Abstract
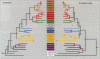
The SNF2 family of proteins includes representatives from a variety of species with roles in cellular processes such as transcriptional regulation (e.g. MOT1, SNF2 and BRM), maintenance of chromosome stability during mitosis (e.g. lodestar) and various aspects of processing of DNA damage, including nucleotide excision repair (e.g. RAD16 and ERCC6), recombinational pathways (e.g. RAD54) and post-replication daughter strand gap repair (e.g. RAD5). This family also includes many proteins with no known function. To better characterize this family of proteins we have used molecular phylogenetic techniques to infer evolutionary relationships among the family members. We have divided the SNF2 family into multiple subfamilies, each of which represents what we propose to be a functionally and evolutionarily distinct group. We have then used the subfamily structure to predict the functions of some of the uncharacterized proteins in the SNF2 family. We discuss possible implications of this evolutionary analysis on the general properties and evolution of the SNF2 family.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ahne F., Baur M., Eckardt-Schupp F. The REV2 gene of Saccharomyces cerevisiae: cloning and DNA sequence. Curr Genet. 1992 Oct;22(4):277–282. doi: 10.1007/BF00317921. [DOI] [PubMed] [Google Scholar]
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. Basic local alignment search tool. J Mol Biol. 1990 Oct 5;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Auble D. T., Hansen K. E., Mueller C. G., Lane W. S., Thorner J., Hahn S. Mot1, a global repressor of RNA polymerase II transcription, inhibits TBP binding to DNA by an ATP-dependent mechanism. Genes Dev. 1994 Aug 15;8(16):1920–1934. doi: 10.1101/gad.8.16.1920. [DOI] [PubMed] [Google Scholar]
- Ayres M. D., Howard S. C., Kuzio J., Lopez-Ferber M., Possee R. D. The complete DNA sequence of Autographa californica nuclear polyhedrosis virus. Virology. 1994 Aug 1;202(2):586–605. doi: 10.1006/viro.1994.1380. [DOI] [PubMed] [Google Scholar]
- Bang D. D., Verhage R., Goosen N., Brouwer J., van de Putte P. Molecular cloning of RAD16, a gene involved in differential repair in Saccharomyces cerevisiae. Nucleic Acids Res. 1992 Aug 11;20(15):3925–3931. doi: 10.1093/nar/20.15.3925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow P. N., Luisi B., Milner A., Elliott M., Everett R. Structure of the C3HC4 domain by 1H-nuclear magnetic resonance spectroscopy. A new structural class of zinc-finger. J Mol Biol. 1994 Mar 25;237(2):201–211. doi: 10.1006/jmbi.1994.1222. [DOI] [PubMed] [Google Scholar]
- Barton A. B., Kaback D. B. Molecular cloning of chromosome I DNA from Saccharomyces cerevisiae: analysis of the genes in the FUN38-MAK16-SPO7 region. J Bacteriol. 1994 Apr;176(7):1872–1880. doi: 10.1128/jb.176.7.1872-1880.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bootsma D., Hoeijmakers J. H. DNA repair. Engagement with transcription. Nature. 1993 May 13;363(6425):114–115. doi: 10.1038/363114a0. [DOI] [PubMed] [Google Scholar]
- Bork P., Koonin E. V. An expanding family of helicases within the 'DEAD/H' superfamily. Nucleic Acids Res. 1993 Feb 11;21(3):751–752. doi: 10.1093/nar/21.3.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson M., Laurent B. C. The SNF/SWI family of global transcriptional activators. Curr Opin Cell Biol. 1994 Jun;6(3):396–402. doi: 10.1016/0955-0674(94)90032-9. [DOI] [PubMed] [Google Scholar]
