, et al. 2006. Structural characterization of Set1 RNA recognition motifs and their role in histone H3 lysine 4 methylation. J Mol Biol 359: 1170–1181 [DOI] [PubMed] [Google Scholar]
- Trievel RC, Shilatifard A 2009. WDR5, a complexed protein. Nat Struct Mol Biol 16: 678–680 [DOI] [PubMed] [Google Scholar]
- Vanderzwan-Butler CJ, Prazak LM, Gergen JP 2007. The HMG-box protein Lilliputian is required for Runt-dependent activation of the pair-rule gene fushi-tarazu. Dev Biol 301: 350–360 [DOI] [PubMed] [Google Scholar]
- Vermeulen M, Mulder KW, Denissov S, Pijnappel WW, van Schaik FM, Varier RA, Baltissen MP, Stunnenberg HG, Mann M, Timmers HT 2007. Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell 131: 58–69 [DOI] [PubMed] [Google Scholar]
- Wada T, Takagi T, Yamaguchi Y, Ferdous A, Imai T, Hirose S, Sugimoto S, Yano K, Hartzog GA, Winston F, et al. 1998. DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes Dev 12: 343–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YL, Faiola F, Xu M, Pan S, Martinez E 2008. Human ATAC Is a GCN5/PCAF-containing acetylase complex with a novel NC2-like histone fold module that interacts with the TATA-binding protein. J Biol Chem 283: 33808–33815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Lin C, Smith ER, Guo H, Sanderson BW, Wu M, Gogol M, Alexander T, Seidel C, Wiedemann LM, et al. 2009. Global analysis of H3K4 methylation defines MLL family member targets and points to a role for MLL1-mediated H3K4 methylation in the regulation of transcriptional initiation by RNA polymerase II. Mol Cell Biol 29: 6074–6085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Cui B, Gorovsky MA 2009a. Histone H2B ubiquitylation is not required for histone H3 methylation at lysine 4 in tetrahymena. J Biol Chem 284: 34870–34879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Zang C, Cui K, Schones DE, Barski A, Peng W, Zhao K 2009b. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell 138: 1019–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston F, Chaleff DT, Valent B, Fink GR 1984. Mutations affecting Ty-mediated expression of the HIS4 gene of Saccharomyces cerevisiae. Genetics 107: 179–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittwer F, van der Straten A, Keleman K, Dickson BJ, Hafen E 2001. Lilliputian: an AF4/FMR2-related protein that controls cell identity and cell growth. Development 128: 791–800 [DOI] [PubMed] [Google Scholar]
- Wu M, Wang PF, Lee JS, Martin-Brown S, Florens L, Washburn M, Shilatifard A 2008. Molecular regulation of H3K4 trimethylation by Wdr82, a component of human Set1/COMPASS. Mol Cell Biol 28: 7337–7344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wysocka J, Swigut T, Milne TA, Dou Y, Zhang X, Burlingame AL, Roeder RG, Brivanlou AH, Allis CD 2005. WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development. Cell 121: 859–872 [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y, Takagi T, Wada T, Yano K, Furuya A, Sugimoto S, Hasegawa J, Handa H 1999. NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell 97: 41–51 [DOI] [PubMed] [Google Scholar]
- Yan Q, Moreland RJ, Conaway JW, Conaway RC 1999. Dual roles for transcription factor IIF in promoter escape by RNA polymerase II. J Biol Chem 274: 35668–35675 [DOI] [PubMed] [Google Scholar]
- Yang Z, Yik JH, Chen R, He N, Jang MK, Ozato K, Zhou Q 2005. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol Cell 19: 535–545 [DOI] [PubMed] [Google Scholar]
- Yap KL, Zhou MM 2010. Keeping it in the family: diverse histone recognition by conserved structural folds. Crit Rev Biochem Mol Biol 45: 488–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasukawa T, Kamura T, Kitajima S, Conaway RC, Conaway JW, Aso T 2008. Mammalian Elongin A complex mediates DNA-damage-induced ubiquitylation and degradation of Rpb1. EMBO J 27: 3256–3266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama A, Cleary ML 2008. Menin critically links MLL proteins with LEDGF on cancer-associated target genes. Cancer Cell 14: 36–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama A, Kitabayashi I, Ayton PM, Cleary ML, Ohki M 2002. Leukemia proto-oncoprotein MLL is proteolytically processed into 2 fragments with opposite transcriptional properties. Blood 100: 3710–3718 [DOI] [PubMed] [Google Scholar]
- Yokoyama A, Wang Z, Wysocka J, Sanyal M, Aufiero DJ, Kitabayashi I, Herr W, Cleary ML 2004. Leukemia proto-oncoprotein MLL forms a SET1-like histone methyltransferase complex with menin to regulate Hox gene expression. Mol Cell Biol 24: 5639–5649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu BD, Hess JL, Horning SE, Brown GA, Korsmeyer SJ 1995. Altered Hox expression and segmental identity in Mll-mutant mice. Nature 378: 505–508 [DOI] [PubMed] [Google Scholar]
- Zeitlinger J, Stark A, Kellis M, Hong JW, Nechaev S, Adelman K, Levine M, Young RA 2007. RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat Genet 39: 1512–1516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S, Wyrick JJ, Reese JC 2010. Novel trans-tail regulation of H2B ubiquitylation and H3K4 methylation by the N terminus of histone H2A. Mol Cell Biol 30: 3635–3645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Yik JH 2006. The Yin and Yang of P-TEFb regulation: implications for human immunodeficiency virus gene expression and global control of cell growth and differentiation. Microbiol Mol Biol Rev 70: 646–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Spicuglia S, Hsieh JJ, Mitsiou DJ, Hoiby T, Veenstra GJ, Korsmeyer SJ, Stunnenberg HG 2006. Uncleaved TFIIA is a substrate for taspase 1 and active in transcription. Mol Cell Biol 26: 2728–2735 [DOI] [PMC free article] [PubMed] [Google Scholar]