Abstract
Engulfment of apoptotic cells by phagocytosis ensures the removal of unwanted and defective cells. We developed a genetically encoded marker for cell engulfment, pHMA, which consists of the pH-sensitive derivative of GFP, pHluorin, fused to the actin-binding domain of Moesin. In healthy cells of Drosophila embryos and cultured cells, pHMA resides at the cell cortex. In dying cells, pHMA loses its cortical localization and reports a modest decrease in pH. In embryos, the dying cells lose their apical contacts, then move basally and are ultimately engulfed by neighboring cells or macrophages. The cell corpse material is strongly acidified soon after engulfment and persists in the phagocytic cell for several hours. Changes in the pHMA signal correlate well with increases or decreases in apoptosis. These data show that pHMA is a useful reporter for cell engulfment and can be used in screening for mutations that affect cell engulfment.
INTRODUCTION
Numerous cells die during embryogenesis. To ensure that these effete cells do not interfere with development, they must be cleared by engulfment. Much has been learned about the process of cell engulfment from studies in C. elegans that relied on DIC microscopy to visualize un-engulfed cells (Mangahas and Zhou, 2005). More recent studies in C. elegans utilized time-lapse imaging to identify novel cell engulfment and corpse processing genes and to establish their temporal relationships (Yu et al., 2006; Venegas and Zhou, 2007; Yu et al., 2008; Zou et al., 2009). Mammalian cell culture has been critical to our understanding of cell engulfment in mammals (Erwig and Henson, 2007). However, there is a need to study cell engulfment in vivo, particularly in the context of animals that undergo regulative development. This report describes a new reagent for time-lapse monitoring of cell engulfment in Drosophila embryos, tissue and cells. In Drosophila embryos, epithelial cells are mostly engulfed by neighboring cells, while cells dying within internal organs, such as the brain, are engulfed by macrophages (Pazdera et al., 1998; Mergliano and Minden, 2003; Robertson et al., 2003; Sears et al., 2003). The molecular connection between cell death and cell engulfment within the context of the living Drosophila embryo is poorly understood. Therefore, it is important to be able to monitor cell death and cell engulfment events in real time.
Until recently, injection of the vital dye, acridine orange (AO), was the primary method for monitoring apoptosis in live Drosophila embryos. Newly developed genetically expressed reporters of apoptosis include: Apoliner and SCAT, which detect caspase 3 activation (Takemoto et al., 2003; Takemoto et al., 2007; Bardet et al., 2008), and CIETDY, which detects the activity of the initiator Caspase 8, DED, (Mazzalupo and Cooley, 2006). These reporters eliminate the need for AO and allow one to observe cell death in different tissues and developmental stages. The engulfment of dead cells in Drosophila embryos has been monitored by injecting VGAL, a reporter for cell engulfment that becomes fluorescent when the cytoplasm of a dying cell is mixed with the lysosomal compartment of the engulfing cell (Mergliano and Minden, 2003). Since VGAL is membrane impermeable, it has to be loaded into cells by injection into syncytial stage embryos, where all cells receive equal amounts of VGAL. Thus, VGAL can only be used to visualize the engulfment of embryonic cells. Moreover, VGAL cannot be used to monitor the engulfment of specific cell types since it is uniformly loaded into all cells. More importantly, because VGAL can only be introduced by injection, it is impractical to use VGAL in large screens for mutations that affect cell engulfment. Recently, a pH-sensitive fluorescent dye, pHrodo, has been used to assess cell engulfment by FACS analysis (Miksa et al., 2009). To circumvent the limitations of dyes such as VGAL, we designed a genetically encoded reporter for cell engulfment that will allow one to study the clearance of apoptotic corpses in live animals.
A hallmark of apoptotic cell engulfment is the fusion of the phagosome with the lysosomal compartment (Odaka and Mizuochi, 1999). We have taken advantage of the acidic environment of the lysosome and employed the pH-sensitive ratiometric derivative of GFP, pHluorin, to report the movement of dying cell’s contents into the acidic milieu of the engulfing cell’s lysosome. To aid in the visualization of pHluorin in healthy cells, we chose to tether it to the cell cortex via the Moesin actin-binding domain. The cortical localization of pHluorin was hypothesized to reveal morphological changes in apoptotic cells. This pHluorin::moesin actin-binding-domain chimera is referred to as pHMA. Here we show that when pHMA is expressed in cultured Drosophila cells, its ratiometric signal reports that the pH of healthy cells drops slightly from 7.4-7.0 to 6.8-6.6 prior to engulfment, and then drops rapidly to as low as 5 after cell engulfment. As predicted, the number of acidified pHMA bodies increases in embryos with increased cell death and is not present in embryos lacking cell death. Time-lapse analysis of pHMA-expressing embryos reveals a sequence of characteristic morphological changes in apoptotic, epithelial cells prior to, and after their engulfment. Thus, pHMA serves as a robust engulfment reporter that will be a useful reagent to study the dynamics of apoptosis and to screen for new mutations that affect apoptotic cell engulfment.
RESULTS AND DISCUSSION
Design of pHMA
To generate a dynamic, genetically encoded reporter for cell engulfment, we relied on pHluorin and the actin-binding domain of Moesin. pHluorin is a pH-sensitive GFP derivative that emits in the usual GFP spectrum, but has a bimodal excitation spectrum; it is maximally excited at 475 nm in acidic environments, and at 395 nm in neutral surroundings (Miesenbock et al., 1998). Therefore, local changes in pH can be detected by taking a ratio of emission intensities at excitations of 470 nm and 410 nm (R470/410). pHluorin has been used to determine the acidification of intracellular compartments and neurotransmitter uptake (Li et al., 2005). To localize pHluorin cortically, we engineered a fusion of pHluorin and the actin-binding domain of Moesin to generate pHMA. The actin-binding domain of Moesin was chosen because has been successfully used in the Drosophila embryo in a fusion known as GMA, which consists of GFP fused to the 140 amino acid C-terminal actin-binding domain of Moesin (Edwards et al., 1997; Dutta et al., 2002). GMA reveals the cortical actin network and allows one to visualize cell geometry and morphogenetic events (Edwards et al., 1997). GMA has also been used to examine the role of apoptosis in the squamous epithelial cells of the Drosophila amnioserosa during dorsal closure (Toyama et al., 2008). To allow for tissue-specific and inducible expression in Drosophila, pHMA was placed downstream of the UAS promoter (Brand and Perrimon, 1993). pHMA was also placed under the control of polyubiquitin promoter (Ubi) (Davis et al., 1995) promoter for uniform expression (Fig. 1A).
Fig. 1.
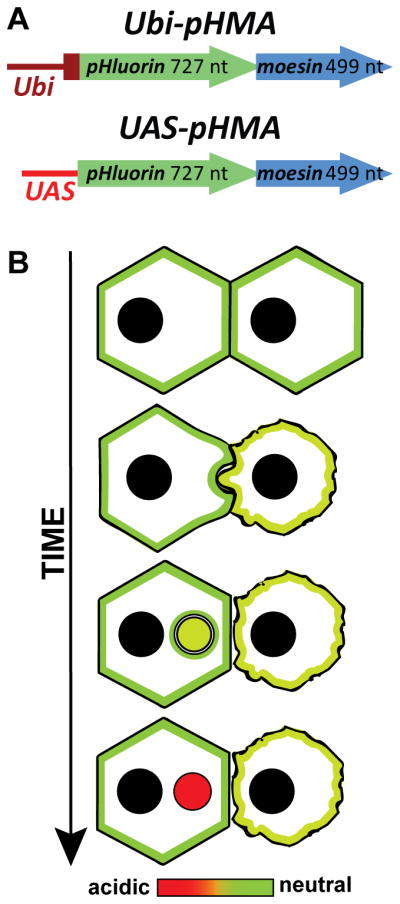
Cartoon of pHMA, a genetic reporter for cell engulfment. A: Ubi-pHMA and UAS-pHMA constructs that were transfected into S2 cells or were used to generate transgenic Drosophila. B: Hypothetical reporter activity of pHMA in cells undergoing apoptosis/cell engulfment. The engulfing cell (left) phagocytoses a part of the apoptotic cell (right) that shows membrane blebbing. The color scale from green (neutral) to red (acidic) represents progressive acidification of apoptotic phagosome. The acidified apoptotic phagosome is represented in red.
We hypothesized that pHMA would reveal the cell-shape changes that take place during apoptosis. Some studies suggest that apoptotic cells become slightly acidified prior to engulfment (Barry and Eastman, 1992). pHMA will be useful for determining if this is also the case for Drosophila cells. When an apoptotic cell is engulfed, the cortically localized actin, along with pHMA, will be internalized into the apoptotic phagosome of the engulfing cell where they undergo further acidification as they first fuse with the endocytic compartment and eventually fuse with the lysosomes of the engulfing cell (Fig. 1B).
