Abstract
Chromosomal instability leading to aneuploidy occurs in most sporadic colorectal cancers (CRCs) and is believed to be an early driving force in disease progression. Despite this observation, the cellular advantages conferred by these cytogenetic alterations are poorly understood. Here, we provide evidence that serum-free passage of originally diploid, immortalized human colonic epithelial cells (HCECs) gave rise to the acquisition of trisomy 7 (+7), an aneuploidy detected in more than 40% of colorectal adenomas. These cells remain diploid under long-term growth in 2% serum conditions. Analysis by GTG banding and fluorescent in situ hybridization detected no rare preexisting +7 cell in the original population, suggesting a conversion of diploid cells to an aneuploid state. The acquisition of +7 also precedes loss or truncation of the adenomatosis polyposis coli gene as both diploid and +7 cells express full-length, functional protein. Coculturing of fluorescent-labeled cells demonstrate that +7 HCECs have a growth advantage over diploid cells in serum-free conditions. Defects in cell migration and aberrant regulation of the epidermal growth factor receptor, located on chromosome 7p, are also detected in +7 HCECs. Interestingly, knockdown of TP53 and expression of K-RasV12 in +7 HCECs resulted in the emergence of trisomy 20, another nonrandom aneuploidy observed in ∼85% of CRC. In summary, we describe isogenic colonic epithelial cells that represent cytogenetic changes occurring frequently in sporadic CRC. The emergence and characterization of trisomy 7 and 20 demonstrate that these HCECs may serve as unique human cell-based models to examine the effects of chromosomal instability in CRC progression.
Introduction
Sporadic colorectal cancer (CRC) arises from the sequential accumulation of nonrandom genetic and epigenetic alterations that drive normal colonic epithelium toward neoplastic transformation [1]. A hallmark observed in most epithelial cancers is the development of chromosomal instability (CIN) [2]. CIN is prominent in sporadic CRC patients [3] and describes the process by which cells accrue increasing amounts of aneuploidy, an abnormal number of chromosomes [4]. As cells develop alterations in critical cell cycle control pathways, an increased incidence of aneuploidy can be detected as cells progressively become more transformed [5]. This suggests that aneuploidy may be a key factor in the early events of CRC initiation that predisposes cells toward tumorigenesis [6]. Although CIN can be observed in ∼85% of sporadic CRC cases [7], the causes and consequences of CIN and aneuploidy remain largely unknown [8].
Improved human cellular reagents modeling early CIN events in CRC pathogenesis are required to more clearly characterize the biologic effects of aneuploidy and to identify novel strategies for chemoprevention. We have recently developed a method to isolate and immortalize human colonic epithelial cells (HCECs) derived from noncancerous tissues of patients undergoing routine screening or surveillance colonoscopy [9]. Immortalized HCECs (termed HCEC CT for transduction with Cdk4 and hTERT) remain diploid under long-term propagation in 2% serum growth conditions and do not exhibit any malignant phenotypes (e.g., anchorage-independent growth, invasion through extracellular matrices, tumor formation in immunocompromised mice). However, we have observed one of the earliest known events in CRC initiation on long-term serum-free cultivation of one HCEC line: the development of CIN.
The pattern of CIN leading to aneuploidy in CRC seems to be at least partially nonrandom. Specific chromosomal changes (such as trisomies or loss of chromosome arms) become apparent during the adenoma stage, whereas other changes become evident in more advanced lesions [10]. One of the earliest alterations that occur in up to ∼40%of colonic adenomas is the development of trisomy for chromosome 7 (+7) [10–13]. An HCEC line propagated in our laboratory (HCEC 1CT) was derived from a patient with a previous history of sigmoid adenocarcinoma. After immortalization, these noncancerous cells were placed in serum-free culture for an extended period and the emerged cells contained +7 as the sole cytogenetic abnormality. We have initiated experiments with these +7 cells (HCEC 1CT+7) to elucidate if and how this single but common chromosomal alteration may confer a selective advantage to cells [14]. Recent experiments from yeast studies show that aneuploidy can be beneficial under a range of stress conditions by providing a growth advantage [15], although these findings have not been replicated using human cells.
We report here the isolation and initial characterization of 1CT+7 HCECs. We provide evidence that these aneuploid cells possess a measurable growth advantage under serum-deprived conditions and obtain other changes (e.g., defective cell migration) that may affect normal colonic physiology and promote tumorigenesis. In addition, we also describe the emergence of trisomy 20 (+20) HCEC subpopulations after introducing defined oncogenic changes into 1CT+7 HCECs. Because +7 and +20 are common and recurrent chromosomal abnormalities frequently detected throughout various CRC stages [16–18], these isogenically derived HCEC lines provide a valuable cell-based model to further dissect the biologic consequences behind aneuploidy-driven CRC and may serve as useful reagents in the discovery of novel chemopreventive strategies.
Materials and Methods
Cell Culture
The culture conditions of HCECs have been reported elsewhere [9,19]. Briefly, HCECs are maintained under 2% oxygen and 5% carbon dioxide on Primaria (BD Biosciences, San Jose, CA) plates in 4:1 high-glucose Dulbecco modified Eagle medium/medium 199 with 2% cosmic calf serum (Hyclone, Logan, UT) plus growth supplements: epidermal growth factor (EGF; 20 ng/ml; Peprotech, Rocky Hill, NJ), hydrocortisone (1 mg/ml), insulin (10 mg/ml), transferrin (2 mg/ml), and sodium selenite (5 nM) (all Sigma, St Louis, MO).
Karyotype and Fluorescent In Situ Hybridization
HCECs at 70% confluency were treated with 0.01 µg/ml colcemid (KaryoMax; Gibco/Invitrogen, Carlsbad, CA) for 2 hours, and metaphase chromosomes were harvested under standard protocols. Slides were dropped using a Thermotron chamber (Thermotron, Inc, Holland, MI) and stained with G bands by trypsin using Giemsa. The images of each metaphase were captured with a Zeiss Axioskop 2 microscope (Photometrics, Tucson, AZ), and Applied Imaging CytoVision Software (Applied Imaging Corp., Spring Valley, NY) was used to analyze these metaphase cells, and each chromosome analysis was performed on at least 20 metaphases at each PD. DNA probes specific for human centromeres of chromosomes 7 and 20 (CEP7 and CEP20) were obtained from Abbott Molecular, Inc (Des Plaines, IL) for fluorescent in situ hybridization (FISH) analysis using a Zeiss Axioskop 2 fluorescent microscope with CCD camera (Photometrics). At least 500 interphase cells at each PD were examined.
