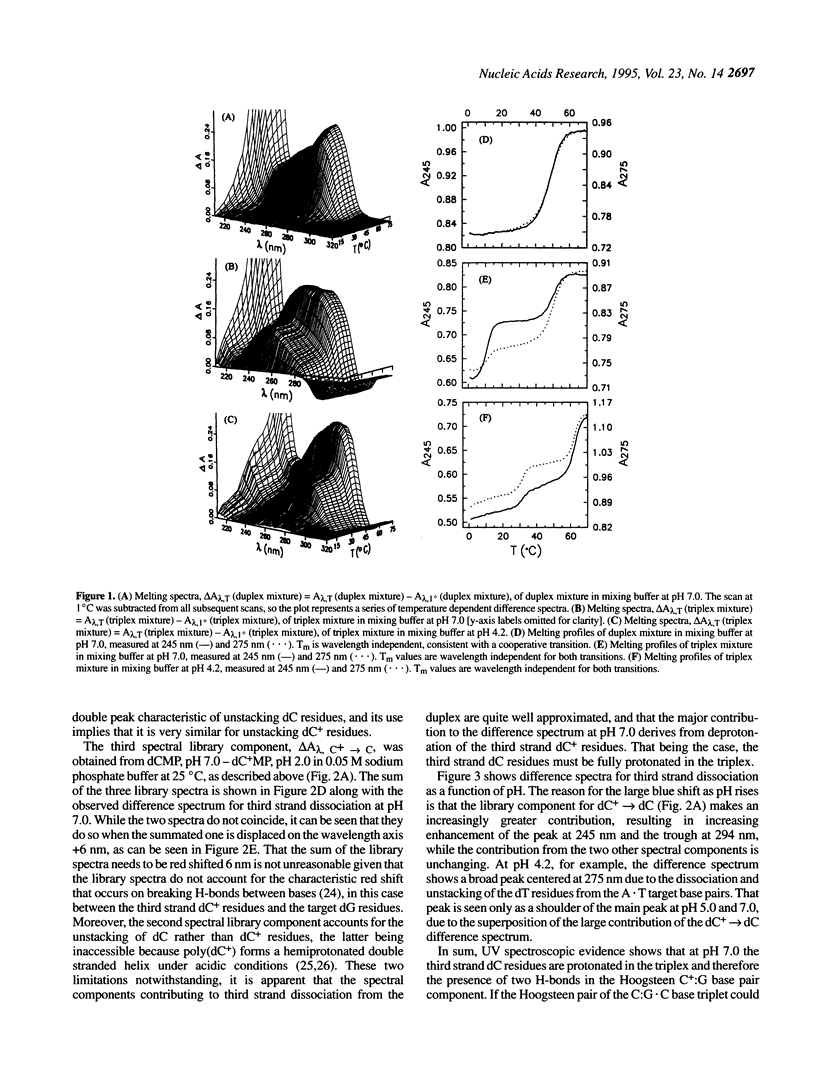
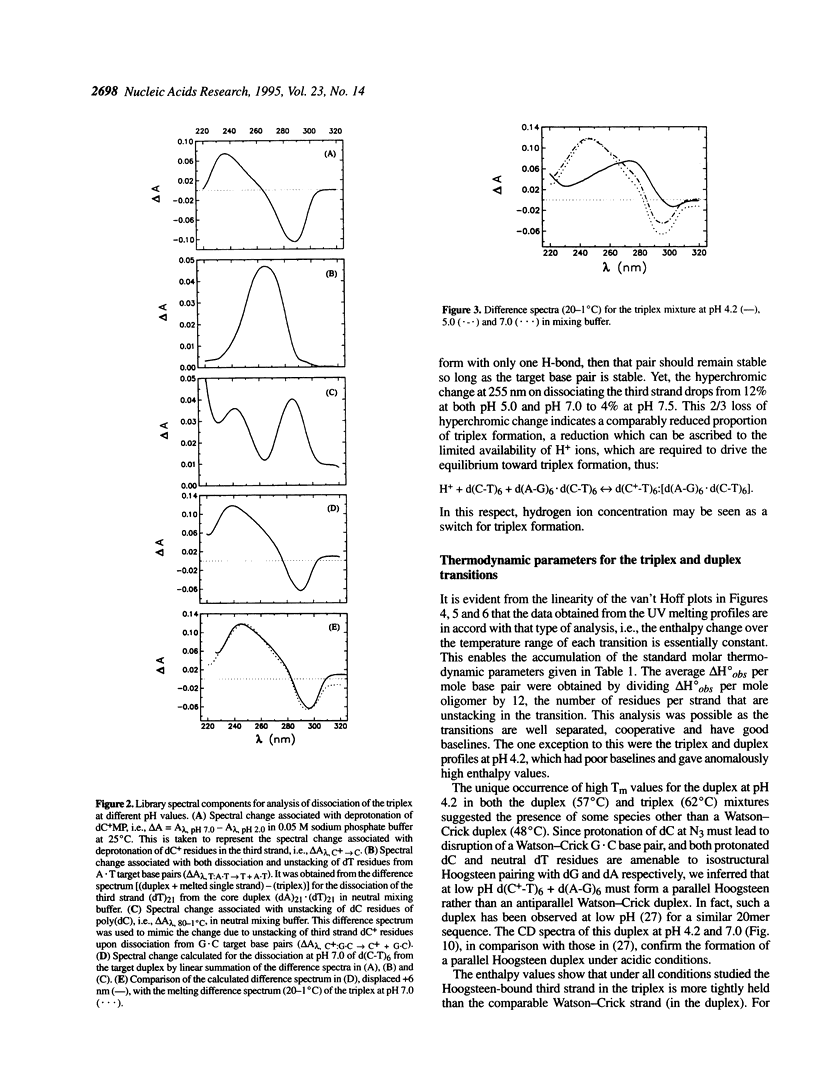
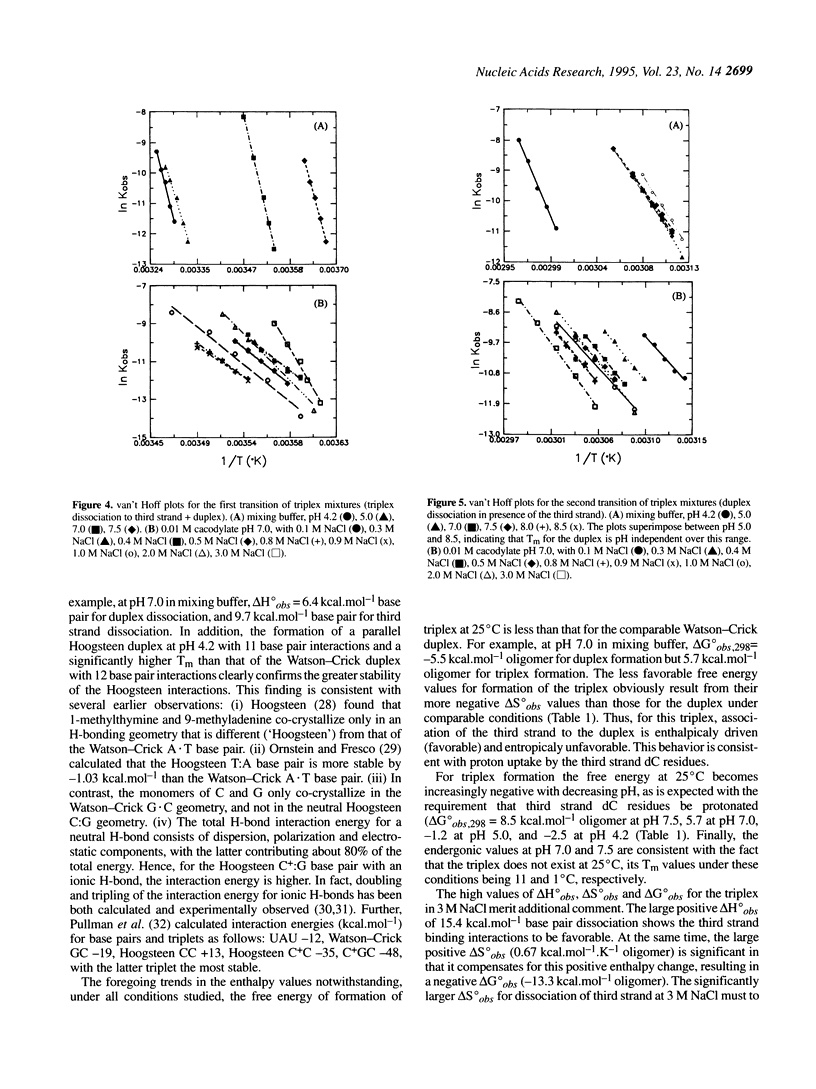
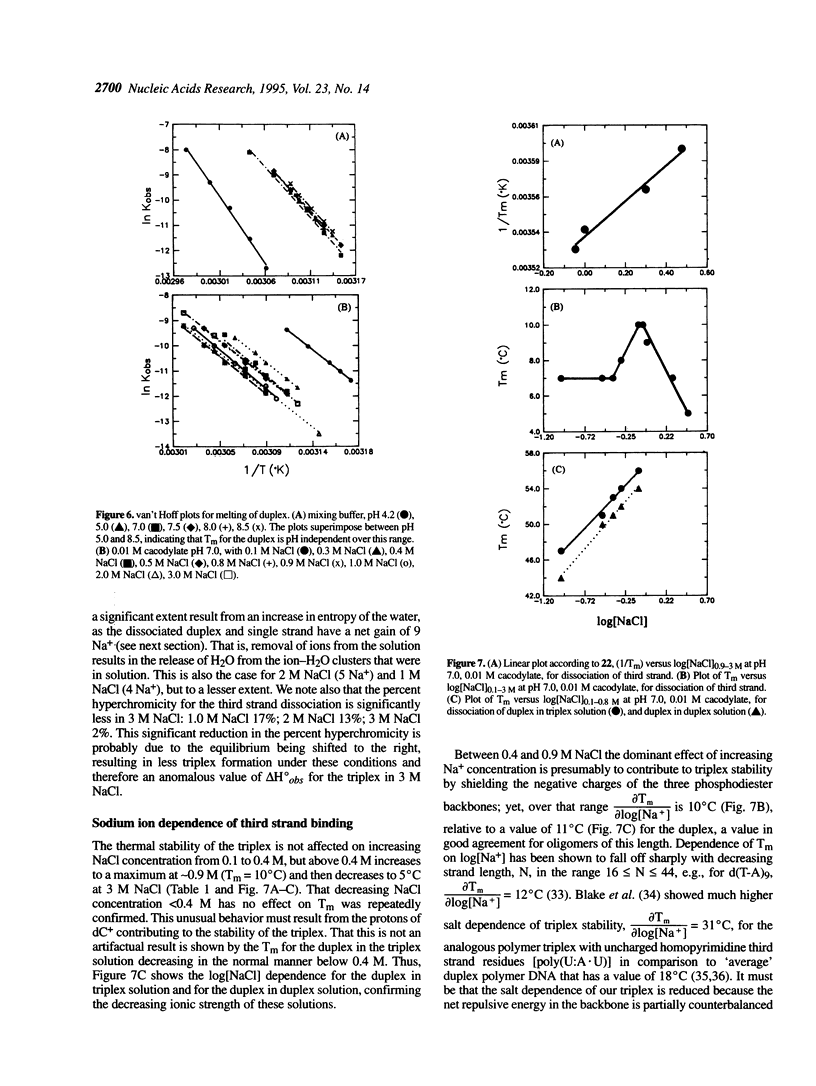
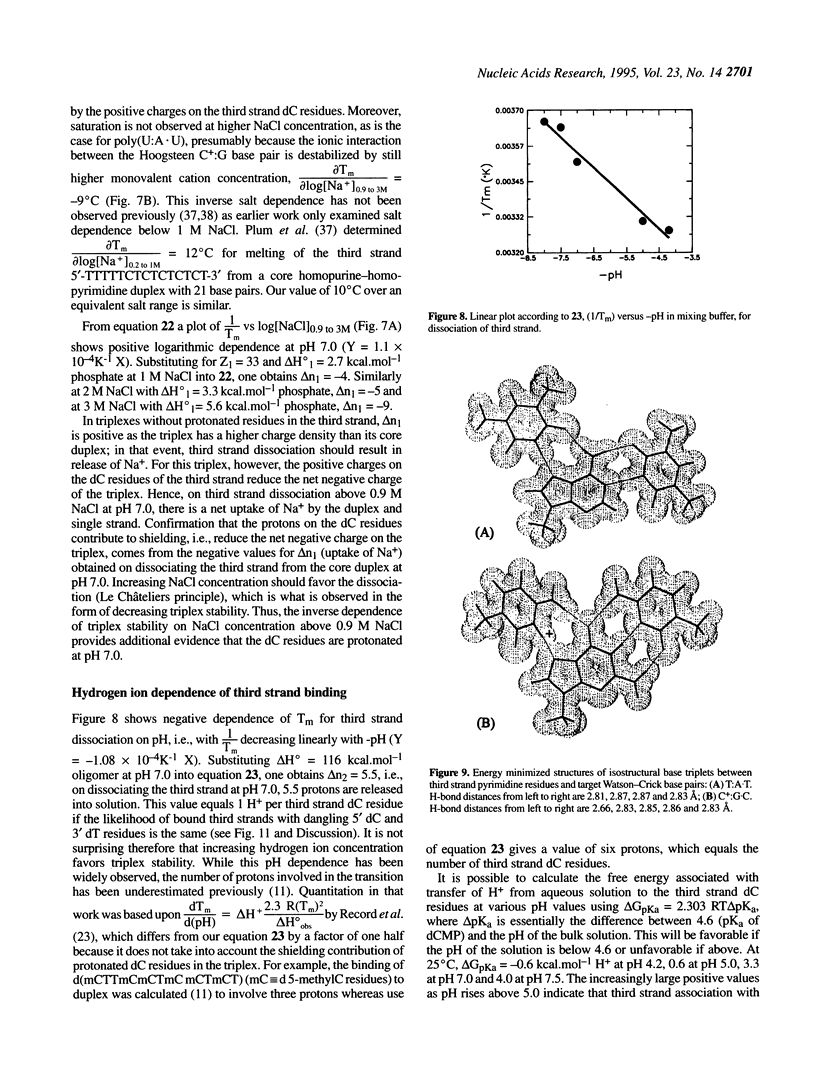
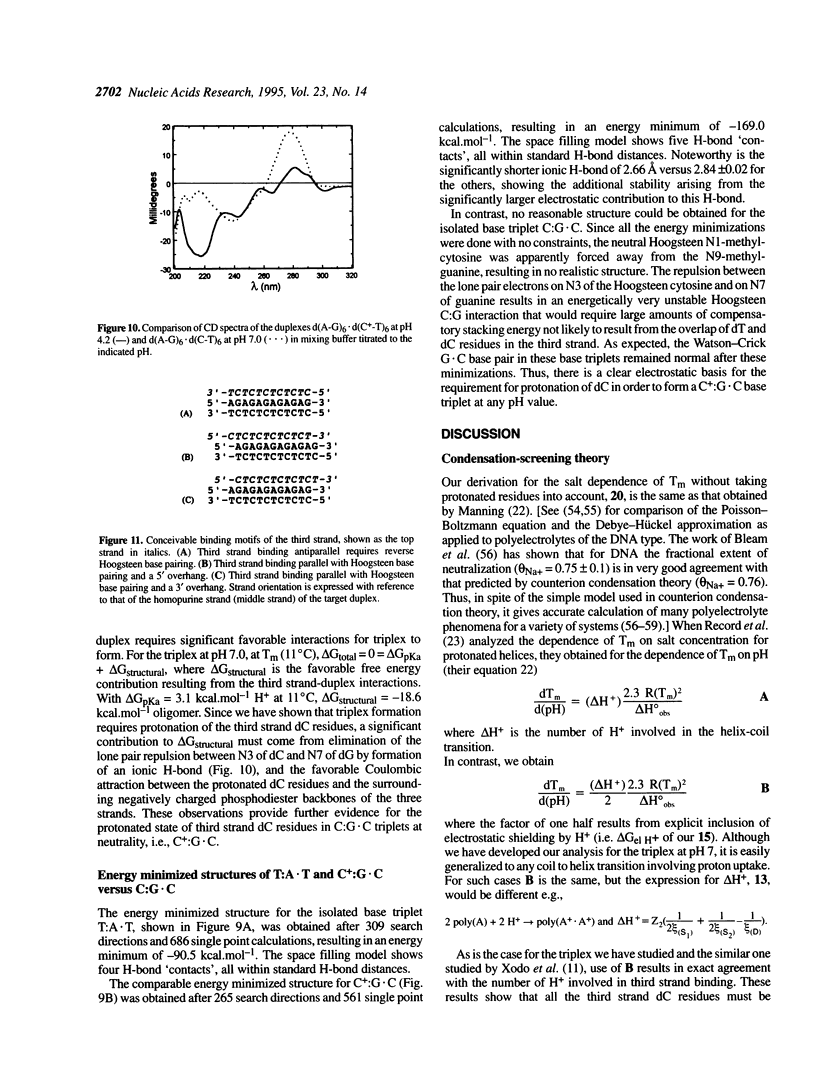
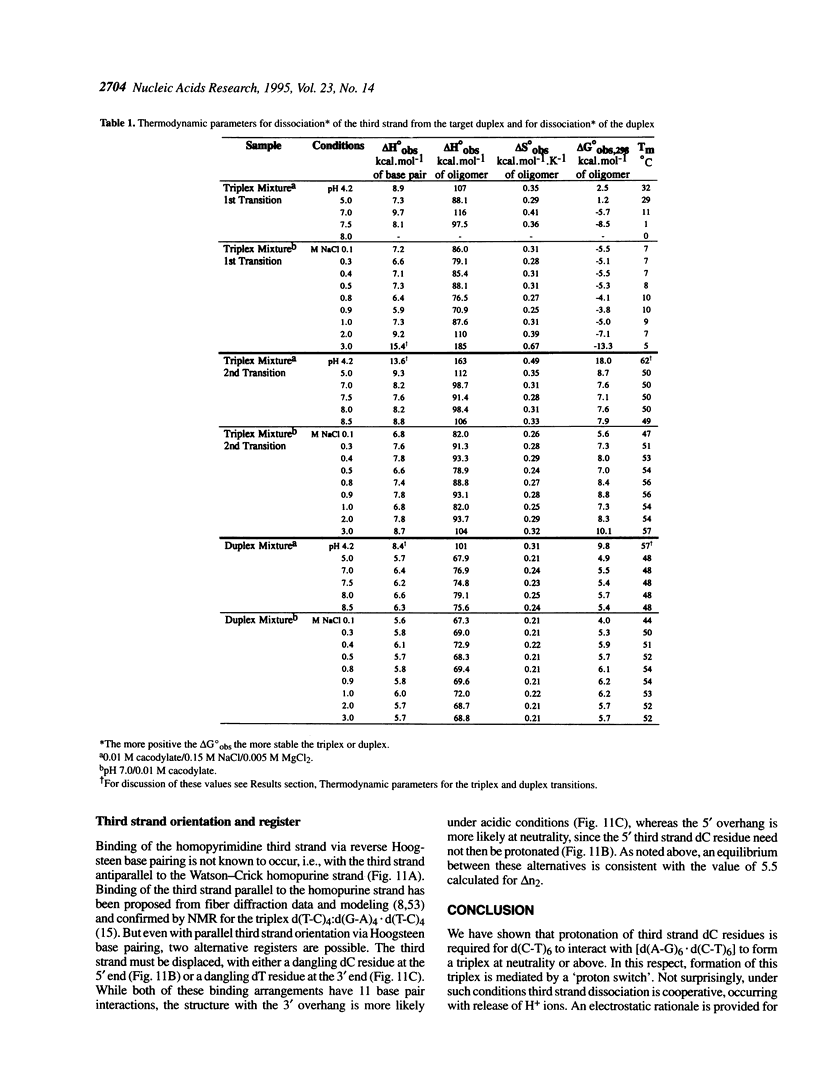
Abstract
Near-UV difference spectral analysis of the triplex formed from d(C-T)6 and d(A-G)6.d(C-T)6 in neutral and acidic solution shows that the third strand dC residues are protonated at pH 7.0, far above their intrinsic pKa. Additional support for ion-dipole interactions between the third strand dC residues and the G.C target base pairs comes from reduced positive dependence of triplet stability on ionic strength below 0.9 M Na+, inverse dependence above 0.9 M Na+ and strong positive dependence on hydrogen ion concentration. Molecular modeling (AMBER) of C:G.C and C+:G.C base triplets with the third strand base bound in the Hoogsteen geometry shows that only the C+:G.C triplet is energetically feasible. van't Hoff analysis of the melting of the triplex and target duplex shows that between pH 5.0 and 8.5 in 0.15 M NaCl/0.005 M MgCl2 the enthalpy of melting (delta H degree obs) varies from 5.7 to 6.6 kcal.mol-1 for the duplex in a duplex mixture and from 7.3 to 9.7 kcal.mol-1 for third strand dissociation in the triplex mixture. We have extended the condensation-screening theory of Manning to pH-dependent third strand binding. In this development we explicitly include the H+ contribution to the electrostatic free energy and obtain [formula: see text]. The number of protons released in the dissociation of the third strand from the target duplex at pH 7.0, delta n2, is thereby calculated to be 5.5, in good agreement with approximately six third strand