pHMA expression in S2 cells reveals pH changes of living, dying and engulfed cells
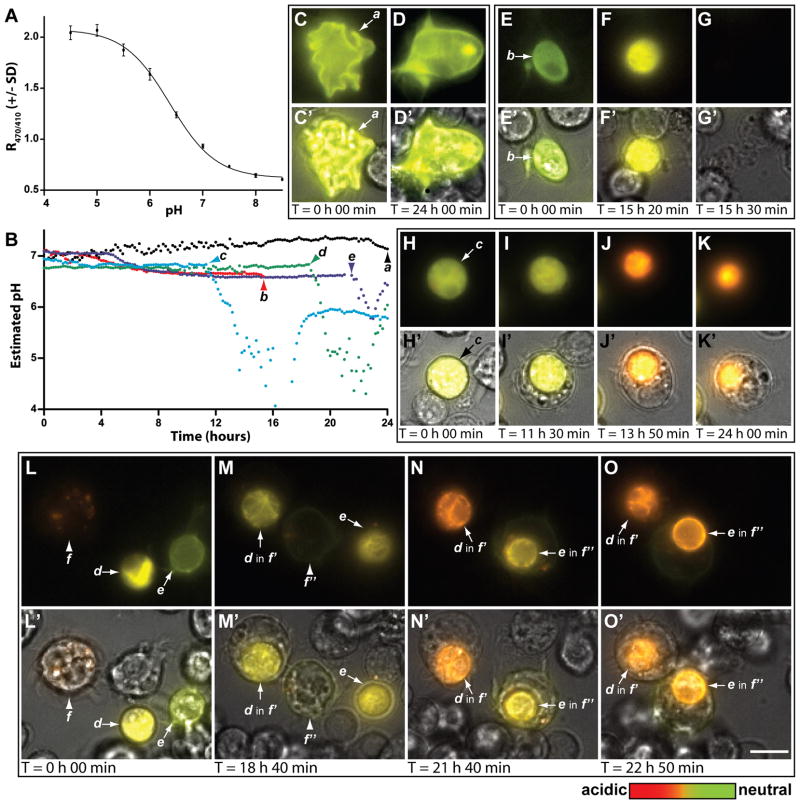
As proof of principle, pHMA was expressed in Drosophila cultured cells and its localization and fluorescence characteristics were monitored over the life-cycle of the cells. S2 and S2R+ cells, which are hemocyte-like, phagocytic cells derived from late-stage Drosophila embryos, were used for this analysis (Stroschein-Stevenson et al., 2006). To establish the relationship between intracellular pH and pHMA’s fluorescence response, pHMA-expressing cells were fixed and permeabilized and the pH was adjusted with phosphate buffers ranging from pH 4.5 to 8.5, in 0.5 pH intervals. The ratio of pHMA emission when excited by 470 and 410 nm light (R470/410) was determined at each pH interval (Fig. 2A). Nonlinear regression analysis was used to generate a standard curve of pHMA R470/410 relative to pH (r2 = 0.998). The resulting model was used to approximate the pH of individual living, dying and engulfed cells observed during time-lapse recordings (Fig. 2B). Time-lapse recordings of S2 cells that were transfected with Ubi-pHMA for two days were made over 24 hour periods with images taken at ten minute intervals. Over four hundred S2 cells transiently expressing pHMA were analyzed. During the 24-hour imaging period, 48 pHMA-expressing cells (11%) were engulfed, while 30 (7%) suddenly disappeared, suggesting a necrotic demise. The number of engulfed pHMA-positive cells did not appreciably accumulate from the beginning 31/418 (7%) to the end of the recording period 40/443 (9%), indicating that the corpses were completely processed over time, and that the exposure conditions of the ten minute time-lapse cycle did not induce photo-toxic apoptosis. The pH of cells that were morphologically healthy stayed between 7.0–7.4 (cell a: Fig. 2B, Fig. 2C and D, C′ and D′, and Suppl. Movie 1). Most, but not all, of these cells had cortically localized pHMA. Healthy cells displayed variable morphology that ranged from highly crenated to relatively round with long processes. Cells that were eventually engulfed or ruptured displayed decreased motility, rounded up, and the expressed pHMA frequently transitioned from cortical to cytoplasmic. These changes were accompanied by slight acidification to 6.8-6.6. Acidification of cytoplasm has previously been described as a component of apoptosis (Barry and Eastman, 1992). For example, in U937 cells the pH has been observed to vary from 7.2 in normal to 6.8 in pre-apoptotic to 5.7 in apoptotic cells (Nilsson et al., 2003; Nilsson et al., 2006). About 40% of the slightly acidified cells rapidly and completely lost their pHMA signal (cell b: Fig. 2B, Fig. 2E–G, E′-G′, and Suppl. Movie 2); this loss is presumed to result from the loss of membrane integrity since the non-fluorescent, poorly refractile cell corpses were still visible after the loss of pHMA signal, indicating necrosis. The frequency of cell rupture was greatly increased by adding cell death inducing agents such as 10 μM actinomycin D (data not shown). Cells that were engulfed, acidified over 30–60 minutes to a much greater degree, pH <6, than pre-engulfment. Most engulfed corpses remained acidified at pH 5 or below for several hours, while the phago-lysosome decreased in size over time (cell c: Fig. 2B and Fig. 2H-K, H′-K′, Suppl. Movie 3). The pH of the phago-lysosomes appeared to rebound–most cell corpses partially re-neutralized (cells d and e: Fig. 2B and Fig. 2L-O, L′-O′, Suppl. Movie 4). The biological significance of this phagosome pH recovery is not clear at this point, but will require further examination. All of dying cells were engulfed whole. Over the 24 hour recording period, we observed approximately 140 partial engulfment events that possibly involved the pruning of cell processes from healthy cells or the engulfment of detached cell fragments. As will be described in the in vivo analysis of pHMA, this wholesale engulfment of dying cells by S2 cells differs from the piecemeal engulfment of epithelial cells seen in live embryos. This may be due to the experimental conditions or to the hemocytic nature of S2 cells. These data clearly show that pHMA-expressing S2 cells provide a useful platform for studying cell engulfment in vitro.
Fig. 2.
pHMA acidifies after cell engulfment in S2 cells. A: Standard R470/410 curve generated by fixing and buffering S2R+ pHMA-expressing cells. The average R470/410 and standard deviations (see methods) are plotted against pH. B: pH estimates for cells “a” to “e” derived from the curve fit in A. C–O: Wide-field fluorescence images of cells from time-lapse movies; emission at excitation of 410 nm is green and at excitation of 470 nm is red. C′–O′: Fluorescence images are overlaid with transmitted light images to reveal unlabeled cells in the field. C–D, C′–D′: Morphologically healthy cell has cortical expression of pHMA, cell “a” (arrow). E–G, E′–G′: Dying cell “b” (arrow) acidifies slightly (green->yellow); fluorescence disappears. H–K, H′–K′: Cell “c” acidifies slightly and then acidifies substantially after engulfment and persists for many hours as a strongly acidified, but shrinking vesicle. L–O, L′–O′: Cells “d” and “e” acidify slightly and then acidify substantially after engulfment by “ f′ ” and “ f″ ”, the daughter cells of “ f ”. Notice that the pH of d and e’s engulfed corpses rebounds over time. Scale bar is 10 μm.
The in vivo pattern of pHMA acidification mirrors known apoptosis and engulfment reporters
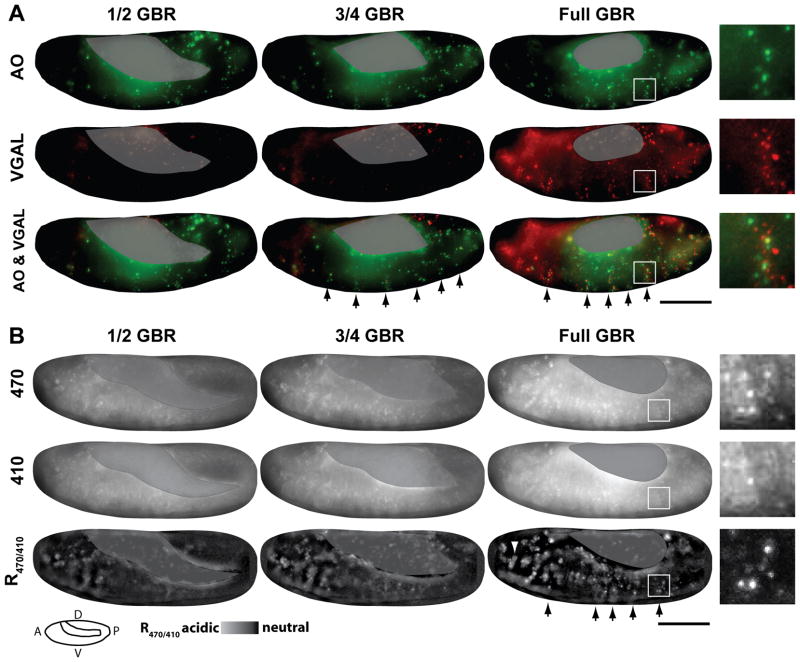
To confirm that pHMA can serve as an in vivo cell engulfment marker in Drosophila embryos, the pattern of pHMA acidification was compared to the known injectable cell death and engulfment reporters, acridine orange (AO) and a fluorogenic marker, VGAL, respectively (Fig. 3A) (Namba et al., 1997; Pazdera et al., 1998; Mergliano and Minden, 2003). Cell death and engulfment initiate during embryonic stage 12, when retraction of the germ band begins (Schock and Perrimon, 2002). As in mammals, Drosophila embryonic-cell engulfment is performed by both professional phagocytic cells, macrophages, and neighboring cells in the epithelium (Abrams et al., 1993; Pazdera et al., 1998; Mergliano and Minden, 2003). Time-lapse analysis has shown that epithelial cell death and engulfment occurs in a characteristic segmented pattern during germ band retraction and dorsal closure, stages 12–14 (Fig. 3A, arrows and Supp. Movie 5) (Abrams et al., 1993; Pazdera et al., 1998). Epithelial cell engulfment is performed mostly by neighboring cells rather than dedicated phagocytes (Pazdera et al., 1998). The apoptotic AO signal appears first, followed by the cell engulfment signal from VGAL (Fig. 3A) and (Mergliano and Minden, 2003). Phagocytosis of apoptotic cells is piecemeal as evidenced by the limited overlap between AO- and VGAL-positive vesicles (Figure 3A, inset). This indicates that different phagocytic compartments contain variable amounts of apoptotic nuclear and cytoplasmic material. Time-lapse analysis allows one to distinguish between AO and VGAL signals emanating from epithelial cells and macrophages based on their size and motility. Epithelial cells have smaller and less motile AO and VGAL bodies (Suppl. Movie 5).
Fig. 3.
The pattern of pHMA acidification mimics VGAL, a known cell engulfment marker. Shown here are images from time-lapse recordings of embryos at ½, ¾, and full germ band retraction (GBR). A: Shows a wild-type embryo injected with AO (green) and VGAL (red). The images are maximum intensity projections of three 5 μm optical sections. B: Shows a UAS-pHMA embryo that was injected with GAL4VP16 protein to induce uniform pHMA expression. Emission of pHMA collected with an FITC filter, when excited by 470 nm and 410 nm, and ratio image of the above signals (R470/410). These images are maximum intensity projections of ten 1 μm optical sections. The R470/410 ratio images were generated by subtracting the background from the 470 and 410 nm images as described in the methods section. Yolk auto-fluorescence is masked with a grey overlay. The segmentally repeated pattern of cell death is indicated by the arrows. Enlarged views of a single segment of dying cells are shown as insets. A cluster of macrophages that contains acidified pHMA is indicated by an arrowhead. Wide-field images were acquired with a 20X lens; lateral view; anterior, left; ventral, down; scale bar is 100 μm.