Coculturing Competition Assays
HCECs were labeled with dsRed by transduction with a pSSI-8018 lentiviral vector followed by blasticidin selection. Two hundred thousand nonlabeled HCEC 1CT cells were mixed with equal numbers of dsRed-labeled HCEC 1CT+7 cells in a 10-cm2 plate. Cells were cultured for at least 38 days in serum-free medium with growth supplements and passaged every 8 days. Phase/fluorescence pictures are representative images from the center of triplicate culture plates. Control experiments represent the coculturing of nonlabeled and labeled-HCEC 1CT+7 cells.
Western Blot Analysis
Total cell lysates were prepared by harvesting cells in Laemmli SDS reducing buffer. Protein concentrations were measured using a Pierce BCA protein assay kit (Thermo Scientific, Rockford, IL), resolved on an 8% to 10% polyacrylamide gel, and transferred to polyvinylidine fluoride. Ultracentrifugation was used to fractionate lysates as specified. Table W1 lists the primary antibodies used. HRP-conjugated goat antimouse or antirabbit ( Jackson ImmunoResearch, West Grove, PA) were used as secondary antibodies at 1:5000 and detected with SuperSignal West Pico or Femto Chemiluminescent Substrate Kit (Thermo Scientific). Bands were quantified using a ChemiDoc XRS+ Imager with Image Lab software (Bio-Rad, Hercules, CA) and normalized to β-actin.
Response to EGF and Cetuximab
For cell counting experiments, 8.0 x 104 cells were cultured in the presence of 0, 1, 2.5, or 5 ng/ml EGF in a six-well dish in serum-free medium with growth supplements with either 50 µg/ml cetuximab (Erbitux; ImClone Systems, Inc, Branchburg, NJ) or saline. Cell numbers were obtained 5 days later. For Western blots with cetuximab, serum/EGF-starved cells were treated with 0 to 50 µg/ml cetuximab for 1 hour before stimulation with 5 ng/ml EGF for 15 minutes. Lysates were collected immediately and analyzed as previously described.
Colony Formation Assay
Cells were seeded in triplicate 10-cm2 culture plates at three different clonal densities (50–400 cells per plates) in either serum or serum-free medium containing growth supplements. After 10 days, colonies were washed with PBS, stained for 30 minutes with 6% glutaraldehyde/0.5% crystal violet solution, and counted.
Migration Assays
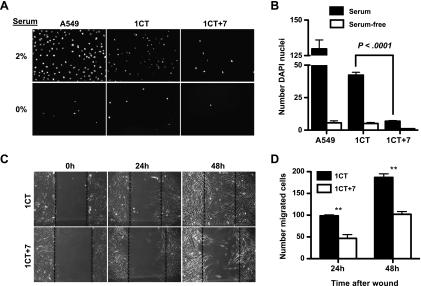
Transwell migration assays were performed using a modified Boyden chamber. Briefly, 4.0 x 104 cells were serum-starved for 6 hours and plated into serum-free and GF-free medium onto 8.0-µm pore transwell PET membranes (BD Biosciences, Bedford, MA). Five hundred microliters of medium containing 2% serum and growth supplements was added to the bottom well. Nonmigratory cells were scraped off 24 hours later, and migratory cells were fixed in 100% methanol, washed, and stained with 4′,6-diamidino-2-phenylindole. Experiments were performed in triplicate transwells and quantified by averaging the number of DAPI nuclei per 20x field of view counting five fields per chamber (n = 15). Wound migration assays were performed by plating cells to confluency in a six-well plate in serum-free medium with growth supplements. After 24 hours, a wound was created by scratching the confluent monolayer with a P200 pipette tip and debris was removed by PBS washes. The wound was imaged by phase microscopy at the middle of the scratch line at different time points. Cells migrated into the wound were counted and averaged from triplicates.
Results
Emergence of HCEC 1CT+7 from Diploid Populations
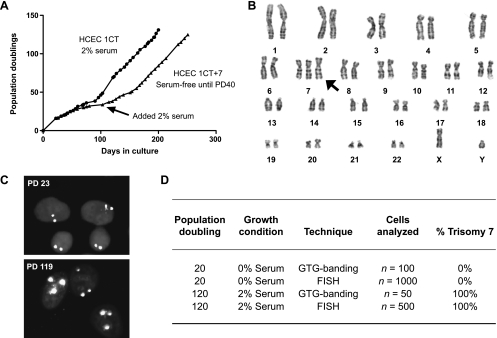
The HCEC 1CT line used in these experiments were derived from a patient with a previous history of CRC and maintain a normal karyotype (46,XY) when continuously propagated in 2% oxygen and medium containing 2% serum [9]. As a separate experiment, a subset of HCEC 1CTs were cultured and passaged under serum-free conditions for a prolonged period and underwent a phase of slow growth. At approximately 40 population doublings (PDs), 2% serum was added to the culture medium (Figure 1A). Cells that emerged from this population contained +7 as the sole cytogenetic change (Figure 1B). No distinct differences were observed in cell morphology between the two cell types (Figure W1A). Although we cannot rigorously prove that an extremely rare +7 cell did not preexist in the population during the time of serum-free culture, GTG-banding and FISH analysis of the original population at early PD showed no evidence of a rare subpopulation of cells containing +7 (all 1000 interphase cells were diploid for chromosome 7 by FISH using a chromosome 7-specific probe) (Figure 1, C–D). Emerged HCEC 1CT+7 populations were confirmed by analyzing metaphase cells by GTG banding (50/50 cells analyzed) and FISH (500/500 cells analyzed) (Figure 1D). Array CGH analyses (NCBI GEO accession numbers: GSE24092 and GSM593069-GSM593070) also revealed no other large amplifications or deletions other than whole chromosome 7 amplifications (Figure W1B).
Figure 1.
Trisomy 7 HCECs (HCEC 1CT+7) appeared under long-term culture in serum-free conditions. (A) Growth curve of originally diploid cells propagated in either serum or serum-free conditions. Long-term culture of HCEC 1CT in serum maintains a diploid karyotype. Serum-free HCEC 1CT underwent a period of slow growth around PD30 in which case 2% serum was added to the culture at PD40. (B) The emerged population of cells originally maintained under serum-free conditions displayed a trisomy for chromosome 7 by karyotypic analysis as the sole cytogenetic alteration. (C) FISH analysis on early and late PD cells using a chromosome 7-specific probe. (D) Trisomy 7 cells emerged from originally diploid HCECs with no early traces of a small aneuploid population.