dc residues per mole of triplex. This work shows that when third strand binding requires protonated residues that would otherwise be neutral, triplex formation and dissociation are mediated by proton uptake and release, i.e., a proton switch. As a by-product of this study, we have found that at low pH the Watson-Crick duplex d(A-G)6.d(C-T)6 undergoes a transition to a parallel Hoogsteen duplex d(A-G)6.d(C(+)-T)6.
Full text
PDF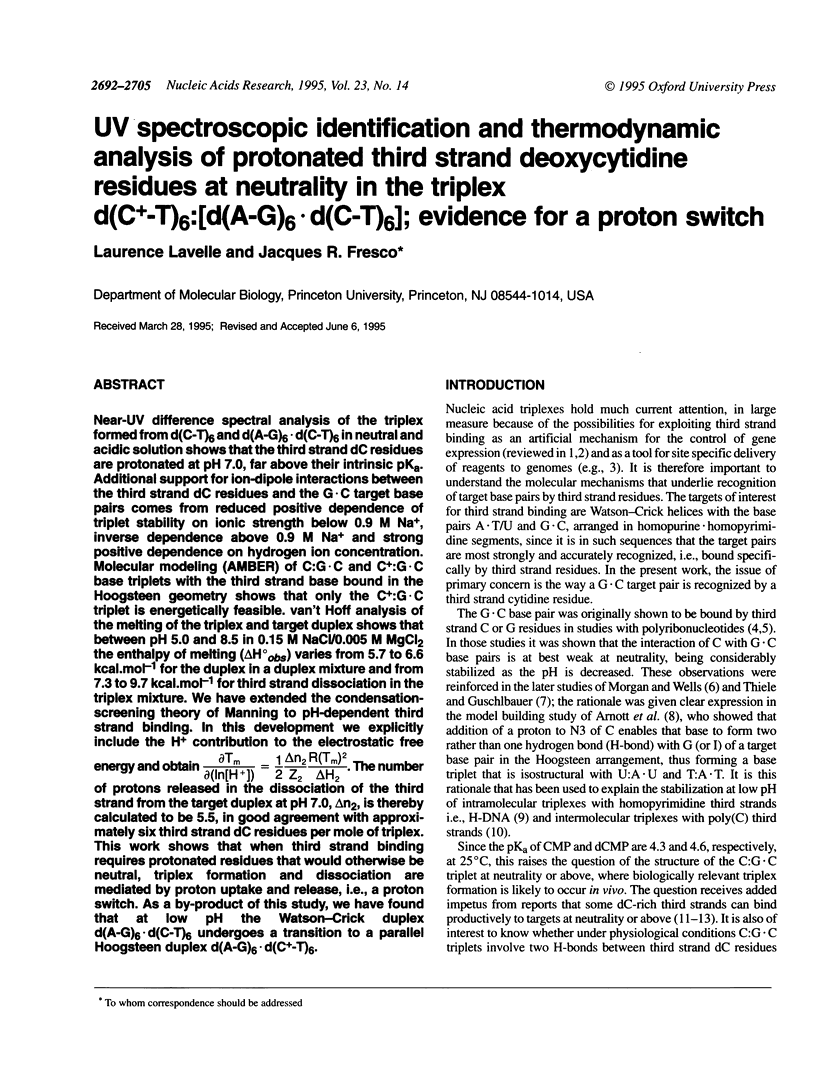






Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- AKINRIMISI E. O., SANDER C., TS'O P. O. Properties of helical polycytidylic acid. Biochemistry. 1963 Mar-Apr;2:340–344. doi: 10.1021/bi00902a028. [DOI] [PubMed] [Google Scholar]
- Adams A., Lindahl T., Fresco J. R. Conformational differences between the biologically active and inactive forms of a transfer ribonucleic acid. Proc Natl Acad Sci U S A. 1967 Jun;57(6):1684–1691. doi: 10.1073/pnas.57.6.1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson C. F., Record M. T., Jr Ion distributions around DNA and other cylindrical polyions: theoretical descriptions and physical implications. Annu Rev Biophys Biophys Chem. 1990;19:423–465. doi: 10.1146/annurev.bb.19.060190.002231. [DOI] [PubMed] [Google Scholar]
- Antao V. P., Gray D. M. CD spectral comparisons of the acid-induced structures of poly[d(A)], poly[r(A)], poly[d(C)], and poly[r(C)]. J Biomol Struct Dyn. 1993 Apr;10(5):819–839. doi: 10.1080/07391102.1993.10508677. [DOI] [PubMed] [Google Scholar]
- Arnott S., Bond P. J., Selsing E., Smith P. J. Models of triple-stranded polynucleotides with optimised stereochemistry. Nucleic Acids Res. 1976 Oct;3(10):2459–2470. doi: 10.1093/nar/3.10.2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnott S., Selsing E. Structures for the polynucleotide complexes poly(dA) with poly (dT) and poly(dT) with poly(dA) with poly (dT). J Mol Biol. 1974 Sep 15;88(2):509–521. doi: 10.1016/0022-2836(74)90498-7. [DOI] [PubMed] [Google Scholar]