To generate strong, uniform pHMA expression, UAS-pHMA embryos were injected with purified Gal4VP16 protein during the syncytial blastoderm stage (Fig. 3B) (Cambridge et al., 1997). Since pHMA is based on the original GFP allele, not the enhanced GFP derivative, it requires about two hours to mature to its fluorescent form (Heim et al., 1995). Thus, pHMA that is expressed in the cellular blastoderm, stage 5, begins to fluoresce around stage 12. The majority of the experiments reported here were done on a wide-field, deconvolution microscope system. Relatively low, 20X magnification recordings were used to assess the pattern of pHMA expression and acidification. This magnification was not sufficient to visualize the cortical localization of pHMA. Acidified pHMA is more strongly excited by 470 nm light, which showed a stronger increase in fluorescence of both diffuse and punctate structures during germ band retraction (Fig. 3B, 470 nm row). Excitation at 410 nm, which preferentially excites pHMA at neutral pH displayed a gradual and slightly mottled increase in fluorescence over time (Fig. 3B, 410 nm row). This diffuse fluorescence seen both in the 410 nm and 470 nm excitation wavelengths may be due to variation in cell density or expressivity. Ratioing the 470 nm and 410 nm images, which biases toward acidified pHMA, cancelled out the diffuse signal and revealed a punctate pattern that closely resembled the spatial and temporal patterns of the AO apoptotic pattern and VGAL engulfment pattern (Fig. 3B, R470/410 row). Notice that the segmental pattern of pHMA R470/410 is similar to the segmental patterns of AO and VGAL (Fig. 3A and B, arrows, Suppl. Movie 6). Further, the size of AO-positive, VGAL-positive and acidic pHMA-positive vesicles are also similar (Fig. 3A and B, insets). These data show that pHMA behaves as expected in wild-type embryos consistent with the engulfment of apoptotic cells.
Increasing cell death increases the number of acidified pHMA bodies
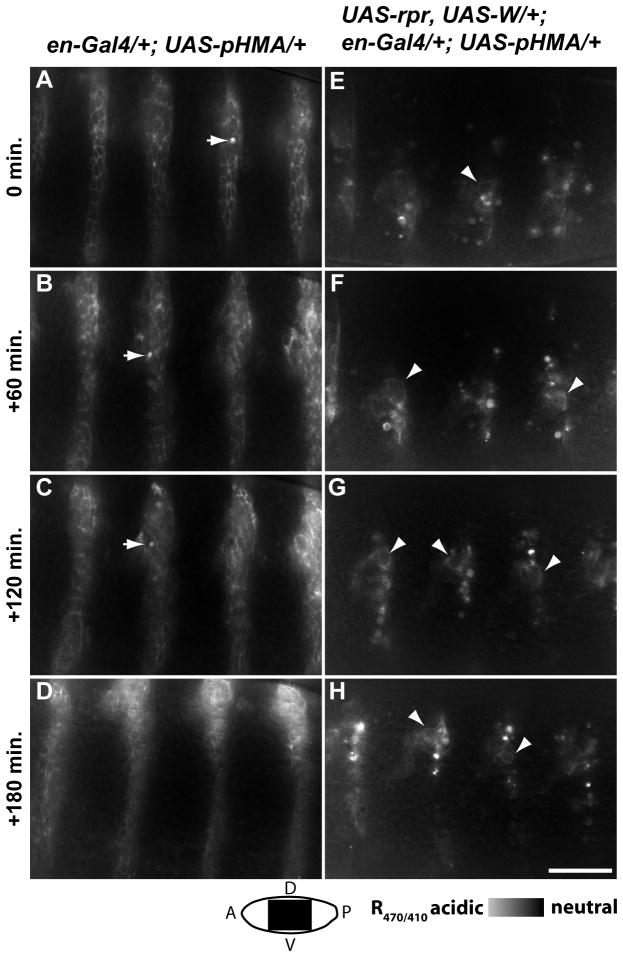
To validate the use of pHMA as an in vivo engulfment reporter, we tested if pHMA is able to indicate both increases and decreases in cell engulfment. To determine if pHMA can detect increases in cell engulfment, apoptosis was induced in a subset of embryonic cells by over-expressing UAS-reaper (UAS-rpr) and UAS-Wrinkled (UAS-W), along with UAS-pHMA, under the control of engrailed-GAL4. At 60X magnification, the cell cortices of wild-type, engrailed-positive cells that express pHMA are visible when excited with both 410 nm and 470 nm light (Fig. 4A–D and Suppl. Movie 7). It is important to note that the background threshold setting for these ratio images was set to that of the non-engrailed expressing cells. Cell cortices appear as mid-grey, while acidified pHMA vesicles appear as white (Fig. 4A–D, arrows). R470/410 images of engrailed cells expressing rpr, W and pHMA reveal that pHMA appears in diffuse, irregular stripes with numerous, bright vesicles (Fig. 4E–H and Suppl. Movie 8). The number of acidified pHMA-containing bodies in engrailed cells expressing rpr and W was much greater than the number observed in wild-type engrailed cells. The acidified pHMA bodies in rpr, W-expressing cells were similar in morphology to vesicles seen in wild-type embryos, however they were slightly larger, indicating that the removal of this larger than usual population of dying cells may be perturbed. There was also a significant increase in the number of macrophages that contained acidified-pHMA bodies in embryos with increased apoptosis (data not shown). Only a few of the rpr, W, pHMA cells retained their normal cortical pHMA localization (Fig. 4E–H, arrowheads). Since the rpr, W and pHMA genes are turned on simultaneously, most of the affected cells are dead or dying by the time pHMA becomes strongly fluorescent.
Fig. 4.
Induction of apoptosis increases the number of acidified pHMA vesicles. Shown here are R470/410 ratio images of en-Gal4;UAS-pHMA (A–D) and en-Gal4;UAS-pHMA,UAS-rpr,UAS-W (E–H) embryos during stages 13–14, dorsal closure. Notice the clearly outlined cells in the en-Gal4;UAS-pHMA embryo, with the occasional apoptotic cell (arrows), and the numerous acidified vesicles in the en-Gal4;UAS-pHMA,UAS-rpr,UAS-W embryo, with only a few cells showing cortical pHMA (arrowheads). These are single optical sections taken by a wide-field microscopy with a 60X lens; lateral view; anterior, left; ventral, down; scale bar is 30 μm.
Loss of cell death abrogates acidified pHMA bodies
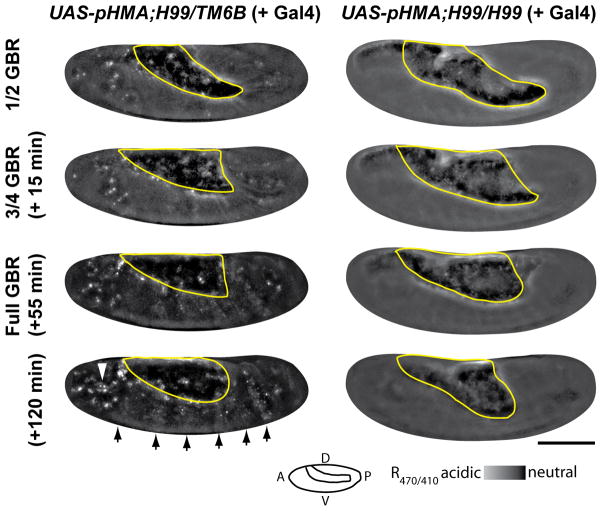
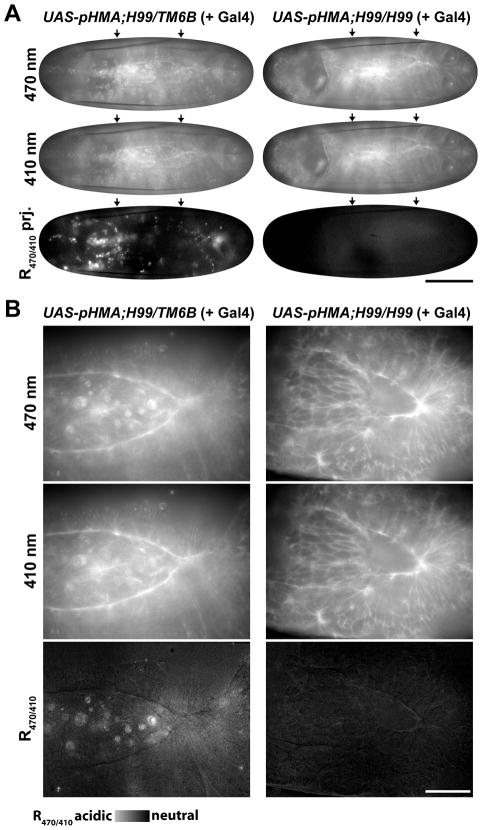
To assess if pHMA can detect decreases in cell death, we utilized the H99 mutation, which is missing the pro-apoptotic genes rpr, W and grim (White et al., 1994). H99 homozygous embryos exhibit virtually no apoptosis (White et al., 1994). Embryos heterozygous for this mutation develop normally and do not have any cell death defects. Ratio images of UAS-pHMA; H99/TM6B embryos injected with GAL4VP16 showed that these heterozygous embryos had wild-type patterns of acidified pHMA signal in the epithelium and macrophages (Fig. 5, left column, arrows and arrowhead). UAS-pHMA;H99/H99 embryos injected with GAL4VP16 had normal levels of pHMA expression in the individual pHMA channels (data not shown), but the R470/410 ratio images showed an almost total lack of acidified pHMA in epithelial cells and macrophages (Fig. 5, right column). The lack of pHMA signal in H99 homozygous embryos supports the hypothesis that pHMA will detect events that are associated with apoptosis. Taken together, these data clearly demonstrate that pHMA will be useful for detecting mutations and treatments that alter the normal pattern of cell death and engulfment.
Fig. 5.