Acquisition of Trisomy 7 Precedes Loss or Truncation of Adenomatosis Polyposis Coli
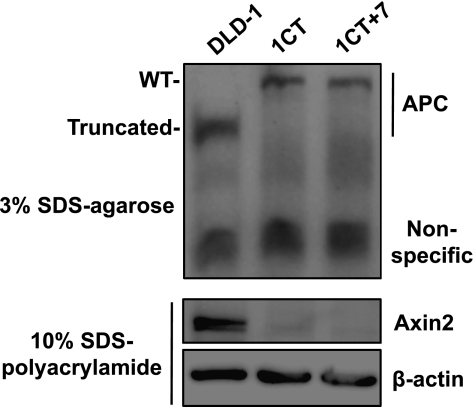
Loss or truncation of the adenomatosis polyposis coli (APC) gene is believed to be the earliest genetic lesion responsible for the generation of CIN and initiation toward the adenoma stage [20]. HCEC 1CT and 1CT+7 both express abundant levels of full-length APC, whereas DLD-1 cancer cells express the truncated form (Figure 2). Furthermore, we show that Axin2 is not detected in HCECs using the same lysates on a polyacrylamide gel, suggesting that APC is fully functional in these cells (Figure 2). Axin2, a negative feedback regulator of the Wnt signaling pathway, is upregulated in the presence of Wnt ligands and nuclear β-catenin [21]. With functional APC, downstream components of β-catenin (such as Axin2) are not expressed. Therefore, the generation of CIN and subsequent acquisition of +7 appears before loss or truncation of APC in the HCEC 1CT line.
Figure 2.
Expression of full-length, functional APC in both diploid and trisomy 7 HCECs. The CRC cell line DLD-1 is used as a positive control for truncated APC. The same lysates were also probed for Axin2, a protein upregulated in the presence of Wnt ligands and subsequent nuclear β-catenin.
Diploid HCECs Lose Growth Advantages in Serum-Free Conditions
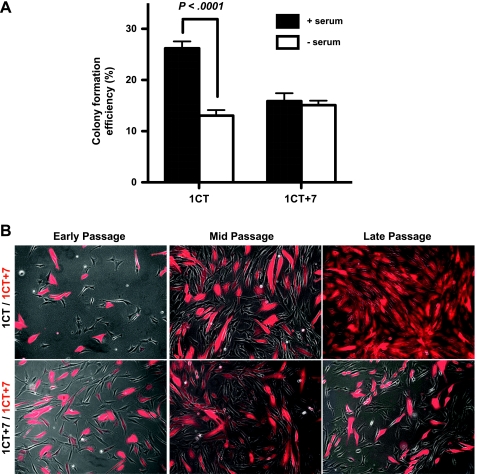
We next tested whether serum affects the clonogenicity of these cells (Figure 3A). Colony formation assays indicate that, in the presence of serum, diploid cells have a higher percentage of colony forming cells compared with 1CT+7. However, in serum-depleted conditions, the clonogenicity of 1CT HCECs significantly decreases although there are no effects on 1CT+7 cells, suggesting that diploid cells lose proliferative advantages when shifting from more optimal (serum) conditions to more stressful (serum-free) conditions.
Figure 3.
Diploid HCECs lose growth advantages when cultured in serum-free medium. (A) HCECs were plated at clonal density in serum or serum-free medium for 10 days. The absence of serum significantly reduces the clonogenicity of 1CT but has no effect on 1CT+7 cells. (B) DsRed-labeled 1CT+7 cells were mixed with an equal number of nonlabeled 1CT cells and passaged in serum-free growth medium. Late passage represents approximately 38 days in culture, splitting the cells once every 8 days. Control experiments with nonlabeled 1CT+7 show no differences in the ratio of dsRed-positive to negative cells. Columns represent mean ± SEM. P values obtained by two-tailed, unpaired Student's t test.
In agreement with previous reports using other cell systems [22,23], we observed that diploid 1CT HCECs proliferate more rapidly compared to 1CT+7 HCECs in the presence of 2% serum and growth supplements (0.9 PD/d [26.6 h/doubling] compared with 0.7 PD/d [34.3 h/doubling], respectively). However, in defined medium only supplemented with growth factors, both cell types divide at approximately equal rates (0.3 PD/d [80 h/doubling]). To differentiate the nearly identical growth rates between 1CT and 1CT+7 HCECs in defined medium, we cocultured equal numbers of nonlabeled diploid cells with dsRed-labeled 1CT+7 cells for more than a month in serum-free medium. After 38 days, most cells in culture were dsRed-positive 1CT+7 HCECs (Figure 3B). In control assays, coculturing nonlabeled 1CT+7 cells with dsRed-labeled 1CT+7 cells showed no apparent differences after several weeks in culture (Figure 3B). In experiments with 2% serum, diploid cells quickly took over the culture (data not shown). Reversing the labeled cells also had no effect on these observations (data not shown).
Aberrant Epidermal Growth Factor Receptor Regulation in HCEC 1CT+7
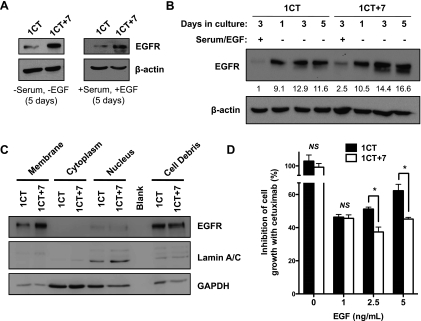
Because the epidermal growth factor receptor (EGFR) is located on chromosome 7, we hypothesized that EGFR misregulation may be a critical alteration in +7 HCECs [24]. We found a larger-than-expected increase in EGFR protein levels in 1CT+7 cells compared with diploid HCECs in both serum and serum/EGF-free conditions after 5 days in culture (Figure 4A). Next, we measured EGFR levels in both cell types when serum/EGF-starved during a period of 1, 3, and 5 days. Although both cell types express similar levels of EGFR after 1 day of passage, 1CT+7 cells exhibits increased EGFR levels during a period of 5 days, whereas EGFR in diploid HCECs remain at more constant levels throughout (Figure 4B). Analysis of fractionated lysates by Western blot also indicates the localization of EGFR to the cell membrane in both 1CT and 1CT+7 cells (Figure 4C).
Figure 4.