- Blake R. D., Massoulié J., Fresco J. R. Polynucleotides. 8. A spectral approach to the equilibria between polyriboadenylate and polyribouridylate and their complexes. J Mol Biol. 1967 Dec 14;30(2):291–308. [PubMed] [Google Scholar]
- Bleam M. L., Anderson C. F., Record M. T. Relative binding affinities of monovalent cations for double-stranded DNA. Proc Natl Acad Sci U S A. 1980 Jun;77(6):3085–3089. doi: 10.1073/pnas.77.6.3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braunlin W. H., Anderson C. F., Record M. T., Jr Competitive interactions of Co(NH3)6(3+) and Na+ with helical B-DNA probed by 59Co and 23Na NMR. Biochemistry. 1987 Dec 1;26(24):7724–7731. doi: 10.1021/bi00398a028. [DOI] [PubMed] [Google Scholar]
- Cheng Y. K., Pettitt B. M. Stabilities of double- and triple-strand helical nucleic acids. Prog Biophys Mol Biol. 1992;58(3):225–257. doi: 10.1016/0079-6107(92)90007-s. [DOI] [PubMed] [Google Scholar]
- Dolinnaya N. G., Braswell E. H., Fossella J. A., Klump H., Fresco J. R. Molecular and thermodynamic properties of d(A(+)-G)10, a single-stranded nucleic acid helix without paired or stacked bases. Biochemistry. 1993 Sep 28;32(38):10263–10270. doi: 10.1021/bi00089a049. [DOI] [PubMed] [Google Scholar]
- Elson E. L., Scheffler I. E., Baldwin R. L. Helix formation by d(TA) oligomers. 3. Electrostatic effects. J Mol Biol. 1970 Dec 28;54(3):401–415. doi: 10.1016/0022-2836(70)90118-x. [DOI] [PubMed] [Google Scholar]