- Chiba H., Muramatsu M., Nomoto A., Kato H. Two human homologues of Saccharomyces cerevisiae SWI2/SNF2 and Drosophila brahma are transcriptional coactivators cooperating with the estrogen receptor and the retinoic acid receptor. Nucleic Acids Res. 1994 May 25;22(10):1815–1820. doi: 10.1093/nar/22.10.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark M. W., Zhong W. W., Keng T., Storms R. K., Barton A., Kaback D. B., Bussey H. Identification of a Saccharomyces cerevisiae homolog of the SNF2 transcriptional regulator in the DNA sequence of an 8.6 kb region in the LTE1-CYS1 interval on the left arm of chromosome I. Yeast. 1992 Feb;8(2):133–145. doi: 10.1002/yea.320080208. [DOI] [PubMed] [Google Scholar]
- Côté J., Quinn J., Workman J. L., Peterson C. L. Stimulation of GAL4 derivative binding to nucleosomal DNA by the yeast SWI/SNF complex. Science. 1994 Jul 1;265(5168):53–60. doi: 10.1126/science.8016655. [DOI] [PubMed] [Google Scholar]
- Delmas V., Stokes D. G., Perry R. P. A mammalian DNA-binding protein that contains a chromodomain and an SNF2/SWI2-like helicase domain. Proc Natl Acad Sci U S A. 1993 Mar 15;90(6):2414–2418. doi: 10.1073/pnas.90.6.2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doe C. L., Murray J. M., Shayeghi M., Hoskins M., Lehmann A. R., Carr A. M., Watts F. Z. Cloning and characterisation of the Schizosaccharomyces pombe rad8 gene, a member of the SNF2 helicase family. Nucleic Acids Res. 1993 Dec 25;21(25):5964–5971. doi: 10.1093/nar/21.25.5964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donahue B. A., Yin S., Taylor J. S., Reines D., Hanawalt P. C. Transcript cleavage by RNA polymerase II arrested by a cyclobutane pyrimidine dimer in the DNA template. Proc Natl Acad Sci U S A. 1994 Aug 30;91(18):8502–8506. doi: 10.1073/pnas.91.18.8502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunaief J. L., Strober B. E., Guha S., Khavari P. A., Alin K., Luban J., Begemann M., Crabtree G. R., Goff S. P. The retinoblastoma protein and BRG1 form a complex and cooperate to induce cell cycle arrest. Cell. 1994 Oct 7;79(1):119–130. doi: 10.1016/0092-8674(94)90405-7. [DOI] [PubMed] [Google Scholar]
- Elfring L. K., Deuring R., McCallum C. M., Peterson C. L., Tamkun J. W. Identification and characterization of Drosophila relatives of the yeast transcriptional activator SNF2/SWI2. Mol Cell Biol. 1994 Apr;14(4):2225–2234. doi: 10.1128/mcb.14.4.2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitch W. M., Margoliash E. Construction of phylogenetic trees. Science. 1967 Jan 20;155(3760):279–284. doi: 10.1126/science.155.3760.279. [DOI] [PubMed] [Google Scholar]
- Funahashi J., Sekido R., Murai K., Kamachi Y., Kondoh H. Delta-crystallin enhancer binding protein delta EF1 is a zinc finger-homeodomain protein implicated in postgastrulation embryogenesis. Development. 1993 Oct;119(2):433–446. doi: 10.1242/dev.119.2.433. [DOI] [PubMed] [Google Scholar]
- Gecz J., Pollard H., Consalez G., Villard L., Stayton C., Millasseau P., Khrestchatisky M., Fontes M. Cloning and expression of the murine homologue of a putative human X-linked nuclear protein gene closely linked to PGK1 in Xq13.3. Hum Mol Genet. 1994 Jan;3(1):39–44. doi: 10.1093/hmg/3.1.39. [DOI] [PubMed] [Google Scholar]
- Girdham C. H., Glover D. M. Chromosome tangling and breakage at anaphase result from mutations in lodestar, a Drosophila gene encoding a putative nucleoside triphosphate-binding protein. Genes Dev. 1991 Oct;5(10):1786–1799. doi: 10.1101/gad.5.10.1786. [DOI] [PubMed] [Google Scholar]
- Glassner B. J., Mortimer R. K. Synergistic interactions between RAD5, RAD16 and RAD54, three partially homologous yeast DNA repair genes each in a different repair pathway. Radiat Res. 1994 Jul;139(1):24–33. [PubMed] [Google Scholar]
- Hanawalt P. C., Donahue B. A., Sweder K. S. Repair and transcription. Collision or collusion? Curr Biol. 1994 Jun 1;4(6):518–521. doi: 10.1016/s0960-9822(00)00112-3. [DOI] [PubMed] [Google Scholar]