HCEC 1CT+7 exhibits aberrant EGFR regulation and cell growth is more potently inhibited by cetuximab. (A) HCEC 1CT+7 expresses higher levels of EGFR in serum and serum-free conditions compared with 1CT as shown by Western blot analyses. (B) Cells were cultured in serum/EGF-free medium for 1, 3, and 5 days, and lysates were probed for EGFR. 1CT+7 cells show increased EGFR levels in the absence of serum/EGF over time. Bands were quantified by densitometric scanning and normalized to loading control. (C) Expression of EGFR is localized to the cell membrane as shown by Western blot analyses on fractionated lysates. Lamin A/C and GAPDH are used as nonmembrane controls. (D) Cetuximab, an EGFR inhibitor, can more significantly reduce 1CT+7 cell proliferation induced by EGF compared with diploid cells. Cells were cultured for 5 days in various concentrations of EGF in the presence of 50 µg/ml cetuximab or saline (n = 3). Columns represent mean ± SEM. *P < .05, by two-tailed, unpaired Student's t test.
Because diverse cell types with various EGFR levels respond differently to EGFR-targeting therapies, we decided to test whether there are different responses to an EGFR inhibitor between HCEC 1CT and 1CT+7. Cetuximab, a monoclonal antibody targeting the extracellular domain of EGFR [25], can effectively inhibit EGFR activation induced by short-term EGF stimulation in both 1CT and 1CT+7 (Figure W2A). Interestingly, 1CT+7 cell proliferation induced by various concentrations of EGF in the growth medium is more potently reduced in vitro in the presence of cetuximab after 5 days in culture compared with diploid cells (Figure 4D), perhaps indicating an augmented dependency on EGFR signaling in +7 HCECs. In addition, no mutations were detected in the tyrosine kinase domains of EGFR by exon sequencing in both cell types (primers listed in Table W2).
HCEC 1CT+7 Have Defects in Cell Migration
To further characterize 1CT and 1CT+7 HCECs, we next investigated if there are any differences in cell motility. Cell migration is an important factor in the rapid turnover of colonic crypts [26], and defects in cell motility could thus provide an initial survival advantage in the early stages of CRC (e.g., retention of epithelial cells within crypts, increased time frame to accumulate mutations). Using a transwell migration assay, 1CT+7 HCECs exhibited defects in cell migration through a porous membrane toward serum-containing medium (Figure 5, A and B). This loss in migration ability is also recapitulated in a two-dimensional wound migration assay (Figure 5, C and D). In contrast to diploid cells, 1CT+7 HCECs have a decreased ability to migrate into the wound area. These results suggest a disruption in the highly regulated process of cell migration [27]. Analysis of matrix metalloproteinases (MMPs) by gelatin zymography also reveals a significant repression of pro-MMP-2 levels in HCEC 1CT+7 compared with HCEC 1CT and 2CT (an immortalized normal cell line from a non-CRC patient) (Figure W2B).
Figure 5.
HCEC 1CT+7 displays defects in cell migration. (A) In a transwell migration assay, cells were seeded in serum-free medium and allowed to migrate through an 8-µm pore membrane toward either 2% or 0% serum medium. Migrated cells were stained with DAPI. A549 lung cells are used as a positive control for cell migration. (B) Quantification of (A) by averaging the number of cells per 20x magnification (n = 15). (C) HCEC 1CT+7 are slower to migrate into a gap created by an experimental wound to the cell monolayer. (D) Quantification of (C) by averaging the number of cells that migrated into the gap compared with 0 hours (n = 3). All columns represent mean ± SEM. **P < .005, by two-tailed, unpaired Student's t test.
Emergence of Trisomy 20 in HCEC 1CT+7 Expressing K-RasV12 and shTP53
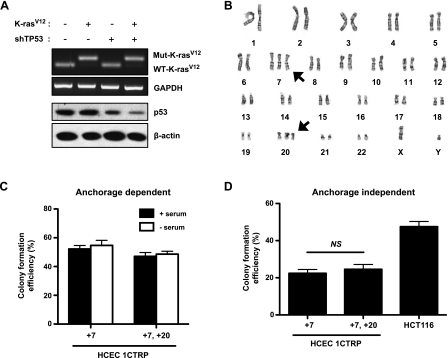
Next, we attempted to experimentally transform these HCECs in vitro by recapitulating commonly observed genetic events in CRC progression. We started by introducing the K-RasV12 oncogene and stably depleting TP53 levels using short hairpin RNA (shRNA) [28] in both 1CT and 1CT+7 HCECs (termed HCEC 1CTRP and 1CTRP+7, respectively) (Figure 6A). Interestingly, metaphase spreads revealed that only 1CTRP+7HCECs (and not diploid 1CTRP cells) displayed a trisomy for chromosome 20 (+20) in 20% of the population (7/35 cells analyzed by GTG banding) (Figure 6B). These results were also confirmed by FISH (Figure W2C). After isolating a clonal population of +20 cells, we determined that +20 had no obvious effects on clonogenicity in anchorage-dependent colony formation assays (Figure 6C) or anchorage-independent assays in soft agar (Figure 6D). Alterations to K-Ras and TP53, however, increased serum-free anchorage-dependent clonogenicity from ∼15% in 1CT and 1CT+7 cells (Figure 3A) to ∼50% to 55% in 1CTRP+7 and 1CTRP+7, +20 cells (Figure 6C). Unlike diploid 1CT cells, it seems that the absence or presence of serum has no effect on the clonogenicity of aneuploid HCECs. These 1CTRP +7, +20 HCECs could further serve as models to study CIN-mediated CRC progression, most likely representing later-staged adenomas. However, these cells still do not fully represent the transformed state because they lack the ability to form tumors in immunocompromised mice (data not shown).
Figure 6.
Introduction of a constitutively active K-RasV12 oncogene and shRNA-mediated depletion of TP53 induces the emergence of trisomy 20. (A) Knockdown of TP53 and expression of K-RasV12 in HCEC 1CT+7. (B) Karyotypic analysis reveals +20 in 7 of 35 cells analyzed by GTG banding. No +20 cells were detected in diploid HCECs under the same conditions. Neither knockdown of TP53 nor expression of K-RasV12 alone was sufficient to induce +20 in HCEC 1CT+7. The addition of an extra chromosome 20 has no effect on (C) anchorage-dependent (adherent culture) and (D) -independent (soft agar) clonogenicity. HCT-116 colon cancer cells were used as a positive control for anchorage independent growth. All columns represent mean ± SEM.
