Abstract
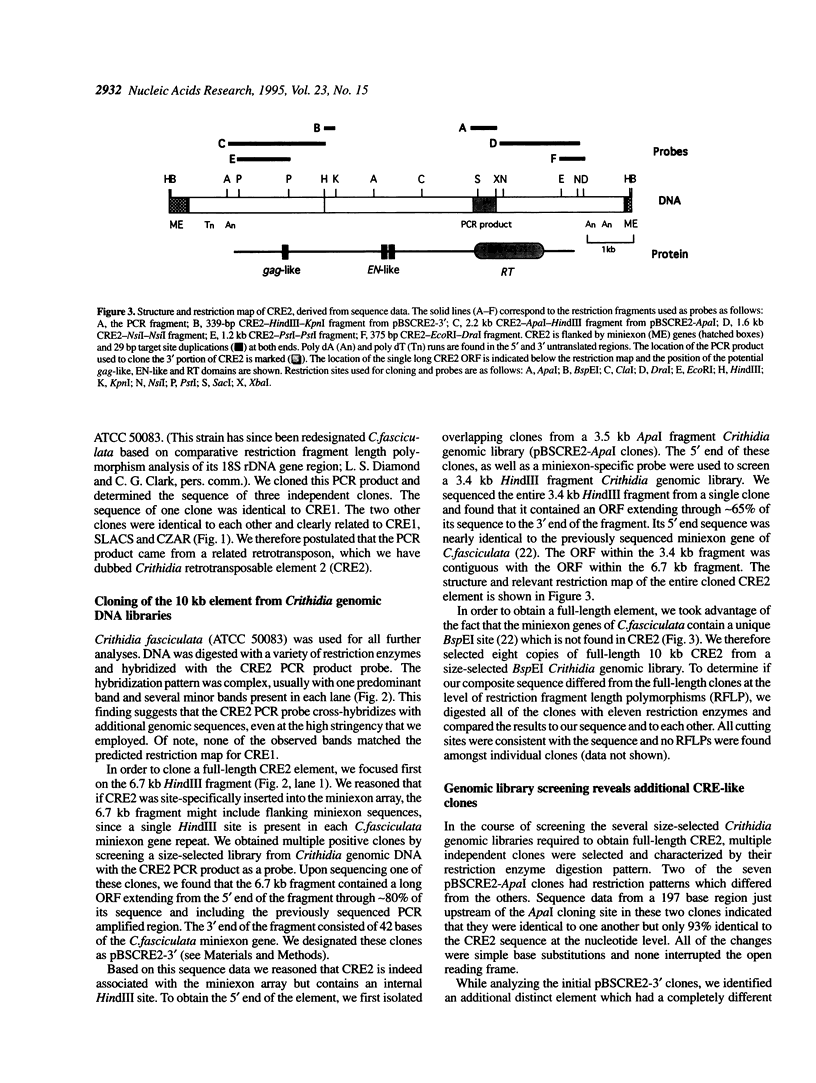
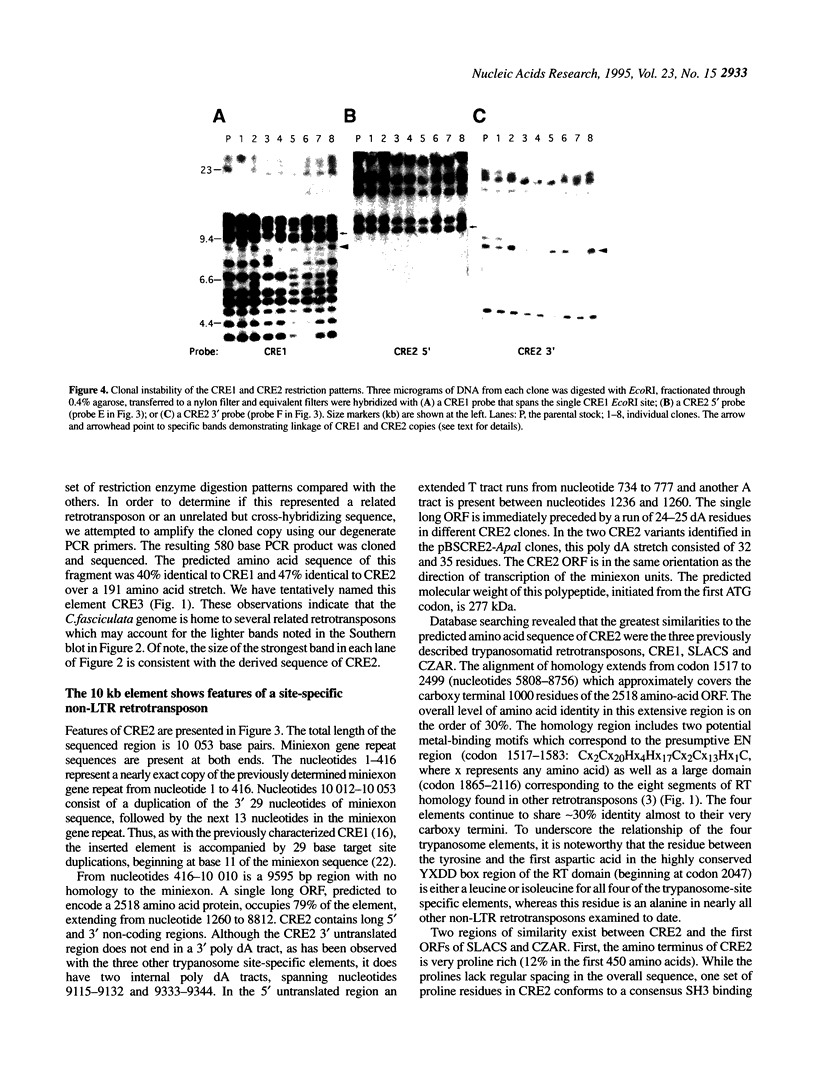
We have identified a new member of the family of trypanosome site-specific retrotransposons, using a degenerate oligonucleotide PCR strategy. The 9595 bp element, termed Crithidia retrotransposable element 2 (CRE2), was cloned and found to be inserted in the tandemly arrayed miniexon genes of Crithidia fasciculata. The element is flanked by 29 bp target site duplications but lacks the 3' poly dA tract characteristic of most other non-long terminal repeat retrotransposons. The amino terminal region of the single 2518-codon open reading frame contains a putative metal-binding motif and a proline-rich region similar to gag-like domains of other retrotransposons. The carboxy terminal region of this open reading frame shares sequence homology with the reverse transcriptase and putative endonuclease regions of three previously described trypanosomatid site-specific retrotransposons. All four of these retrotransposons are specifically inserted between nucleotides 11 and 12 of the highly conserved 39mer sequence of the miniexon gene. Most copies of CRE2 and the previously characterized CRE1 are located on different sized chromosomes. Additional CRE-related sequences were identified by screening Crithidia libraries. These results suggest that a particular sequence in the C. fasciculata miniexon repeat is the target for multiple distinct site-specific retrotransposon insertions.