Cell-death-deficient embryos lack an acidified pHMA signal. Shown here are R470/410 ratio images of a UAS-pHMA;H99/TM6B (or UAS-pHMA;TM6B/TM6B) embryo (left column) and a UAS-pHMA;H99/H99 embryo (right column) injected with GAL4VP16 at ½, ¾, full and 65 minutes post GBR. Arrows indicate the normal pattern of segmental cell engulfment; the arrowhead points to an area with high macrophage density. Notice the lack of pHMA acidification in the H99 homozygous embryo. The yolk mass is outlined in yellow. Wide-field microscopy; 20X lens; lateral view; anterior, left; ventral, down; scale bar is 100 μm.
Autophagy does not result in acidification of pHMA
Since pHMA is acidified in the lysosome after cell engulfment, it is pertinent to ask, if pHMA also detects autophagy, a process that also moves cytoplasmic material into the lysosome. In Drosophila embryos, autophagy is prevalent during dorsal closure, particularly in the amnioserosa. During dorsal closure, this entire extra-embryonic epithelial tissue is removed via apoptosis and autophagy (Toyama et al., 2008). Interestingly, in H99 mutants, the amnioserosa is eventually removed solely by autophagy that is readily detectable from late dorsal closure onward (Mohseni et al., 2009). We monitored pHMA fluorescence in H99 embryos throughout dorsal closure, and detected no acidified pHMA. In embryos past 50% dorsal closure, the acidification of pHMA in amnioserosa cells is clearly visible in H99/TM6B embryos, which have normal levels of apoptosis and autophagy (Fig. 6A and B, left column). In similarly staged homozygous H99 embryos, pHMA acidification was not observed, indicating that pHMA only indicates apoptotic cell engulfment, not autophagy (Fig. 6A and B, right column and Suppl. Movie 9). One possible reason that pHMA does not indicate autophagy may be due to its cortical localization and that autophagy primarily targets non-cortical, cytoplasmic components, such as mitochondria. Other internalization events, such as endocytosis and multi-vesicular body formation, rarely introduce cortical cytoplasm into the lysosomal compartment. If such processes do exist, the flux of cortical cytoplasm into the lysosome is below our limit of detection since healthy, pHMA-expressing S2 cells do not show any internal, acidified pHMA. Thus, pHMA appears to be specific for monitoring changes associated within apoptotic cells and the consequences of their engulfment.
Fig.6.
Cell-death-deficient embryos with normal autophagy show no pHMA acidification. Shown here are images captured with 470 nm and 410 nm excitation and R470/410 images of UAS-pHMA; H99/TM6B (or UAS-pHMA; TM6B/TM6B) (left column) and UAS-pHMA;H99/H99 (right column) embryos. Dorsal view, images from time-lapse series at >50% dorsal closure. A: Single focal planes of images captured with 470 nm and 410 nm excitation. Maximum intensity projections of five focal planes at the surface of the embryo taken 2 μm apart showing R470/410 ratio. Arrows indicate the anterior-posterior limits of the dorsal opening. Images were acquired with 20X lens; scale bar is 100 μm. B. Single focal plane; 60X images; scale bar is 30 μm.
pHMA reveals morphological changes prior to and during apoptotic cell engulfment
Apoptosis and cell engulfment are highly dynamic processes. Understanding the cell morphology changes associated with these processes in the context of the living tissue will be very helpful in determining the timeline of cell elimination. Since pHMA is visible in both healthy and apoptotic cells, we are able to track the morphological changes prior to engulfment by running time-lapse movies in reverse. To study these morphological changes, we recorded high resolution (60X magnification), high time sampling (30 second interval) time-lapse movies of wild-type, UAS-pHMA embryos injected with GAL4VP16. Under these extreme recording conditions, we found that the 410 nm light used to detect neutral pH pHMA had phototoxic effects, on the other hand, embryos were unaffected by the 470 nm light used under these time-lapse conditions to visualize acidified pHMA. Imaging with 410 nm light at lower magnification and longer time-lapse intervals (> 2 minutes) had no effect on embryo development. The lower resolution recordings revealed that engulfed corpses appear as bright, vesicles. To trace the morphological changes that give rise to these bright spots, the time-lapse recordings were played in reverse from the time a pHMA spot first appeared to when it converts back into an intact, normal-sized cell. Since these high spatial- and temporal-resolution recordings were made only with 470 nm excitation light, this section only comments on changes in overall pHMA signal. It is not known if these changes are related to changes in acidification or concentration.
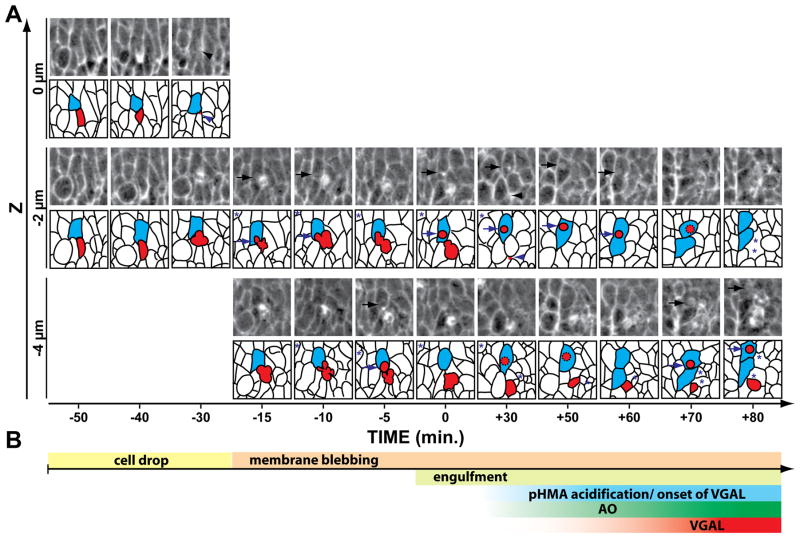
About 50 minutes prior to its engulfment, apoptotic cells appear to “drop” basally by 2–10 μm, but remained within the epithelial cell layer (Fig. 7A, −50 min, red shaded cell, see Suppl. Fig. 1 for a marked version). This cell dropping was marked by an accumulation of cortical pHMA around the dropping cell and an increase in diffuse cytoplasmic pHMA (Fig 7A, −40 min). It is not certain if this cortical pHMA accumulation is in the dying cell or the neighboring cells. These observations are consistent with studies in chick embryos describing a ring of actin and myosin within apoptotic cells that were extruded and within the surrounding cells (Rosenblatt et al., 2001). Once the dying cells had begun to drop, they exhibited membrane blebbing, as suggested by the irregular shape of the dying cell and partial spatial overlap with the engulfing cell (Fig 7A, −15 to −5 min arrows). The blebbing cells accumulated pHMA internally, depleting pHMA from the cortical surfaces of the blebs (Fig. 7A). The first signs of internalization of apoptotic cell fragments by neighboring cells occurred five to fifteen minutes prior to the formation of a discrete vesicle, or phagosome, within the engulfing cell (Fig. 7A, −15 to 0 min, arrows). Initially, pHMA associated with the phagosome appeared both cortically and as internal foci (Fig. 7A, 0 to 30 min, arrows). At this resolution it was not possible to determine if the cortical pHMA surrounding the phagosome was on the cytoplasmic surface or internal to the phagosome. Forty minutes after engulfment the pHMA appeared diffuse and uniformly distributed throughout the phagosome, suggesting that the phagosome had completely fused with the lysosome and that breakdown of the engulfed corpse had begun. pHMA persisted in the engulfed vesicle for many hours after engulfment. The remainder of the apoptotic corpse that was not engulfed by the cell shown in Fig. 7A was eventually removed by other neighbor cells and later by a macrophage (data not shown).
Fig. 7.
Timeline of apoptosis revealed by pHMA. A: Shown here is a series of time-lapse images at different focal planes centering on a single apoptotic cell in the ventral epidermis of a stage 12, UAS-pHMA embryo injected with GAL4VP16 and imaged for acidified pHMA with 470 nm excitation using a 60X lens. Five, 2 μm optical sections were recorded at a 30 second interval. Each frame is a 21.6 × 21.6 μm sub-section of the field-of-view. A cartoon of the cell boundaries appears below each image where the dying cell is highlighted in red and the cell that engulfs a portion of the dying cell is highlighted in blue. Asterisks mark other dying cells in the field. The zero time-point indicates the point at which the engulfing cell is first observed to contain a discrete phagocytic vesicle. Fifty minutes prior to the engulfment event, the apoptotic cell (red) had normal cell morphology and its pHMA was detected in the cell cortex, as expected for healthy cells. A distinguishing feature of this dying cell was the diffuse distribution of pHMA throughout the apical and sub-apical cytoplasm, which increased in intensity over the next ten minutes. Thirty minutes prior to the engulfment event, the dying cell moved basally (arrowhead), which was followed by membrane blebbing and overlap with the engulfing cell (arrows) over the next 30 minutes. The membrane blebbing was accompanied by the appearance of internal foci of pHMA within the dying cell, with lower levels of pHMA at the periphery of the membrane blebs. By time zero, the engulfment of a piece of the apoptotic cell was completed with the appearance of a discrete vesicle (red) seen in an adjacent cell (blue). This initial phagosome appeared to have a non-uniform distribution of pHMA, which became diffuse in the ensuing 40 minutes. The rest of the cell corpse was engulfed by other neighboring cells and macrophages. These events were not traced because of their complexity. Notice that the engulfing cell divided soon after engulfment. B: A timeline of the engulfment events plotted according to the time scale shown in (A).