Discussion
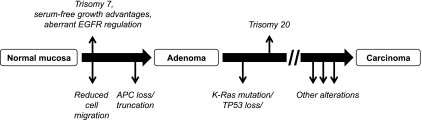
We have isolated isogenic derivatives of HCECs from the same biopsy tissue that are either diploid when maintained in 2% serum or acquire +7 trisomy under serum-free conditions. Analysis of a large sampling of cells by GTG banding and FISH detected no preexisting trisomy cells in the original population, arguing for conversion rather than selection. While we cannot formally prove that rare +7 cells did not preexist in the original diploid population, its frequency is less than 1 of 1000 cells. Our data support the hypothesis that, in the initial stages of culture, a cell from an originally diploid population underwent CIN and subsequent de novo conversion to a +7 cell. This was followed by a selective growth advantage for the rare +7 cell type in serum-deprived culture conditions to eventually out compete diploid cells within the population. This intrinsic defect leading to CIN seems to be specific to the HCEC 1CT line because cells from another patient (HCEC 2CT) do not possess any chromosomal alterations under the same experimental conditions. Figure 7 provides a schematic placing our observations in the context of the current CRC progression paradigm.
Figure 7.
Updated schematic to the current CRC progression paradigm. In the HCEC 1CT model, the appearance of trisomy 7 coinciding with aberrant EGFR regulation and defects in cell migration precedes the loss or truncation of APC. Trisomy 20 also appears after alterations to K-Ras or TP53. Because these cells still do not form tumors in mice, there must be other alterations that drive the progression toward the carcinoma phenotype. The identification of these driver alterations will be the subject of future studies.
Mutation or epigenetic misregulation of the APC gene is one of the earliest known alterations in CRC progression and has been shown to be a significant factor in the generation of CIN[20]. Although APC has many known functions, it is currently unknown exactly how loss of APC leads to the accrual of aneuploidy. Truncation events are the main cause in a majority of APC-inactivated CRC cases [29] followed by promoter hypermethylation [30]. We have shown that both 1CT and 1CT+7 HCECs express full-length APC protein. In addition, DNA sequencing of the APC mutation cluster region reveals no preexisting mutations (primers listed in Table W2). Pyrosequencing was also used to confirm hypomethylated promoters in both the diploid and trisomy lines (0%–2% methylation in 1CT, 1CT+7, and 2CT cells) (data not shown). Contrary to other reports [31–34], our results suggest that APC loss is not required for the onset of CIN in the HCEC 1CT line. In addition, we did not observe any differences in messenger RNA transcript levels for Sgo1 and Bub1 by quantitative polymerase chain reaction (data not shown), two frequently downregulated genes in colon cancer associated with increased levels of CIN [35,36].
Aneuploidy is generally disadvantageous to cells in nonstressful environments. For example, under normal growth conditions, aneuploid mouse embryonic fibroblasts have an overall decreased cellular fitness compared to diploid fibroblasts [22]. This suggests that cells may develop other changes to overcome the initial chromosomal aberration to prevent elimination from the population [37]. This mechanism of compensation for additional chromosomes may confer unknown fitness advantages under specific conditions that allow aneuploid cells to outcompete diploid cells [15]. We hypothesize that under stressful or non-optimal growth conditions, 1CT+7 HCECs gained a growth advantage over their diploid counterparts and this gain may act as a mechanism for cancer initiation [38]. When shifted to serum-free conditions, we show that diploid HCECs lose growth advantages, whereas aneuploid cells gain a slight growth advantage. Thus, the emergence of an extra chromosome could confer a selective advantage to 1CT+7 cells to out-compete normal cells and could provide one explanation for early CRC pathogenesis events in terms of cell competition within colonic crypts. It is currently unknown whether the aberrant EGFR regulation exhibited by 1CT+7 HCECs contributes to the conversion or selection pressures for +7 cells.
Another difference between diploid and trisomy 7 HCECs is a defect in cell migration in 1CT+7 cells. Proper cell migration is a coordinated and dynamic process in the physiological turnover of the colonic epithelium [39]. Stem cells at the bottom of the colon crypts give rise to progenitor cells that differentiate as they migrate toward the lumen of the colon. As differentiated cells reach the apex of the crypt, they undergo apoptosis and are shed into the lumen to make way for newer cells emerging from the base [40]. Thus, the strict regulation in the number of cells gained at the bottom of crypts roughly equals the number of cells lost at the apex [41]. Even partial loss of cell migration within the epithelium could therefore be disruptive to normal colon crypt physiology [42]. A decrease can be seen in the motility of 1CT +7 HCECs compared with their diploid counterparts (Figure 4). These results suggest that, in vivo, +7 HCECs may be slightly impaired in cryptmigration and could thus lead to interference with normal colonic turnover [27]. This may predispose cells to the accumulation of tumorigenic alterations owing to prolonged retention of cells within the crypt. This defect in 1CT+7 cell migration also correlates with a decrease in pro-MMP-2 levels (Figure W1C). MMP-2 is expressed in nontumorigenic mouse colonic epithelial cells [43] and is associated with normal cell migration and physiological wound healing processes [44]. Although aberrant MMP-2 expression has been implicated in disease progression and metastatic potential of cancer cells [45], MMP-2 is required and sufficient to initiate the migration of normal epithelial cells [46]. Our results suggest a possible association between decreased MMP-2 levels and reduced +7 epithelial cell migration that may exist within colonic crypts. In addition, cell models of Down syndrome (human trisomy 21, mouse trisomy 16) also display perturbed migratory abilities [47,48], although the mechanistic link between aneuploidy and cell migration remains unclear.
Introduction of oncogenic K-RasV12 and stable knockdown of TP53 in 1CT+7 HCECs (1CTRP+7) led to the emergence of another nonrandom chromosomal alteration, trisomy 20 (+20), appearing in 20% of the population (1CTRP+7, +20). A clonal isolate was selected and expanded to generate a pure population of 1CTRP+7, +20HCECs. Neither expression of K-RasV12 nor TP53 knockdown alone was sufficient to induce any chromosomal abnormalities. These events also could not be recapitulated in normal diploid 1CT or 2CT HCECs, suggesting an intrinsic defect within +7 HCECs upon experimental manipulation of both K-Ras and TP53. These results also provide additional evidence for the conversion (rather than selection) of preexisting aneuploidy cells in the initial population because there was no evidence of +20 chromosomes unless oncogenic changes were introduced into 1CT+7 cells. Although the EGFR pathway may contribute to the selective advantage of 1CT+7 HCECs over diploid cells, the mechanism by which +7, +20 begins to selectively outcompete +7 in HCEC 1CTRP remains unknown. Interestingly, +7 and +20 events are both frequently detected throughout CRC pathogenesis [49], and these cytogenetic changes seem to be nonrandom. Because it is highly unlikely that such CRC-specific chromosomal changes could be occurring by chance, we believe these 1CTRP+7, +20 HCECs represent a further progressed cell type that may be recapitulating a hierarchy of frequently observed events throughout different stages of CRC. These cells with experimentally introduced oncogenic changes and spontaneous chromosomal abnormalities appearing within the same population of initially diploid cells could also contribute to the cellular heterogeneity generally seen among tumor samples [50].