- Hanawalt P., Mellon I. Stranded in an active gene. Curr Biol. 1993 Jan;3(1):67–69. doi: 10.1016/0960-9822(93)90156-i. [DOI] [PubMed] [Google Scholar]
- Haynes S. R., Dollard C., Winston F., Beck S., Trowsdale J., Dawid I. B. The bromodomain: a conserved sequence found in human, Drosophila and yeast proteins. Nucleic Acids Res. 1992 May 25;20(10):2603–2603. doi: 10.1093/nar/20.10.2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S., Henikoff J. G. Protein family classification based on searching a database of blocks. Genomics. 1994 Jan 1;19(1):97–107. doi: 10.1006/geno.1994.1018. [DOI] [PubMed] [Google Scholar]
- Henikoff S. Sequence analysis by electronic mail server. Trends Biochem Sci. 1993 Jul;18(7):267–268. doi: 10.1016/0968-0004(93)90179-q. [DOI] [PubMed] [Google Scholar]
- Henikoff S. Transcriptional activator components and poxvirus DNA-dependent ATPases comprise a single family. Trends Biochem Sci. 1993 Aug;18(8):291–292. doi: 10.1016/0968-0004(93)90037-n. [DOI] [PubMed] [Google Scholar]
- Higgins D. G., Bleasby A. J., Fuchs R. CLUSTAL V: improved software for multiple sequence alignment. Comput Appl Biosci. 1992 Apr;8(2):189–191. doi: 10.1093/bioinformatics/8.2.189. [DOI] [PubMed] [Google Scholar]
- Ho L., Hanawalt P. C. Gene-specific DNA repair in terminally differentiating rat myoblasts. Mutat Res. 1991 Sep;255(2):123–141. doi: 10.1016/0921-8777(91)90047-s. [DOI] [PubMed] [Google Scholar]
- Huang M. E., Chuat J. C., Galibert F. A possible yeast homolog of human active-gene-repairing helicase ERCC6+. Biochem Biophys Res Commun. 1994 May 30;201(1):310–317. doi: 10.1006/bbrc.1994.1703. [DOI] [PubMed] [Google Scholar]
- Iwasaki H., Ishino Y., Toh H., Nakata A., Shinagawa H. Escherichia coli DNA polymerase II is homologous to alpha-like DNA polymerases. Mol Gen Genet. 1991 Apr;226(1-2):24–33. doi: 10.1007/BF00273583. [DOI] [PubMed] [Google Scholar]
- Johnson R. E., Henderson S. T., Petes T. D., Prakash S., Bankmann M., Prakash L. Saccharomyces cerevisiae RAD5-encoded DNA repair protein contains DNA helicase and zinc-binding sequence motifs and affects the stability of simple repetitive sequences in the genome. Mol Cell Biol. 1992 Sep;12(9):3807–3818. doi: 10.1128/mcb.12.9.3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khavari P. A., Peterson C. L., Tamkun J. W., Mendel D. B., Crabtree G. R. BRG1 contains a conserved domain of the SWI2/SNF2 family necessary for normal mitotic growth and transcription. Nature. 1993 Nov 11;366(6451):170–174. doi: 10.1038/366170a0. [DOI] [PubMed] [Google Scholar]
- Kolstø A. B., Bork P., Kvaløy K., Lindback T., Grønstadt A., Kristensen T., Sander C. Prokaryotic members of a new family of putative helicases with similarity to transcription activator SNF2. J Mol Biol. 1993 Mar 20;230(2):684–688. doi: 10.1006/jmbi.1993.1185. [DOI] [PubMed] [Google Scholar]
- Laurent B. C., Treich I., Carlson M. The yeast SNF2/SWI2 protein has DNA-stimulated ATPase activity required for transcriptional activation. Genes Dev. 1993 Apr;7(4):583–591. doi: 10.1101/gad.7.4.583. [DOI] [PubMed] [Google Scholar]
- Laurent B. C., Treitel M. A., Carlson M. Functional interdependence of the yeast SNF2, SNF5, and SNF6 proteins in transcriptional activation. Proc Natl Acad Sci U S A. 1991 Apr 1;88(7):2687–2691. doi: 10.1073/pnas.88.7.2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurent B. C., Yang X., Carlson M. An essential Saccharomyces cerevisiae gene homologous to SNF2 encodes a helicase-related protein in a new family. Mol Cell Biol. 1992 Apr;12(4):1893–1902. doi: 10.1128/mcb.12.4.1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurent B. C., Yang X., Carlson M. An essential Saccharomyces cerevisiae gene homologous to SNF2 encodes a helicase-related protein in a new family. Mol Cell Biol. 1992 Apr;12(4):1893–1902. doi: 10.1128/mcb.12.4.1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis L. K., Jenkins M. E., Mount D. W. Isolation of DNA damage-inducible promoters in Escherichia coli: regulation of polB (dinA), dinG, and dinH by LexA repressor. J Bacteriol. 