pHMA acidification in vivo occurs after cell corpse engulfment
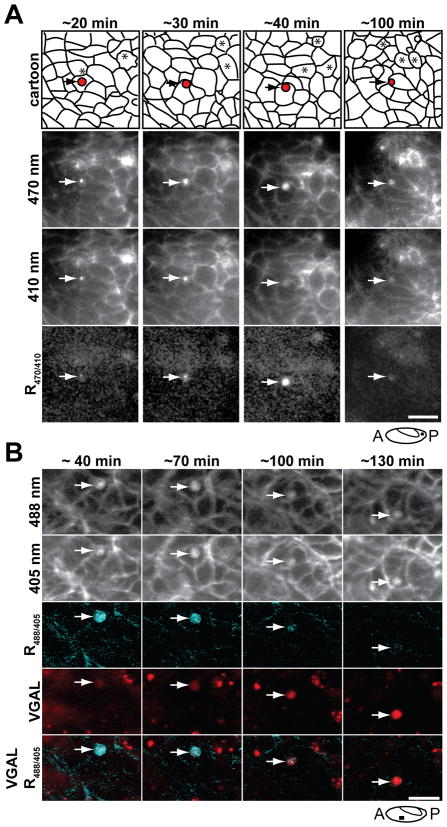
To determine when phagosomes of engulfed cell corpses become acidified, the high-magnification time-lapse recordings were repeated at two minute intervals with 410 nm and 470 nm illumination, conditions that allowed wide-field imaging for several hours without adverse effects. Between the period of apoptotic cell dropping and the final removal of apoptotic corpses, the fluorescence intensity of pHMA increased both in the corpses and the newly engulfed vesicles (Fig. 7A and 8A, asterisks and arrow), but with little change in the ratio of acidified pHMA, indicating this increase in fluorescence was mostly likely due to cell shrinkage rather than acidification (Fig. 8A, R470/410). The signal from yolk auto-fluorescence interferes with our ability to make fine R470/410 ratio calculations so that subtle pH changes prior to engulfment went undetected. The engulfed bodies appeared to become strongly acidified about 30–60 minutes after cell engulfment (Fig. 8A, R470/410, arrows). The time of acidification correlates with the transition from localized pHMA within engulfed vesicles to uniform distribution of pHMA within the phagosomes, indicating that it takes this time for the phagosome to fuse with lysosomes and begin breaking down the contents of the phagosome. Our analysis of pHMA in S2 cells showed that pHMA acidification generally took 30–60 minutes for the apoptotic cell pH to drop from 6.8 to below 6, the pH which we consider to be strongly acidified.
Fig. 8.
Acidification of pHMA in vesicles is followed by the VGAL signal. A: Shown here is series of ratio images of a UAS-pHMA embryo injected with GAL4VP16 to show the acidification of the phagosome. Notice that pHMA diffuses within the vesicle as it becomes acidified (arrows). Wide-field microscopy of posterior lateral epithelium at late GBR imaged with a 60X lens. The indicated time points are relative to the initial engulfment event. The vesicle is shown in red in the cartoon; dying cells are indicated with asterisks. The images are single optical sections extracted from a time-lapse series composed of five 2 μm optical sections acquired at two-minute intervals. Scale bar is 10 μm. B: Shown here are selected time points from a time-lapse recording of a UAS-pHMA embryo injected with GAL4VP16 and VGAL to determine the relationship between phagosome acidification and cleavage of VGAL to its fluorescent form. These images were captured with a confocal microscope. The indicated time-points are relative to the initial engulfment event. Each row of images shows (top to bottom): pHMA fluorescence when excited by 488 nm, 405 nm, ratio image (R488/405) in cyan, VGAL in red, and the overlap of R488/405 with VGAL. Each image is a maximum intensity projection of three 1 μm optical sections. Scale bar is 10 μm.
It is known that wild-type multimeric GFP-like proteins can survive in the lysosomal environment (Katayama et al., 2008). The diffused pHMA signal persisted for several hours within pHMA-positive vesicles. During their maturation, engulfed vesicles became smaller and showed a decrease in the pHMA signal (Fig. 8A, 100 min). While it is difficult to follow individual apoptotic cells and their remnants for long periods of time due to the embryo’s large-scale morphogenetic movements, we have observed acidified pHMA vesicles persist for more than five hours (data not shown). The same longevity of engulfed pHMA was seen in S2 cells.
The delay between apoptotic cell engulfment and acidification of the resulting phagosome is not unexpected and has been observed in other situations. The timeline of cell engulfment and apoptotic phagosome maturation in the epithelium of Drosophila embryos (Fig. 7B) is comparable to the engulfment schedule observed in other systems. In C. elegans, the majority of corpses that appear refractile with DIC microscopy disappear within 30 minutes of their appearance (Yu et al., 2006). Time-lapse, fluorescence imaging studies in C. elegans have shown that lysosomes are recruited to the phagosomal surface within ten minutes of engulfment, that lysosomal fusion occurs twenty minutes later, and that phagosomal degradation takes place 50–70 minutes after engulfment (Yu et al., 2006; Yu et al., 2008; Zhou and Yu, 2008). Mammalian cell culture systems show more rapid acidification of phagosomes. Murine peritoneal and bone marrow macrophages acidify phagosomes to a steady state of pH 5 within fifteen minutes of engulfment (Underhill, 2005; Yates and Russell, 2005; Haggie and Verkman, 2007). Another study observed phagosome acidification rates by non-professional and professional phagocytes, using LysoTracker, with apoptotic cells and various opsonized beads to be on the order of 60–90 minutes (Erwig et al., 2006). These studies show that different cell types process engulfed material, such as apoptotic corpses, bacteria, and beads, at different rates. The engulfment schedule for Drosophila embryonic epithelial cells appears to be within known limits.
Timeline of pHMA acidification with VGAL and AO signaling
Prior to the development of pHMA, VGAL was used to monitor cell engulfment events. To determine the correlation between VGAL and pHMA acidification, VGAL and GAL4VP16 were injected into UAS-pHMA embryos and time-lapse recordings of VGAL fluorescence and pHMA ratios were made. Figure 8B shows a vesicle about 40 minutes after engulfment where the pHMA was strongly acidified. At this time, there was very little VGAL signal. Over the next ninety minutes, the pHMA signal collapsed into a smaller volume and decreased in intensity. There was a concomitant increase in VGAL fluorescence, indicating that the VGAL signal matures more slowly than pHMA acidification. This lag between acidification and VGAL maturation is not surprising since proton diffusion is very rapid compared to the enzymatic degradation of VGAL to its fluorescent form by lysosomal β-galactosidase (Minden, 1996). Like pHMA, the VGAL signal persisted for long periods of time in the developing embryo. VGAL fluorescence persists much longer than pHMA because its fluorescent moiety, resorufin, is not biodegradable.
AO has been prominently used in Drosophila to identify apoptotic corpses. Our previous study determined that there is a 15–45 minute delay between the initial appearance of AO and subsequent VGAL fluorescence (Mergliano and Minden, 2003). It is desirable to also determine the temporal relationship between pHMA acidification and AO signaling, however, since pHMA and AO have overlapping fluorescence emission wavelengths, it was not possible to record pHMA acidification and AO fluorescence simultaneously. We surmise that if VGAL fluorescence peaks about one to two hours minutes after the initial appearance of engulfment vesicles and that AO precedes VGAL by 15–45 minutes, then one can conclude that AO fluorescence appears well after initial engulfment events. This estimate contradicts previous claims that AO fluoresces in un-engulfed apoptotic cells (Abrams et al., 1993). It is important to note that this earlier study used single time-point analysis of AO stained embryos and focused on interstitial cell bodies that may have been either macrophages or sloughed off brain cells. Our time-lapse analysis of the engulfment of apoptotic epithelial cells by neighboring epithelia is more akin to studies in C. elegans where AO stains corpses after they are engulfed (Kinchen et al., 2008). All engulfment in C. elegans is mediated by neighboring epithelia, not macrophages Mutations in C. elegans that prevent phagosomal maturation or acidification lack AO staining. A timeline of the events surrounding the engulfment of a dying cell is diagrammed in Fig. 7B. This timeline was assembled from several different time-lapse experiments as described in the preceding sections. This timeline will be helpful when analyzing mutations or treatments that affect different aspects of apoptosis and engulfment.
pHMA reveals changes in larval tissues
To demonstrate the broad utility of pHMA, it was expressed in larval wing imaginal discs and salivary glands (Fig. 9). The wing imaginal disc is a model system to study and identify apoptotic signals (Umemori et al., 2009) as well as organ size regulation and cell competition (de la Cova et al., 2004; Moreno and Basler, 2004; Li and Baker, 2007; Martin et al., 2009). In wild-type discs apoptosis occurs at low levels and is interspersed throughout the disc (Milan et al., 1997). The absence of fast-moving macrophage and the amenability to genetic manipulation that allows to induce or suppress apoptosis and cell engulfment (Li and Baker, 2007) make an imaginal disc an attractive model for studying cell engulfment.
Fig 9.
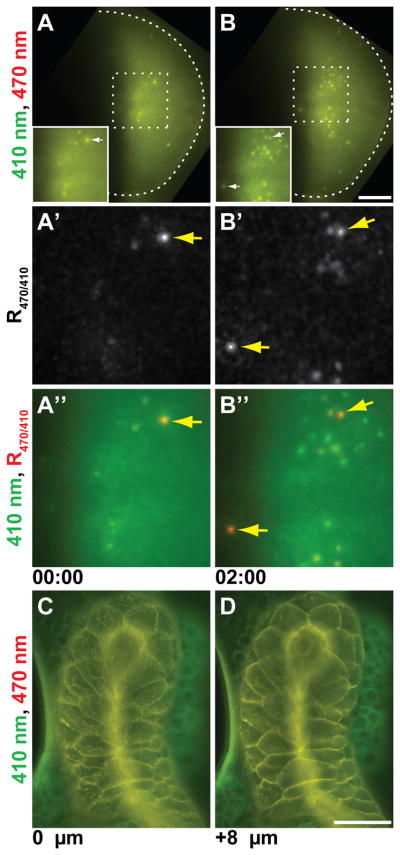
pHMA expression in salivary glands and wing imaginal discs. A and B: pHMA expression in engrailed-positive cells in the wing pouch of a larval imaginal disc. Maximum projection images of ten 2 μm optical sections extracted from a time-lapse movie are shown at two time points, two hours apart. The images captured with 410 nm excitation are displayed in green and 470 nm are in red. The arrows point to cellular material that show strong acidification. The scale bar is 30 μm. A′ and B′: Show magnified R470/410 ratio images of the insets in A and B. A″ and B″: Shows the same region as A′ and B′ with the 410 nm fluorescence signal in green overlayed with the R470/410 image in red. C and D: Shows Ubi-pHMA expression in a larval salivary gland. The images are surface (C) and subsurface (D) sections of a single time point. The 410 nm fluorescence signal is displayed in green and 470 nm is in red. Scale bar is 100 μm.
en-Gal4 was used to drive the expression of UAS-pHMA in the posterior compartment of the developing wing imaginal disc (Fig. 9A and B). Time-lapse imaging of the wing pouch of the dissected disc reveals relatively uniform pHMA expression across the posterior compartment. A few cells displayed increased pHMA concentration, which was represented by an increase in both 410 nm and 470 nm fluorescence signal, R470/410 was unchanged. Soon after the initial increase in neutral pH pHMA, the R470/410 signal increased, indicating acidification. The pHMA signal from the presumably engulfed apoptotic cell was later extinguished (Fig. 9A′, B′, A″, B″, and Suppl. Movie 10). The magnification used for these recordings was not sufficient to resolve individual cells and cell fragments. These time-lapse data show that pHMA will be a useful reporter for cell death and engulfment in the wing disc. The dynamics of the pHMA response appears to be similar between embryos and wing discs. Like the embryo epidermis, the engulfment of dying wing epithelia appears to be neighbor dependent.