In summary, we describe the isolation and initial characterization of +7 and +20 HCEC 1CT lines derived from an originally diploid population. Although the underlying mechanism of CIN in the HCEC 1CT population has yet to be identified, these isogenic cells will be useful in studying CIN-mediated CRC initiation and progression in a genetically tractable human cell-based model. Furthermore, these cells could serve as reagents for the discovery of novel drug targets that can exploit specific vulnerabilities in aneuploid HCECs to inhibit their growth or induce apoptosis for therapeutic purposes.
Supplementary Materials and Methods
Array Comparative Genomics Hybridization
Total DNA was isolated using DNeasy Blood & Tissue Kit (Qiagen, Valencia, CA), and samples were hybridized with Affymetrix (Santa Clara, CA) Genome-Wide Human SNP 5.0 according to the manufacturer's protocol at the UT Southwestern Genomics and Microarray Core Facility. Washing and staining were performed on an Affymetrix Fluidics Station 450, and the array was processed with Affymetrix GeneChip Operating Software (GCOS) and GeneChip Genotyping Analysis Software (GTYPE). Data were analyzed using Nexus CGH (BioDiscovery), and DNA-Chip Analyzer (dChip) was used for background subtraction and normalization.
Gelatin Zymography
Cells were seeded in six-well plates at 4 x 104 in 2 ml of serum-free medium. After 40 hours, 32 µl of conditioned medium was collected and resolved on an 8% polyacrylamide gel containing 0.1% gelatin. The gel was incubated in 2.5% Triton X-100 for 30 minutes two times at room temperature with gentle agitation. The gel was then washed with water and incubated for 18 hours at 37°C in a solution containing 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 10 mM CaCl2, and 0.02% NaN3. The gel was then stained with Coomassie R-250 for 30 m followed by destaining and drying onto a blotting paper.
TP53 Knockdown and Expression of K-RASV12
TP53 shRNA expressing HCEC 1CT+7 cells were described elsewhere [1]. Retroviral pBABE-hyg-K-rasV12 was used to drive oncogenic K-Ras expression. Briefly, 2 µg of retroviral vector was transfected to Phoenix A cells with Effectene (Qiagen). Forty-eight hours after transfection, viral supernatants were collected, cleared through a 0.45-µm filter, and used to infect HCEC 1CT+7 +shTP53 cells with 4 µg/ml polybrene (Sigma). Infected cells were selected in 250 µg/ml hygromycin. Reverse transcription-polymerase chain reaction followed by a diagnostic restriction digestion was used to confirm expression of oncogenic KRAS as described [2].
Acknowledgments
The authors thank C. Nirodi for providing cetuximab and for insightful discussions. The authors also thank C. Cornelius for technical assistance and M. White for providing the pBABE-KRAS construct.
Abbreviations
- +7
trisomy 7
- +20
trisomy 20
- APC
adenomatosis polyposis coli
- CIN
chromosomal instability
- CRC
colorectal cancer
- CT
immortalized with Cdk4 and hTERT
- CTRP
HCEC CT with K-RasV12 expression and TP53 knockdown
- EGFR
epidermal growth factor receptor
- FISH
fluorescent in situ hybridization
- HCEC
human colonic epithelial cell
- MMP
matrix metalloproteinase
Footnotes
This work was supported by NASA grant nos. NNXO8BA54G and NNX09AU95G. All authors disclose no potential conflicts of interest.
This article refers to supplementary materials, which are designated by Tables W1 and W2 and Figures W1 and W2 and are available online at www.neoplasia.com.
References
- 1.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–767. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- 2.Lengauer C, Kinzler KW, Vogelstein B. Genetic instability in colorectal cancers. Nature. 1997;386:623–627. doi: 10.1038/386623a0. [DOI] [PubMed] [Google Scholar]
- 3.Rajagopalan H, Nowak MA, Vogelstein B, Lengauer C. The significance of unstable chromosomes in colorectal cancer. Nat Rev Cancer. 2003;3:695–701. doi: 10.1038/nrc1165. [DOI] [PubMed] [Google Scholar]
- 4.Williams BR, Amon A. Aneuploidy: cancer's fatal flaw? Cancer Res. 2009;69:5289–5291. doi: 10.1158/0008-5472.CAN-09-0944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sieber OM, Heinimann K, Tomlinson IP. Genomic instability—the engine of tumorigenesis? Nat Rev Cancer. 2003;3:701–708. doi: 10.1038/nrc1170. [DOI] [PubMed] [Google Scholar]
- 6.Komarova NL, Lengauer C, Vogelstein B, Nowak MA. Dynamics of genetic instability in sporadic and familial colorectal cancer. Cancer Biol Ther. 2002;1:685–692. doi: 10.4161/cbt.321. [DOI] [PubMed] [Google Scholar]
- 7.Grady WM, Carethers JM. Genomic and epigenetic instability in colorectal cancer pathogenesis. Gastroenterology. 2008;135:1079–1099. doi: 10.1053/j.gastro.2008.07.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weaver BA, Cleveland DW. The aneuploidy paradox in cell growth and tumorigenesis. Cancer Cell. 2008;14:431–433. doi: 10.1016/j.ccr.2008.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roig AI, Eskiocak U, Hight SK, Kim SB, Delgado O, Souza RF, Spechler SJ, Wright WE, Shay JW. Immortalized epithelial cells derived from human colon biopsies express stem cell markers and differentiate in vitro. Gastroenterology. 2010;138:1012–1021. doi: 10.1053/j.gastro.2009.11.052. [DOI] [PubMed] [Google Scholar]
- 10.Habermann JK, Paulsen U, Roblick UJ, Upender MB, McShane LM, Korn EL, Wangsa D, Kruger S, Duchrow M, Bruch HP, et al. Stage-specific alterations of the genome, transcriptome, and proteome during colorectal carcinogenesis. Genes Chromosomes Cancer. 2007;46:10–26. doi: 10.1002/gcc.20382. [DOI] [PubMed] [Google Scholar]