- Gehring K., Leroy J. L., Guéron M. A tetrameric DNA structure with protonated cytosine.cytosine base pairs. Nature. 1993 Jun 10;363(6429):561–565. doi: 10.1038/363561a0. [DOI] [PubMed] [Google Scholar]
- Granot J., Kearns D. R. Interactions of DNA with divalent metal ions. II. Proton relaxation enhancement studies. Biopolymers. 1982 Jan;21(1):203–218. doi: 10.1002/bip.360210116. [DOI] [PubMed] [Google Scholar]
- Guschlbauer W. Protonated polynucleotide structures. I. The thermal denaturation of polycytidylic acid in acid solution. Proc Natl Acad Sci U S A. 1967 May;57(5):1441–1448. doi: 10.1073/pnas.57.5.1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanvey J. C., Williams E. M., Besterman J. M. DNA triple-helix formation at physiologic pH and temperature. Antisense Res Dev. 1991 Winter;1(4):307–317. doi: 10.1089/ard.1991.1.307. [DOI] [PubMed] [Google Scholar]
- Holcomb D. N., Timasheff S. N. Temperature dependence of the hydrogen ion equilibria in poly(riboadenylic acid). Biopolymers. 1968 Apr;6(4):513–529. doi: 10.1002/bip.1968.360060407. [DOI] [PubMed] [Google Scholar]
- Htun H., Dahlberg J. E. Topology and formation of triple-stranded H-DNA. Science. 1989 Mar 24;243(4898):1571–1576. doi: 10.1126/science.2648571. [DOI] [PubMed] [Google Scholar]
- INMAN R. B. TRANSITIONS OF DNA HOMOPOLYMERS. J Mol Biol. 1964 Sep;9:624–637. doi: 10.1016/s0022-2836(64)80171-6. [DOI] [PubMed] [Google Scholar]
- Jaishree T. N., Wang A. H. NMR studies of pH-dependent conformational polymorphism of alternating (C-T)n sequences. Nucleic Acids Res. 1993 Aug 11;21(16):3839–3844. doi: 10.1093/nar/21.16.3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LIPSETT M. N. COMPLEX FORMATION BETWEEN POLYCYTIDYLIC ACID AND GUANINE OLIGONUCLEOTIDES. J Biol Chem. 1964 Apr;239:1256–1260. [PubMed] [Google Scholar]