Another Drosophila tissue that has served as a model for cell death is larval salivary gland. Salivary glands from wall-crawling Ubi-pHMA larvae and white pre-pupae were dissected and mounted for time-lapse microscopy. The pHMA signal in these late larvae/early pupae is mostly cortical (Fig. 9C and D). There appears to be some fibrillar and vesicular accumulation of pHMA. Under the culture conditions used in this experiment, the signal from 470 nm excitation that is slightly higher than the 410 nm signal in the salivary gland relative to the attached fat body, suggesting that the salivary gland is acidified relative to the adjacent fat body. Further studies that reproduce the anterior to posterior wave of ecdysone-induced caspase activation (Takemoto et al., 2007) will be instrumental in relating caspase activity to pHMA acidification and engulfment in the salivary gland.
Autophagy is prevalent during histolysis of the salivary gland (Lee and Baehrecke, 2001), yet it has also been shown that histolysis is mediated by an apoptosome-dependent process that is parallel to or downstream of autophagy (Akdemir et al., 2006). While pHMA does not appear to report autophagy in embryos, further detailed studies of pHMA in salivary glands undergoing histolysis will be interesting for determining the relative contributions of autophagy and engulfment to the histolysis of the salivary glands.
Summary
The expression of pHMA, a fusion of pHluorin to actin-binding domain of Moesin, in cultured Drosophila cells and embryos has proved to be a useful marker for apoptotic cell engulfment. In healthy cultured cells, pHMA resides in the cell cortex and reveals the cell’s dynamic morphological behavior. It also reports on the healthy cell’s neutral pH. When cultured cells begin to die the cells become rounded and quiescent and the cytoplasmic pH drops to about 6.7. Some of these apoptotic cells burst and lose their pHMA signal, while others are engulfed. Thirty to sixty minutes after engulfment, the engulfed pHMA reports the acidification of the phagosome to about pH 5. The acidified pHMA signal persists for several hours, but the pH of the phagosome tends to partially neutralize over time. Expression of pHMA in embryos shows similar changes over the life cycle of the cell. The main difference being that apoptotic cultured cells are engulfed as a single entity by one phagocytic cell, while dying epithelial cells are removed in a piecemeal fashion by multiple neighboring cells, as well as macrophages. pHMA faithfully reports both increases and decreases in embryonic cell death and discriminates between apoptotic cell engulfment and autophagy. Overall, pHMA provides a tool for monitoring several different events during apoptosis in a characteristic time sequence. These features make pHMA a useful reagent for screening for new mutations that affect embryonic apoptosis and cell engulfment. The timeline, which shows that morphological changes occur as early as 50 minutes prior to engulfment and requires several hours to completely degrade the cell corpse, will be useful tool for the detailed characterization of existing and yet-to-be-discovered mutations. Likewise, expression of pHMA in cultured cells will allow one to screen for treatments that affect apoptosis and engulfment in vitro. It will also be interesting to place the other in vivo apoptosis reporters within this timeline to see when they are activated relative to the changes indicated by pHMA. Finally, we anticipate that pHMA will also be a useful reporter for cell death and engulfment in other organisms.
EXPERIMENTAL PROCEDURES
DNA constructs and fly stocks
UAS-pHMA was constructed by replacing the GFP portion of pUAST-GFP::Moesin (GMA) (Bloor and Kiehart, 2001; Dutta et al., 2002), plasmid provided by D.P Kiehart (Duke University) with pHluorin, which was provided by G. Miesenbock (Yale University). A 727 bp fragment of pHluorin was subcloned into UAS-GMA vector, to replace GFP between EcoRI and HindIII sites. To generate the Ubi-pHMA reporter, we replaced the GFP fragment in the pCaSpeR4 Ubi-GFP plasmid (Davis et al., 1995) with the pHluorin::moesin fusion. Transgenic Drosophila were generated using standard procedures (Baek and Ambrosio, 1994) by BestGene Inc. Transgenic stocks with strongest expression were recombined to generate stocks that have two copies of UAS-pHMA (2xUAS-pHMA) on either the second or third chromosomes. Progeny of recombined third chromosome insertion, Cyo/Bl;2xUAS-pHMA/TM6B, were used for Gal4VP16 injections. Engrailed-Gal4 and H99/TM3 (Bloomington Stock Center) were used to generate en-Gal4/Cyo;UAS-pHMA/TM6B and 2xUAS-pHMA; H99/TM6B, respectively. UAS-rpr, UAS-W on × was provided by J.R. Nambu (University of Massachusetts, Amherst).
S2 cell culture, transfection and fixation
S2 and S2R+ cells (Drosophila Genomics Resource Center, Bloomington, IN) were grown in Modified Schneider’s Drosophila Medium (Lonza 04-351Q) supplemented with 10% heat inactivated Fetal Bovine Serum (Lonza 14-503E) and pen/strep (HyClone SV30010). One million cells per well, in a 6-well plate, were transiently transfected with Ubi-pHMA, using Effectene transfection Reagent and adherent cells protocol (QIAGEN). For each transfection, 1 μg of DNA, 8 μl of Enhancer, and 25 μl of Effectene were used. Using this approach about 1/10 of cells expressed pHMA within two days of transfection. Cells retained pHMA for up to two weeks.
S2R+ cells were fixed with 4% paraformaldehyde in PBS for 20 minutes at 4°C with gentle rocking. The fixed cells were pelleted, resuspended in PBS, and permeabilized with 30 μg/ml digitonin in PBS for 5 minutes at room temperature. The pH was adjusted to 4.5 – 8.5, in 0.5 unit increments, by diluting the cell suspension in two volumes of 100 mM sodium phosphate buffer.
pHMA fluorescence versus pH standard curve construction
Fixed and pH adjusted S2R+ cells were imaged with 410 nm and 470 nm excitation. For each pH value, twenty fields of cells, each containing an average of 5 cells per field, were recorded. Three focal planes 2 μm apart were imaged by wide-filed microscopy with a 40X lens for each field of view. A MATLAB script was created to calculate the R470/470 ratios for each field of view. The exposure was adjusted to avoid pixel saturation. The median value for each image was used as background. To calculate the ratio, the background value at the corresponding excitation wavelength was subtracted from each image, and R470/410 of pixel values that were greater than three times the background but less than saturation were computed. The average R470/410 and standard deviation was calculated for each set of images. The data points were plotted using GraphPad software and fitted with a nonlinear curve pH = 6.379-log10(((0.6173–2.077)/(R470/410-2.077))−1), r2 = 0.9981.
Live S2 and S2R+ cells were imaged using microscope exposure settings that were identical to fixed cells. These cells were exposed with 410 nm and 470 nm at 10-minute intervals. Areas that contained individual cells were cropped. The average of the background was calculated for entire movie before cropping. This background was used for all cropped images in the movie. The pH of individual cells was calculated at each time point using the computed R470/410 and the above equation.
Embryo microinjection
Dechorionated embryos were prepared for injection as previously described (Li et al., 1999). Stage 3 embryos were injected with AO (0.5 mg/ml), VGAL (~1.8 mg/ml), or purified Gal4VP16 protein (0.2 mg/ml) as previously described (Mergliano and Minden, 2003). Post injection, embryos were incubated at 25°C for 5–6 hours or at 15°C for 18–20 hours prior to time-lapse imaging.
Salivary gland and wing imaginal disc preparation
The wing discs were collected from wall-crawling larvae, while the salivary glands were collected from either wall-crawling larvae or white pre-pupae. Wing imaginal discs and larval salivary glands were dissected in supplemented Schneider’s medium that was also used for S2 cell culture. The discs were mounted in Schneider’s medium on the gas permeable membranes of petriPERM dishes. The samples were surrounded with halocarbon oil to limit desiccation and covered with a No. 1 coverslip. Time-lapse recordings were made with a wide-field microscope system.
Microscopy
Wide-field, time-lapse images were collected on a Delta Vision imaging system controlled by softWoRx software (Applied Precision, Issaquah, WA) configured around an Olympus IX70 inverted microscope, using a filtered Xenon white light source. Images of VGAL (resorufin-β-galactoside-polyethylene glycol 1,900) and AO images were obtained with a standard “quad” dichroic filter set, using FITC and TRITC emission filters, respectively. The 410 and 470 nm excitation wavelength images were collected using a custom dichotic filter set (Chroma Technology, Rockingham, VT); the images were collected with a FITC emission filter. Confocal images were collected with a Carl Zeiss LSM 510 Meta/UV DuoScan Spectral Confocal Microscope mounted on an inverted Axio Observer Z1 microscope frame equipped with a 405 nm laser diode (30 mW); 458, 477, 488, and 514 nm Argon laser (30 mW all lines); and 561 nm DPSS laser (15 mW). We found that the light intensity of the wide-field microscope system was less phototoxic than the confocal microscope. The confocal was used for the pHMA plus VGAL experiments because this system was capable of near instantaneous line-by-line automatic switching of each excitation laser via an accusto-optical tunable filter (AOTF) in conjunction with a quad-line dichroic mirror.