- 11.Ried T, Knutzen R, Steinbeck R, Blegen H, Schrock E, Heselmeyer K, du Manoir S, Auer G. Comparative genomic hybridization reveals a specific pattern of chromosomal gains and losses during the genesis of colorectal tumors. Genes Chromosomes Cancer. 1996;15:234–245. doi: 10.1002/(SICI)1098-2264(199604)15:4<234::AID-GCC5>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 12.Bomme L, Lothe RA, Bardi G, Fenger C, Kronborg O, Heim S. Assessments of clonal composition of colorectal adenomas by FISH analysis of chromosomes 1, 7, 13 and 20. Int J Cancer. 2001;92:816–823. doi: 10.1002/ijc.1275. [DOI] [PubMed] [Google Scholar]
- 13.Grade M, Becker H, Liersch T, Ried T, Ghadimi BM. Molecular cytogenetics: genomic imbalances in colorectal cancer and their clinical impact. Cell Oncol. 2006;28:71–84. doi: 10.1155/2006/173815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johansson B, Heim S, Mandahl N, Mertens F, Mitelman F. Trisomy 7 in nonneoplastic cells. Genes Chromosomes Cancer. 1993;6:199–205. doi: 10.1002/gcc.2870060402. [DOI] [PubMed] [Google Scholar]
- 15.Pavelka N, Rancati G, Zhu J, Bradford WD, Saraf A, Florens L, Sanderson BW, Hattem GL, Li R. Aneuploidy confers quantitative proteome changes and phenotypic variation in budding yeast. Nature. 2010;468:321–325. doi: 10.1038/nature09529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoglund M, Gisselsson D, Hansen GB, Sall T, Mitelman F, Nilbert M. Dissecting karyotypic patterns in colorectal tumors: two distinct but overlapping pathways in the adenoma-carcinoma transition. Cancer Res. 2002;62:5939–5946. [PubMed] [Google Scholar]
- 17.Lassmann S, Weis R, Makowiec F, Roth J, Danciu M, Hopt U, Werner M. Array CGH identifies distinct DNA copy number profiles of oncogenes and tumor suppressor genes in chromosomal- and microsatellite-unstable sporadic colorectal carcinomas. J Mol Med. 2007;85:293–304. doi: 10.1007/s00109-006-0126-5. [DOI] [PubMed] [Google Scholar]
- 18.Meijer GA, Hermsen MA, Baak JP, van Diest PJ, Meuwissen SG, Belien JA, Hoovers JM, Joenje H, Snijders PJ, Walboomers JM. Progression from colorectal adenoma to carcinoma is associated with non-random chromosomal gains as detected by comparative genomic hybridisation. J Clin Pathol. 1998;51:901–909. doi: 10.1136/jcp.51.12.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roig AI, Shay JW. Immortalization of adult human colonic epithelial cells extracted from normal tissues obtained via colonoscopy. [February 16, 2010];Nat Protoc. 2010 Available at: http://www.nature.com/natureprotocols/prne/633.html. [Google Scholar]
- 20.Rusan NM, Peifer M. Original CIN: reviewing roles for APC in chromosome instability. J Cell Biol. 2008;181:719–726. doi: 10.1083/jcb.200802107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leung JY, Kolligs FT, Wu R, Zhai Y, Kuick R, Hanash S, Cho KR, Fearon ER. Activation of AXIN2 expression by β-catenin-T cell factor. A feedback repressor pathway regulating Wnt signaling. J Biol Chem. 2002;277:21657–21665. doi: 10.1074/jbc.M200139200. [DOI] [PubMed] [Google Scholar]
- 22.Williams BR, Prabhu VR, Hunter KE, Glazier CM, Whittaker CA, Housman DE, Amon A. Aneuploidy affects proliferation and spontaneous immortalization in mammalian cells. Science. 2008;322:703–709. doi: 10.1126/science.1160058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Torres EM, Sokolsky T, Tucker CM, Chan LY, Boselli M, Dunham MJ, Amon A. Effects of aneuploidy on cellular physiology and cell division in haploid yeast. Science. 2007;317:916–924. doi: 10.1126/science.1142210. [DOI] [PubMed] [Google Scholar]
- 24.Upender MB, Habermann JK, McShane LM, Korn EL, Barrett JC, Difilippantonio MJ, Ried T. Chromosome transfer induced aneuploidy results in complex dysregulation of the cellular transcriptome in immortalized and cancer cells. Cancer Res. 2004;64:6941–6949. doi: 10.1158/0008-5472.CAN-04-0474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wong SF. Cetuximab: an epidermal growth factor receptor monoclonal antibody for the treatment of colorectal cancer. Clin Ther. 2005;27:684–694. doi: 10.1016/j.clinthera.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 26.Boman BM, Huang E. Human colon cancer stem cells: a new paradigm in gastrointestinal oncology. J Clin Oncol. 2008;26:2828–2838. doi: 10.1200/JCO.2008.17.6941. [DOI] [PubMed] [Google Scholar]
- 27.Frey MR, Golovin A, Polk DB. Epidermal growth factor-stimulated intestinal epithelial cell migration requires Src family kinase-dependent p38 MAPK signaling. J Biol Chem. 2004;279:44513–44521. doi: 10.1074/jbc.M406253200. [DOI] [PubMed] [Google Scholar]
- 28.Eskiocak U, Kim SB, Roig AI, Kitten E, Batten K, Cornelius C, Zou YS, Wright WE, Shay JW. CDDO-Me protects against space radiation-induced transformation of human colon epithelial cells. Radiat Res. 2010;174(1):27–36. doi: 10.1667/RR2155.1. [DOI] [PubMed] [Google Scholar]
- 29.Tighe A, Johnson VL, Taylor SS. Truncating APC mutations have dominant effects on proliferation, spindle checkpoint control, survival and chromosome stability. J Cell Sci. 2004;117:6339–6353. doi: 10.1242/jcs.01556. [DOI] [PubMed] [Google Scholar]
- 30.Esteller M, Sparks A, Toyota M, Sanchez-Cespedes M, Capella G, Peinado MA, Gonzalez S, Tarafa G, Sidransky D, Meltzer SJ, et al. Analysis of adenomatous polyposis coli promoter hypermethylation in human cancer. Cancer Res. 2000;60:4366–4371. [PubMed] [Google Scholar]