- Leroy J. L., Gehring K., Kettani A., Guéron M. Acid multimers of oligodeoxycytidine strands: stoichiometry, base-pair characterization, and proton exchange properties. Biochemistry. 1993 Jun 15;32(23):6019–6031. doi: 10.1021/bi00074a013. [DOI] [PubMed] [Google Scholar]
- Letai A. G., Palladino M. A., Fromm E., Rizzo V., Fresco J. R. Specificity in formation of triple-stranded nucleic acid helical complexes: studies with agarose-linked polyribonucleotide affinity columns. Biochemistry. 1988 Dec 27;27(26):9108–9112. doi: 10.1021/bi00426a007. [DOI] [PubMed] [Google Scholar]
- Liu K., Miles H. T., Frazier J., Sasisekharan V. A novel DNA duplex. A parallel-stranded DNA helix with Hoogsteen base pairing. Biochemistry. 1993 Nov 9;32(44):11802–11809. doi: 10.1021/bi00095a008. [DOI] [PubMed] [Google Scholar]
- Manning G. S. On the application of polyelectrolyte "limiting laws" to the helix-coil transition of DNA. I. Excess univalent cations. Biopolymers. 1972;11(5):937–949. doi: 10.1002/bip.1972.360110502. [DOI] [PubMed] [Google Scholar]
- Manzini G., Xodo L. E., Gasparotto D., Quadrifoglio F., van der Marel G. A., van Boom J. H. Triple helix formation by oligopurine-oligopyrimidine DNA fragments. Electrophoretic and thermodynamic behavior. J Mol Biol. 1990 Jun 20;213(4):833–843. doi: 10.1016/S0022-2836(05)80267-0. [DOI] [PubMed] [Google Scholar]
- Marky L. A., Breslauer K. J. Calculating thermodynamic data for transitions of any molecularity from equilibrium melting curves. Biopolymers. 1987 Sep;26(9):1601–1620. doi: 10.1002/bip.360260911. [DOI] [PubMed] [Google Scholar]
- Michelson A. M., Massoulié J., Guschlbauer W. Synthetic polynucleotides. Prog Nucleic Acid Res Mol Biol. 1967;6:83–141. doi: 10.1016/s0079-6603(08)60525-5. [DOI] [PubMed] [Google Scholar]