1992 May;174(10):3377–3385. doi: 10.1128/jb.174.10.3377-3385.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovering R., Hanson I. M., Borden K. L., Martin S., O'Reilly N. J., Evan G. I., Rahman D., Pappin D. J., Trowsdale J., Freemont P. S. Identification and preliminary characterization of a protein motif related to the zinc finger. Proc Natl Acad Sci U S A. 1993 Mar 15;90(6):2112–2116. doi: 10.1073/pnas.90.6.2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannhaupt G., Stucka R., Ehnle S., Vetter I., Feldmann H. Molecular analysis of yeast chromosome II between CMD1 and LYS2: the excision repair gene RAD16 located in this region belongs to a novel group of double-finger proteins. Yeast. 1992 May;8(5):397–408. doi: 10.1002/yea.320080507. [DOI] [PubMed] [Google Scholar]
- Mellon I., Hanawalt P. C. Induction of the Escherichia coli lactose operon selectively increases repair of its transcribed DNA strand. Nature. 1989 Nov 2;342(6245):95–98. doi: 10.1038/342095a0. [DOI] [PubMed] [Google Scholar]
- Mellon I., Spivak G., Hanawalt P. C. Selective removal of transcription-blocking DNA damage from the transcribed strand of the mammalian DHFR gene. Cell. 1987 Oct 23;51(2):241–249. doi: 10.1016/0092-8674(87)90151-6. [DOI] [PubMed] [Google Scholar]
- Muchardt C., Yaniv M. A human homologue of Saccharomyces cerevisiae SNF2/SWI2 and Drosophila brm genes potentiates transcriptional activation by the glucocorticoid receptor. EMBO J. 1993 Nov;12(11):4279–4290. doi: 10.1002/j.1460-2075.1993.tb06112.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okabe I., Bailey L. C., Attree O., Srinivasan S., Perkel J. M., Laurent B. C., Carlson M., Nelson D. L., Nussbaum R. L. Cloning of human and bovine homologs of SNF2/SWI2: a global activator of transcription in yeast S. cerevisiae. Nucleic Acids Res. 1992 Sep 11;20(17):4649–4655. doi: 10.1093/nar/20.17.4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paro R., Hogness D. S. The Polycomb protein shares a homologous domain with a heterochromatin-associated protein of Drosophila. Proc Natl Acad Sci U S A. 1991 Jan 1;88(1):263–267. doi: 10.1073/pnas.88.1.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson W. R., Lipman D. J. Improved tools for biological sequence comparison. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson W. R. Rapid and sensitive sequence comparison with FASTP and FASTA. Methods Enzymol. 1990;183:63–98. doi: 10.1016/0076-6879(90)83007-v. [DOI] [PubMed] [Google Scholar]
- Peterson S. N., Hu P. C., Bott K. F., Hutchison C. A., 3rd A survey of the Mycoplasma genitalium genome by using random sequencing. J Bacteriol. 1993 Dec;175(24):7918–7930. doi: 10.1128/jb.175.24.7918-7930.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randazzo F. M., Khavari P., Crabtree G., Tamkun J., Rossant J. brg1: a putative murine homologue of the Drosophila brahma gene, a homeotic gene regulator. Dev Biol. 1994 Jan;161(1):229–242. doi: 10.1006/dbio.1994.1023. [DOI] [PubMed] [Google Scholar]
- Saitou N., Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987 Jul;4(4):406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Schild D., Glassner B. J., Mortimer R. K., Carlson M., Laurent B. C. Identification of RAD16, a yeast excision repair gene homologous to the recombinational repair gene RAD54 and to the SNF2 gene involved in transcriptional activation. Yeast. 1992 May;8(5):385–395. doi: 10.1002/yea.320080506. [DOI] [PubMed] [Google Scholar]