Processing of pHMA-expressing embryo images
Wide-field ratio images were generated by dividing individual pixel values and rescaling the output minimum-maximum. Images of embryos acquired with a 20X lens were processed using softWoRx software to subtract local background, which included autofluorescence that was generated by the yolk during 410 nm excitation. To approximate the background, the images were blurred using a mean 2D Filter with kernel size of 45 pixels. The reference images were smoothed with a median 2D Filter with a three pixel kernel size. The background-subtracted images were generated by subtracting blurred images from the reference images. A constant value was added to the background-subtracted images to eliminate negative pixel intensities. The resulting images were then divided, 470 nm excitation by 410 nm excitation, to generate a ratio image. The images in Fig. 7 were processed to enhance phagocytic vesicles in ImageJ using an FFT bandpass filter set to “Filter Large Structures Down” to 15 pixels and “Filter Small Structures Up” to 0 pixels. The products were subsequently smoothed to suppress graininess. In Fig. 9, to determine R470/410 only for the posterior region of wing disc that expressed pHMA, the ratio images were calculated using the MATLAB script that was generated for S2 cell analysis. Ratios for points with signal at 470 nm excitation that was greater than four times the background level was used. To remove noise, the ratio images of the wing imaginal disc in Fig. 9 were median filtered with a radius of two pixels.
Supplementary Material
Supplemental Figure 1. Timeline of apoptosis revealed by pHMA. Shown here is a version of the images shown in Fig. 7A with a semi-transparent overlay. The dying cell is shaded red and the engulfing cell is shaded blue in the fluorescence images. The dashed lines in the cartoons indicate vesicles that are out of focus.
Supplemental Movie 1 Shows pHMA fluorescence overlayed with transmitted light time-lapse recording of cell a in Fig. 2C and D cell with cortical pHMA localization and neutral pH.
Supplemental Movie 2. Shows pHMA fluorescence overlayed with transmitted light time-lapse recording of cell b in Fig. 2E–G that is likely necrotic.
Supplemental Movie 3. Shows pHMA fluorescence overlayed with transmitted light time-lapse recording of engulfed cell c in Fig. 2H–K.
Supplemental Movie 4. Shows pHMA fluorescence overlayed with transmitted light time-lapse recording of pHMA-expressing cells d and e in Fig. 2L–O. The cell that has some red has already engulfed something. Two other cells acidify slightly (green → yellow) and are engulfed by the daughters of the red cell. They quickly acidify after engulfment (yellow → red).
Supplemental Movie 5. Shows the time-lapse recording of the wild-type embryo injected with AO and VGAL that was used for Fig. 3A.
Supplemental Movie 6. Shows the time-lapse recording of the UAS-pHMA embryo injected with GAL4VP16 that was used for Fig. 3B.
Supplemental Movie 7. Shows the time-lapse recording of the en-GAL4/+; UAS-pHMA/+ embryo that was used for Fig. 4A–D.
Supplemental Movie 8. Shows the time-lapse recording of the UAS-rpr, UAS-W/+; en-GAL4/+; UAS-pHMA/+ embryo that was used for Fig. 4E–H.
Supplemental Movie 9. Shows the time-lapse recording of the dorsal closure in UAS-pHMA;H99/H99 embryo that was used for Fig. 6B, right column. The R470/410 signal remains uniform throughout dorsal closure, indicating that pHMA is not acidified.
Supplemental Movie 10. Shows the time-lapse recording of the en-GAL4, UAS-pHMA wing disc from late third instar larva that was used for Fig. 9A-A″ and 9B-B″. A five-hour recoding was captured at ten-minute intervals.
Acknowledgments
We thank Olga Strachna for technical assistance; Brooke McCartney and the Minden laboratory for their critical reading of the manuscript. We are indebted to Dr. Bino John for help with MATLAB programming and high throughput image analysis. This work was supported by a grant from the National Institutes of Health (1R21 HD049554) to J.S.M. and an American Cancer Society Postdoctoral Fellowship (PF-07-118-01-DDC) to E.F. J.F. also acknowledges financial support through the NIH National Technology Center for Networks and Pathways (5U54 RR022241) funded through the NIH Roadmap for Medical Research.
References
- Abrams JM, White K, Fessler LI, Steller H. Programmed cell death during Drosophila embryogenesis. Development. 1993;117:29–43. doi: 10.1242/dev.117.1.29. [DOI] [PubMed] [Google Scholar]
- Akdemir F, Farkas R, Chen P, Juhasz G, Medved’ova L, Sass M, Wang L, Wang X, Chittaranjan S, Gorski SM, Rodriguez A, Abrams JM. Autophagy occurs upstream or parallel to the apoptosome during histolytic cell death. Development. 2006;133:1457–1465. doi: 10.1242/dev.02332. [DOI] [PubMed] [Google Scholar]
- Baek KH, Ambrosio L. An efficient method for microinjection of mRNA into Drosophila embryos. Biotechniques. 1994;17:1024–1026. [PubMed] [Google Scholar]
- Bardet PL, Kolahgar G, Mynett A, Miguel-Aliaga I, Briscoe J, Meier P, Vincent JP. A fluorescent reporter of caspase activity for live imaging. Proc Natl Acad Sci U S A. 2008;105:13901–13905. doi: 10.1073/pnas.0806983105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry MA, Eastman A. Endonuclease activation during apoptosis: the role of cytosolic Ca2+ and pH. Biochem Biophys Res Commun. 1992;186:782–789. doi: 10.1016/0006-291x(92)90814-2. [DOI] [PubMed] [Google Scholar]
- Bloor JW, Kiehart DP. zipper Nonmuscle myosin-II functions downstream of PS2 integrin in Drosophila myogenesis and is necessary for myofibril formation. Dev Biol. 2001;239:215–228. doi: 10.1006/dbio.2001.0452. [DOI] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Cambridge SB, Davis RL, Minden JS. Drosophila mitotic domain boundaries as cell fate boundaries. Science. 1997;277:825–828. doi: 10.1126/science.277.5327.825. [DOI] [PubMed] [Google Scholar]
- Davis I, Girdham CH, O’Farrell PH. A nuclear GFP that marks nuclei in living Drosophila embryos; maternal supply overcomes a delay in the appearance of zygotic fluorescence. Dev Biol. 1995;170:726–729. doi: 10.1006/dbio.1995.1251. [DOI] [PubMed] [Google Scholar]
- de la Cova C, Abril M, Bellosta P, Gallant P, Johnston LA. Drosophila myc regulates organ size by inducing cell competition. Cell. 2004;117:107–116. doi: 10.1016/s0092-8674(04)00214-4. [DOI] [PubMed] [Google Scholar]
- Dutta D, Bloor JW, Ruiz-Gomez M, VijayRaghavan K, Kiehart DP. Real-time imaging of morphogenetic movements in Drosophila using Gal4-UAS-driven expression of GFP fused to the actin-binding domain of moesin. Genesis. 2002;34:146–151. doi: 10.1002/gene.10113. [DOI] [PubMed] [Google Scholar]
- Edwards KA, Demsky M, Montague RA, Weymouth N, Kiehart DP. GFP-moesin illuminates actin cytoskeleton dynamics in living tissue and demonstrates cell shape changes during morphogenesis in Drosophila. Dev Biol. 1997;191:103–117. doi: 10.1006/dbio.1997.8707. [DOI] [PubMed] [Google Scholar]
- Erwig LP, Henson PM. Clearance of apoptotic cells by phagocytes. Cell Death Differ. 2007 doi: 10.1038/sj.cdd.4402184. [DOI] [PubMed] [Google Scholar]
- Erwig LP, McPhilips KA, Wynes MW, Ivetic A, Ridley AJ, Henson PM. Differential regulation of phagosome maturation in macrophages and dendritic cells mediated by Rho GTPases and ezrin-radixin-moesin (ERM) proteins. Proc Natl Acad Sci U S A. 2006;103:12825–12830. doi: 10.1073/pnas.0605331103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haggie PM, Verkman AS. Cystic fibrosis transmembrane conductance regulator-independent phagosomal acidification in macrophages. J Biol Chem. 2007;282:31422–31428. doi: 10.1074/jbc.M705296200. [DOI] [PubMed] [Google Scholar]
- Heim R, Cubitt AB, Tsien RY. Improved green fluorescence. Nature. 1995;373:663–664. doi: 10.1038/373663b0. [DOI] [PubMed] [Google Scholar]
- Katayama H, Yamamoto A, Mizushima N, Yoshimori T, Miyawaki A. GFP-like proteins stably accumulate in lysosomes. Cell Struct Funct. 2008;33:1–12. doi: 10.1247/csf.07011. [DOI] [PubMed] [Google Scholar]
- Kinchen JM, Doukoumetzidis K, Almendinger J, Stergiou L, Tosello-Trampont A, Sifri CD, Hengartner MO, Ravichandran KS. A pathway for phagosome maturation during engulfment of apoptotic cells. Nat Cell Biol. 2008;10:556–566. doi: 10.1038/ncb1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CY, Baehrecke EH. Steroid regulation of autophagic programmed cell death during development. Development. 2001;128:1443–1455. doi: 10.1242/dev.128.8.1443. [DOI] [PubMed] [Google Scholar]
- Li QJ, Pazdera TM, Minden JS. Drosophila embryonic pattern repair: how embryos respond to cyclin E-induced ectopic division. Development. 1999;126:2299–2307. doi: 10.1242/dev.126.10.2299. [DOI] [PubMed] [Google Scholar]
- Li W, Baker NE. Engulfment is required for cell competition. Cell. 2007;129:1215–1225. doi: 10.1016/j.cell.2007.03.054. [DOI] [PubMed] [Google Scholar]
- Li Z, Burrone J, Tyler WJ, Hartman KN, Albeanu DF, Murthy VN. Synaptic vesicle recycling studied in transgenic mice expressing synaptopHluorin. Proc Natl Acad Sci U S A. 2005;102:6131–6136. doi: 10.1073/pnas.0501145102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangahas PM, Zhou Z. Clearance of apoptotic cells in Caenorhabditis elegans. Semin Cell Dev Biol. 2005;16:295–306. doi: 10.1016/j.semcdb.2004.12.005. [DOI] [PubMed] [Google Scholar]