- Mirkin S. M., Frank-Kamenetskii M. D. H-DNA and related structures. Annu Rev Biophys Biomol Struct. 1994;23:541–576. doi: 10.1146/annurev.bb.23.060194.002545. [DOI] [PubMed] [Google Scholar]
- Mirkin S. M., Lyamichev V. I., Drushlyak K. N., Dobrynin V. N., Filippov S. A., Frank-Kamenetskii M. D. DNA H form requires a homopurine-homopyrimidine mirror repeat. Nature. 1987 Dec 3;330(6147):495–497. doi: 10.1038/330495a0. [DOI] [PubMed] [Google Scholar]
- Morgan A. R., Wells R. D. Specificity of the three-stranded complex formation between double-stranded DNA and single-stranded RNA containing repeating nucleotide sequences. J Mol Biol. 1968 Oct 14;37(1):63–80. doi: 10.1016/0022-2836(68)90073-9. [DOI] [PubMed] [Google Scholar]
- Moser H. E., Dervan P. B. Sequence-specific cleavage of double helical DNA by triple helix formation. Science. 1987 Oct 30;238(4827):645–650. doi: 10.1126/science.3118463. [DOI] [PubMed] [Google Scholar]
- Ornstein R. L., Fresco J. R. Correlation of crystallographically determined and computationally predicted hydrogen-bonded pairing configurations of nucleic acid bases. Proc Natl Acad Sci U S A. 1983 Sep;80(17):5171–5175. doi: 10.1073/pnas.80.17.5171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmanabhan S., Richey B., Anderson C. F., Record M. T., Jr Interaction of an N-methylated polyamine analogue, hexamethonium(2+), with NaDNA: quantitative 14N and 23Na NMR relaxation rate studies of the cation-exchange process. Biochemistry. 1988 Jun 14;27(12):4367–4376. doi: 10.1021/bi00412a025. [DOI] [PubMed] [Google Scholar]