- Schild D., Glassner B. J., Mortimer R. K., Carlson M., Laurent B. C. Identification of RAD16, a yeast excision repair gene homologous to the recombinational repair gene RAD54 and to the SNF2 gene involved in transcriptional activation. Yeast. 1992 May;8(5):385–395. doi: 10.1002/yea.320080506. [DOI] [PubMed] [Google Scholar]
- Selby C. P., Sancar A. Gene- and strand-specific repair in vitro: partial purification of a transcription-repair coupling factor. Proc Natl Acad Sci U S A. 1991 Sep 15;88(18):8232–8236. doi: 10.1073/pnas.88.18.8232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selby C. P., Sancar A. Mechanisms of transcription-repair coupling and mutation frequency decline. Microbiol Rev. 1994 Sep;58(3):317–329. doi: 10.1128/mr.58.3.317-329.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selby C. P., Sancar A. Molecular mechanism of transcription-repair coupling. Science. 1993 Apr 2;260(5104):53–58. doi: 10.1126/science.8465200. [DOI] [PubMed] [Google Scholar]
- Selby C. P., Witkin E. M., Sancar A. Escherichia coli mfd mutant deficient in "mutation frequency decline" lacks strand-specific repair: in vitro complementation with purified coupling factor. Proc Natl Acad Sci U S A. 1991 Dec 15;88(24):11574–11578. doi: 10.1073/pnas.88.24.11574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soininen R., Schoor M., Henseling U., Tepe C., Kisters-Woike B., Rossant J., Gossler A. The mouse Enhancer trap locus 1 (Etl-1): a novel mammalian gene related to Drosophila and yeast transcriptional regulator genes. Mech Dev. 1992 Nov;39(1-2):111–123. doi: 10.1016/0925-4773(92)90030-n. [DOI] [PubMed] [Google Scholar]
- Thompson J. D., Higgins D. G., Gibson T. J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994 Nov 11;22(22):4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troelstra C., van Gool A., de Wit J., Vermeulen W., Bootsma D., Hoeijmakers J. H. ERCC6, a member of a subfamily of putative helicases, is involved in Cockayne's syndrome and preferential repair of active genes. Cell. 1992 Dec 11;71(6):939–953. doi: 10.1016/0092-8674(92)90390-x. [DOI] [PubMed] [Google Scholar]
- Tsuchiya E., Uno M., Kiguchi A., Masuoka K., Kanemori Y., Okabe S., Mikayawa T. The Saccharomyces cerevisiae NPS1 gene, a novel CDC gene which encodes a 160 kDa nuclear protein involved in G2 phase control. EMBO J. 1992 Nov;11(11):4017–4026. doi: 10.1002/j.1460-2075.1992.tb05495.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Aart Q. J., Barthe C., Doignon F., Aigle M., Crouzet M., Steensma H. Y. Sequence analysis of a 31 kb DNA fragment from the right arm of Saccharomyces cerevisiae chromosome II. Yeast. 1994 Jul;10(7):959–964. doi: 10.1002/yea.320100711. [DOI] [PubMed] [Google Scholar]
- Weinmann R. The basic RNA polymerase II transcriptional machinery. Gene Expr. 1992;2(2):81–91. [PMC free article] [PubMed] [Google Scholar]
- Wolffe A. P. Transcriptional activation. Switched-on chromatin. Curr Biol. 1994 Jun 1;4(6):525–528. doi: 10.1016/s0960-9822(00)00114-7. [DOI] [PubMed] [Google Scholar]
- Yoshimoto H., Yamashita I. The GAM1/SNF2 gene of Saccharomyces cerevisiae encodes a highly charged nuclear protein required for transcription of the STA1 gene. Mol Gen Genet. 1991 Aug;228(1-2):270–280. doi: 10.1007/BF00282476. [DOI] [PubMed] [Google Scholar]
- van Gool A. J., Verhage R., Swagemakers S. M., van de Putte P., Brouwer J., Troelstra C., Bootsma D., Hoeijmakers J. H. RAD26, the functional S. cerevisiae homolog of the Cockayne syndrome B gene ERCC6. EMBO J. 1994 Nov 15;13(22):5361–5369. doi: 10.1002/j.1460-2075.1994.tb06871.x. [DOI] [PMC free article] [PubMed] [Google Scholar]