- Martin FA, Herrera SC, Morata G. Cell competition, growth and size control in the Drosophila wing imaginal disc. Development. 2009;136:3747–3756. doi: 10.1242/dev.038406. [DOI] [PubMed] [Google Scholar]
- Mazzalupo S, Cooley L. Illuminating the role of caspases during Drosophila oogenesis. Cell Death Differ. 2006;13:1950–1959. doi: 10.1038/sj.cdd.4401892. [DOI] [PubMed] [Google Scholar]
- Mergliano J, Minden JS. Caspase-independent cell engulfment mirrors cell death pattern in Drosophila embryos. Development. 2003;130:5779–5789. doi: 10.1242/dev.00824. [DOI] [PubMed] [Google Scholar]
- Miesenbock G, De Angelis DA, Rothman JE. Visualizing secretion and synaptic transmission with pH-sensitive green fluorescent proteins. Nature. 1998;394:192–195. doi: 10.1038/28190. [DOI] [PubMed] [Google Scholar]
- Miksa M, Komura H, Wu R, Shah KG, Wang P. A novel method to determine the engulfment of apoptotic cells by macrophages using pHrodo succinimidyl ester. J Immunol Methods. 2009;342:71–77. doi: 10.1016/j.jim.2008.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milan M, Campuzano S, Garcia-Bellido A. Developmental parameters of cell death in the wing disc of Drosophila. Proc Natl Acad Sci U S A. 1997;94:5691–5696. doi: 10.1073/pnas.94.11.5691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minden JS. Synthesis of a new substrate for detection of lacZ gene expression in live Drosophila embryos. Biotechniques. 1996;20:122–129. doi: 10.2144/96201rr02. [DOI] [PubMed] [Google Scholar]
- Mohseni N, McMillan SC, Chaudhary R, Mok J, Reed BH. Autophagy promotes caspase-dependent cell death during Drosophila development. Autophagy. 2009;5:329–338. doi: 10.4161/auto.5.3.7444. [DOI] [PubMed] [Google Scholar]
- Moreno E, Basler K. dMyc transforms cells into super-competitors. Cell. 2004;117:117–129. doi: 10.1016/s0092-8674(04)00262-4. [DOI] [PubMed] [Google Scholar]
- Namba R, Pazdera TM, Cerrone RL, Minden JS. Drosophila embryonic pattern repair: how embryos respond to bicoid dosage alteration. Development. 1997;124:1393–1403. doi: 10.1242/dev.124.7.1393. [DOI] [PubMed] [Google Scholar]
- Nilsson C, Johansson U, Johansson AC, Kagedal K, Ollinger K. Cytosolic acidification and lysosomal alkalinization during TNF-alpha induced apoptosis in U937 cells. Apoptosis. 2006;11:1149–1159. doi: 10.1007/s10495-006-7108-5. [DOI] [PubMed] [Google Scholar]
- Nilsson C, Kagedal K, Johansson U, Ollinger K. Analysis of cytosolic and lysosomal pH in apoptotic cells by flow cytometry. Methods Cell Sci. 2003;25:185–194. doi: 10.1007/s11022-004-8228-3. [DOI] [PubMed] [Google Scholar]
- Odaka C, Mizuochi T. Role of macrophage lysosomal enzymes in the degradation of nucleosomes of apoptotic cells. J Immunol. 1999;163:5346–5352. [PubMed] [Google Scholar]
- Pazdera TM, Janardhan P, Minden JS. Patterned epidermal cell death in wild-type and segment polarity mutant Drosophila embryos. Development. 1998;125:3427–3436. doi: 10.1242/dev.125.17.3427. [DOI] [PubMed] [Google Scholar]
- Robertson K, Mergliano J, Minden JS. Dissecting Drosophila embryonic brain development using photoactivated gene expression. Dev Biol. 2003;260:124–137. doi: 10.1016/s0012-1606(03)00220-3. [DOI] [PubMed] [Google Scholar]
- Rosenblatt J, Raff MC, Cramer LP. An epithelial cell destined for apoptosis signals its neighbors to extrude it by an actin- and myosin-dependent mechanism. Curr Biol. 2001;11:1847–1857. doi: 10.1016/s0960-9822(01)00587-5. [DOI] [PubMed] [Google Scholar]
- Schock F, Perrimon N. Cellular processes associated with germ band retraction in Drosophila. Dev Biol. 2002;248:29–39. doi: 10.1006/dbio.2002.0698. [DOI] [PubMed] [Google Scholar]
- Sears HC, Kennedy CJ, Garrity PA. Macrophage-mediated corpse engulfment is required for normal Drosophila CNS morphogenesis. Development. 2003;130:3557–3565. doi: 10.1242/dev.00586. [DOI] [PubMed] [Google Scholar]
- Stroschein-Stevenson SL, Foley E, O’Farrell PH, Johnson AD. Identification of Drosophila gene products required for phagocytosis of Candida albicans. PLoS Biol. 2006;4:e4. doi: 10.1371/journal.pbio.0040004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemoto K, Kuranaga E, Tonoki A, Nagai T, Miyawaki A, Miura M. Local initiation of caspase activation in Drosophila salivary gland programmed cell death in vivo. Proc Natl Acad Sci U S A. 2007;104:13367–13372. doi: 10.1073/pnas.0702733104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemoto K, Nagai T, Miyawaki A, Miura M. Spatio-temporal activation of caspase revealed by indicator that is insensitive to environmental effects. J Cell Biol. 2003;160:235–243. doi: 10.1083/jcb.200207111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyama Y, Peralta XG, Wells AR, Kiehart DP, Edwards GS. Apoptotic force and tissue dynamics during Drosophila embryogenesis. Science. 2008;321:1683–1686. doi: 10.1126/science.1157052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umemori M, Habara O, Iwata T, Maeda K, Nishinoue K, Okabe A, Takemura M, Takahashi K, Saigo K, Ueda R, Adachi-Yamada T. RNAi-Mediated Knockdown Showing Impaired Cell Survival in Drosophila Wing Imaginal Disc. Gene Regul Syst Bio. 2009;3:11–20. doi: 10.4137/grsb.s2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underhill DM. Phagosome maturation: steady as she goes. Immunity. 2005;23:343–344. doi: 10.1016/j.immuni.2005.09.011. [DOI] [PubMed] [Google Scholar]
- Venegas V, Zhou Z. Two alternative mechanisms that regulate the presentation of apoptotic cell engulfment signal in Caenorhabditis elegans. Mol Biol Cell. 2007;18:3180–3192. doi: 10.1091/mbc.E07-02-0138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White K, Grether ME, Abrams JM, Young L, Farrell K, Steller H. Genetic control of programmed cell death in Drosophila. Science. 1994;264:677–683. doi: 10.1126/science.8171319. [DOI] [PubMed] [Google Scholar]
- Yates RM, Russell DG. Phagosome maturation proceeds independently of stimulation of toll-like receptors 2 and 4. Immunity. 2005;23:409–417. doi: 10.1016/j.immuni.2005.09.007. [DOI] [PubMed] [Google Scholar]
- Yu X, Lu N, Zhou Z. Phagocytic receptor CED-1 initiates a signaling pathway for degrading engulfed apoptotic cells. PLoS Biol. 2008;6:e61. doi: 10.1371/journal.pbio.0060061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Odera S, Chuang CH, Lu N, Zhou Z. C. elegans Dynamin mediates the signaling of phagocytic receptor CED-1 for the engulfment and degradation of apoptotic cells. Dev Cell. 2006;10:743–757. doi: 10.1016/j.devcel.2006.04.007. [DOI] [PubMed] [Google Scholar]
- Zhou Z, Yu X. Phagosome maturation during the removal of apoptotic cells: receptors lead the way. Trends Cell Biol. 2008;18:474–485. doi: 10.1016/j.tcb.2008.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou W, Lu Q, Zhao D, Li W, Mapes J, Xie Y, Wang X. Caenorhabditis elegans myotubularin MTM-1 negatively regulates the engulfment of apoptotic cells. PLoS Genet. 2009;5:e1000679. doi: 10.1371/journal.pgen.1000679. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. Timeline of apoptosis revealed by pHMA. Shown here is a version of the images shown in Fig. 7A with a semi-transparent overlay. The dying cell is shaded red and the engulfing cell is shaded blue in the fluorescence images. The dashed lines in the cartoons indicate vesicles that are out of focus.
Supplemental Movie 1 Shows pHMA fluorescence overlayed with transmitted light time-lapse recording of cell a in Fig. 2C and D cell with cortical pHMA localization and neutral pH.
Supplemental Movie 2. Shows pHMA fluorescence overlayed with transmitted light time-lapse recording of cell b in Fig. 2E–G that is likely necrotic.
Supplemental Movie 3. Shows pHMA fluorescence overlayed with transmitted light time-lapse recording of engulfed cell c in Fig. 2H–K.
Supplemental Movie 4. Shows pHMA fluorescence overlayed with transmitted light time-lapse recording of pHMA-expressing cells d and e in Fig. 2L–O. The cell that has some red has already engulfed something. Two other cells acidify slightly (green → yellow) and are engulfed by the daughters of the red cell. They quickly acidify after engulfment (yellow → red).
Supplemental Movie 5. Shows the time-lapse recording of the wild-type embryo injected with AO and VGAL that was used for Fig. 3A.
Supplemental Movie 6. Shows the time-lapse recording of the UAS-pHMA embryo injected with GAL4VP16 that was used for Fig. 3B.
Supplemental Movie 7. Shows the time-lapse recording of the en-GAL4/+; UAS-pHMA/+ embryo that was used for Fig. 4A–D.
Supplemental Movie 8. Shows the time-lapse recording of the UAS-rpr, UAS-W/+; en-GAL4/+; UAS-pHMA/+ embryo that was used for Fig. 4E–H.
Supplemental Movie 9. Shows the time-lapse recording of the dorsal closure in UAS-pHMA;H99/H99 embryo that was used for Fig. 6B, right column. The R470/410 signal remains uniform throughout dorsal closure, indicating that pHMA is not acidified.
Supplemental Movie 10. Shows the time-lapse recording of the en-GAL4, UAS-pHMA wing disc from late third instar larva that was used for Fig. 9A-A″ and 9B-B″. A five-hour recoding was captured at ten-minute intervals.