- Pilch D. S., Brousseau R., Shafer R. H. Thermodynamics of triple helix formation: spectrophotometric studies on the d(A)10.2d(T)10 and d(C+3T4C+3).d(G3A4G3).d(C3T4C3) triple helices. Nucleic Acids Res. 1990 Oct 11;18(19):5743–5750. doi: 10.1093/nar/18.19.5743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plum G. E., Park Y. W., Singleton S. F., Dervan P. B., Breslauer K. J. Thermodynamic characterization of the stability and the melting behavior of a DNA triplex: a spectroscopic and calorimetric study. Proc Natl Acad Sci U S A. 1990 Dec;87(23):9436–9440. doi: 10.1073/pnas.87.23.9436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pullman B., Claverie P., Caillet J. Interaction energies in hydrogen-bonded purine-pyrimidine triplets. Proc Natl Acad Sci U S A. 1967 Jun;57(6):1663–1669. doi: 10.1073/pnas.57.6.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopal P., Feigon J. NMR studies of triple-strand formation from the homopurine-homopyrimidine deoxyribonucleotides d(GA)4 and d(TC)4. Biochemistry. 1989 Sep 19;28(19):7859–7870. doi: 10.1021/bi00445a048. [DOI] [PubMed] [Google Scholar]
- Rajagopal P., Feigon J. Triple-strand formation in the homopurine:homopyrimidine DNA oligonucleotides d(G-A)4 and d(T-C)4. Nature. 1989 Jun 22;339(6226):637–640. doi: 10.1038/339637a0. [DOI] [PubMed] [Google Scholar]
- Record M. T., Jr Electrostatic effects on polynucleotide transitions. I. Behavior at neutral pH. Biopolymers. 1967;5(10):975–992. doi: 10.1002/bip.1967.360051010. [DOI] [PubMed] [Google Scholar]
- Record M. T., Jr, Woodbury C. P., Lohman T. M. Na+ effects on transition of DNA and polynucleotides of variable linear charge density. Biopolymers. 1976 May;15(5):893–915. doi: 10.1002/bip.1976.360150507. [DOI] [PubMed] [Google Scholar]
- Rougée M., Faucon B., Mergny J. L., Barcelo F., Giovannangeli C., Garestier T., Hélène C. Kinetics and thermodynamics of triple-helix formation: effects of ionic strength and mismatches. Biochemistry. 1992 Sep 29;31(38):9269–9278. doi: 10.1021/bi00153a021. [DOI] [PubMed] [Google Scholar]