- 31.Aoki K, Aoki M, Sugai M, Harada N, Miyoshi H, Tsukamoto T, Mizoshita T, Tatematsu M, Seno H, Chiba T, et al. Chromosomal instability by β-catenin/TCF transcription in APC or β-catenin mutant cells. Oncogene. 2007;26:3511–3520. doi: 10.1038/sj.onc.1210141. [DOI] [PubMed] [Google Scholar]
- 32.Fodde R, Kuipers J, Rosenberg C, Smits R, Kielman M, Gaspar C, van Es JH, Breukel C, Wiegant J, Giles RH, et al. Mutations in the APC tumour suppressor gene cause chromosomal instability. Nat Cell Biol. 2001;3:433–438. doi: 10.1038/35070129. [DOI] [PubMed] [Google Scholar]
- 33.Hadjihannas MV, Bruckner M, Jerchow B, Birchmeier W, Dietmaier W, Behrens J. Aberrant Wnt/β-catenin signaling can induce chromosomal instability in colon cancer. Proc Natl Acad Sci USA. 2006;103:10747–10752. doi: 10.1073/pnas.0604206103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kaplan KB, Burds AA, Swedlow JR, Bekir SS, Sorger PK, Nathke IS. A role for the adenomatous polyposis coli protein in chromosome segregation. Nat Cell Biol. 2001;3:429–432. doi: 10.1038/35070123. [DOI] [PubMed] [Google Scholar]
- 35.Iwaizumi M, Shinmura K, Mori H, Yamada H, Suzuki M, Kitayama Y, Igarashi H, Nakamura T, Suzuki H, Watanabe Y, et al. Human Sgo1 downregulation leads to chromosomal instability in colorectal cancer. Gut. 2009;58:249–260. doi: 10.1136/gut.2008.149468. [DOI] [PubMed] [Google Scholar]
- 36.Pino MS, Chung DC. The chromosomal instability pathway in colon cancer. Gastroenterology. 2010;138:2059–2072. doi: 10.1053/j.gastro.2009.12.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Torres EM, Dephoure N, Panneerselvam A, Tucker CM, Whittaker CA, Gygi SP, Dunham MJ, Amon A. Identification of aneuploidy-tolerating mutations. Cell. 2010;143:71–83. doi: 10.1016/j.cell.2010.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang J, Wang X, Zhao Y, Chen B, Suo G, Dai J. Neoplastic transformation of human diploid fibroblasts after long-term serum starvation. Cancer Lett. 2006;243:101–108. doi: 10.1016/j.canlet.2005.11.022. [DOI] [PubMed] [Google Scholar]
- 39.Humphries A, Wright NA. Colonic crypt organization and tumorigenesis. Nat Rev Cancer. 2008;8:415–424. doi: 10.1038/nrc2392. [DOI] [PubMed] [Google Scholar]
- 40.Stappenbeck TS, Wong MH, Saam JR, Mysorekar IU, Gordon JI. Notes from some crypt watchers: regulation of renewal in the mouse intestinal epithelium. Curr Opin Cell Biol. 1998;10:702–709. doi: 10.1016/s0955-0674(98)80110-5. [DOI] [PubMed] [Google Scholar]
- 41.Nowak MA, Komarova NL, Sengupta A, Jallepalli PV, Shih Ie M, Vogelstein B, Lengauer C. The role of chromosomal instability in tumor initiation. Proc Natl Acad Sci USA. 2002;99:16226–16231. doi: 10.1073/pnas.202617399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lamprecht SA, Lipkin M. Migrating colonic crypt epithelial cells: primary targets for transformation. Carcinogenesis. 2002;23:1777–1780. doi: 10.1093/carcin/23.11.1777. [DOI] [PubMed] [Google Scholar]
- 43.Fenton JI, Wolff MS, Orth MW, Hord NG. Membrane-type matrix metalloproteinases mediate curcumin-induced cell migration in non-tumorigenic colon epithelial cells differing in Apc genotype. Carcinogenesis. 2002;23:1065–1070. doi: 10.1093/carcin/23.6.1065. [DOI] [PubMed] [Google Scholar]
- 44.Murphy G, Gavrilovic J. Proteolysis and cell migration: creating a path? Curr Opin Cell Biol. 1999;11:614–621. doi: 10.1016/s0955-0674(99)00022-8. [DOI] [PubMed] [Google Scholar]
- 45.Seiki M. The cell surface: the stage for matrix metalloproteinase regulation of migration. Curr Opin Cell Biol. 2002;14:624–632. doi: 10.1016/s0955-0674(02)00363-0. [DOI] [PubMed] [Google Scholar]
- 46.Giannelli G, Falk-Marzillier J, Schiraldi O, Stetler-Stevenson WG, Quaranta V. Induction of cell migration by matrix metalloprotease-2 cleavage of laminin-5. Science. 1997;277:225–228. doi: 10.1126/science.277.5323.225. [DOI] [PubMed] [Google Scholar]
- 47.Delom F, Burt E, Hoischen A, Veltman J, Groet J, Cotter FE, Nizetic D. Transchromosomic cell model of Down syndrome shows aberrant migration, adhesion and proteome response to extracellular matrix. Proteome Sci. 2009;7:31. doi: 10.1186/1477-5956-7-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Leffler A, Ludwig M, Schmitt O, Busch LC. Germ cell migration and early development of the gonads in the trisomy 16 mouse—an animal model for Down's syndrome. Ann Anat. 1999;181:247–252. doi: 10.1016/S0940-9602(99)80039-9. [DOI] [PubMed] [Google Scholar]
- 49.Tsafrir D, Bacolod M, Selvanayagam Z, Tsafrir I, Shia J, Zeng Z, Liu H, Krier C, Stengel RF, Barany F, et al. Relationship of gene expression and chromosomal abnormalities in colorectal cancer. Cancer Res. 2006;66:2129–2137. doi: 10.1158/0008-5472.CAN-05-2569. [DOI] [PubMed] [Google Scholar]
- 50.Shackleton M, Quintana E, Fearon ER, Morrison SJ. Heterogeneity in cancer: cancer stem cells versus clonal evolution. Cell. 2009;138:822–829. doi: 10.1016/j.cell.2009.08.017. [DOI] [PubMed] [Google Scholar]
Supplementary References
- 1.Eskiocak U, Kim SB, Roig AI, Kitten E, Batten K, Cornelius C, Zou YS, Wright WE, Shay JW. CDDO-Me protects against space radiation-induced transformation of human colon epithelial cells. Radiat Res. 2010;174(1):27–36. doi: 10.1667/RR2155.1. [DOI] [PubMed] [Google Scholar]
- 2.Sato M, Vaughan MB, Girard L, Peyton M, Lee W, Shames DS, Ramirez RD, Sunaga N, Gazdar AF, Shay JW, et al. Multiple oncogenic changes (K-RAS(V12), p53 knockdown, mutant EGFRs, p16 bypass, telomerase) are not sufficient to confer a full malignant phenotype on human bronchial epithelial cells. Cancer Res. 2006;66:2116–2128. doi: 10.1158/0008-5472.CAN-05-2521. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.