- Singleton S. F., Dervan P. B. Influence of pH on the equilibrium association constants for oligodeoxyribonucleotide-directed triple helix formation at single DNA sites. Biochemistry. 1992 Nov 17;31(45):10995–11003. doi: 10.1021/bi00160a008. [DOI] [PubMed] [Google Scholar]
- Sklenár V., Feigon J. Formation of a stable triplex from a single DNA strand. Nature. 1990 Jun 28;345(6278):836–838. doi: 10.1038/345836a0. [DOI] [PubMed] [Google Scholar]
- Thiele D., Guschlbauer W. Protonated polynucleotide structures. IX. Disproportionation of poly (G)-poly (C) in acid medium. Biopolymers. 1971;10(1):143–157. doi: 10.1002/bip.360100111. [DOI] [PubMed] [Google Scholar]
- Völker J., Klump H. H. Electrostatic effects in DNA triple helices. Biochemistry. 1994 Nov 15;33(45):13502–13508. doi: 10.1021/bi00249a039. [DOI] [PubMed] [Google Scholar]
- Wells R. D., Collier D. A., Hanvey J. C., Shimizu M., Wohlrab F. The chemistry and biology of unusual DNA structures adopted by oligopurine.oligopyrimidine sequences. FASEB J. 1988 Nov;2(14):2939–2949. [PubMed] [Google Scholar]
- Xodo L. E., Manzini G., Quadrifoglio F. Spectroscopic and calorimetric investigation on the DNA triplex formed by d(CTCTTCTTTCTTTTCTTTCTTCTC) and d(GAGAAGAAAGA) at acidic pH. Nucleic Acids Res. 1990 Jun 25;18(12):3557–3564. doi: 10.1093/nar/18.12.3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xodo L. E., Manzini G., Quadrifoglio F., van der Marel G. A., van Boom J. H. Effect of 5-methylcytosine on the stability of triple-stranded DNA--a thermodynamic study. Nucleic Acids Res. 1991 Oct 25;19(20):5625–5631. doi: 10.1093/nar/19.20.5625. [DOI] [PMC free article] [PubMed] [Google Scholar]