Full text
PDF





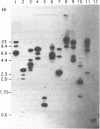
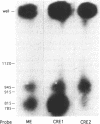
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adams S. E., Mellor J., Gull K., Sim R. B., Tuite M. F., Kingsman S. M., Kingsman A. J. The functions and relationships of Ty-VLP proteins in yeast reflect those of mammalian retroviral proteins. Cell. 1987 Apr 10;49(1):111–119. doi: 10.1016/0092-8674(87)90761-6. [DOI] [PubMed] [Google Scholar]
- Agabian N. Trans splicing of nuclear pre-mRNAs. Cell. 1990 Jun 29;61(7):1157–1160. doi: 10.1016/0092-8674(90)90674-4. [DOI] [PubMed] [Google Scholar]
- Aksoy S. Site-specific retrotransposons of the trypanosomatid protozoa. Parasitol Today. 1991 Oct;7(10):281–285. doi: 10.1016/0169-4758(91)90097-8. [DOI] [PubMed] [Google Scholar]
- Aksoy S., Williams S., Chang S., Richards F. F. SLACS retrotransposon from Trypanosoma brucei gambiense is similar to mammalian LINEs. Nucleic Acids Res. 1990 Feb 25;18(4):785–792. doi: 10.1093/nar/18.4.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg J. M. Potential metal-binding domains in nucleic acid binding proteins. Science. 1986 Apr 25;232(4749):485–487. doi: 10.1126/science.2421409. [DOI] [PubMed] [Google Scholar]
- Besansky N. J., Paskewitz S. M., Hamm D. M., Collins F. H. Distinct families of site-specific retrotransposons occupy identical positions in the rRNA genes of Anopheles gambiae. Mol Cell Biol. 1992 Nov;12(11):5102–5110. doi: 10.1128/mcb.12.11.5102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke W. D., Calalang C. C., Eickbush T. H. The site-specific ribosomal insertion element type II of Bombyx mori (R2Bm) contains the coding sequence for a reverse transcriptase-like enzyme. Mol Cell Biol. 1987 Jun;7(6):2221–2230. doi: 10.1128/mcb.7.6.2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke W. D., Eickbush D. G., Xiong Y., Jakubczak J., Eickbush T. H. Sequence relationship of retrotransposable elements R1 and R2 within and between divergent insect species. Mol Biol Evol. 1993 Jan;10(1):163–185. doi: 10.1093/oxfordjournals.molbev.a039990. [DOI] [PubMed] [Google Scholar]
- Clare J., Farabaugh P. Nucleotide sequence of a yeast Ty element: evidence for an unusual mechanism of gene expression. Proc Natl Acad Sci U S A. 1985 May;82(9):2829–2833. doi: 10.1073/pnas.82.9.2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen G. B., Ren R., Baltimore D. Modular binding domains in signal transduction proteins. Cell. 1995 Jan 27;80(2):237–248. doi: 10.1016/0092-8674(95)90406-9. [DOI] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dombroski B. A., Mathias S. L., Nanthakumar E., Scott A. F., Kazazian H. H., Jr Isolation of an active human transposable element. Science. 1991 Dec 20;254(5039):1805–1808. doi: 10.1126/science.1662412. [DOI] [PubMed] [Google Scholar]
- Eickbush T. H. Transposing without ends: the non-LTR retrotransposable elements. New Biol. 1992 May;4(5):430–440. [PubMed] [Google Scholar]
- Evans J. P., Palmiter R. D. Retrotransposition of a mouse L1 element. Proc Natl Acad Sci U S A. 1991 Oct 1;88(19):8792–8795. doi: 10.1073/pnas.88.19.8792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. "A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity". Addendum. Anal Biochem. 1984 Feb;137(1):266–267. doi: 10.1016/0003-2697(84)90381-6. [DOI] [PubMed] [Google Scholar]
- Gabriel A., Boeke J. D. Reverse transcriptase encoded by a retrotransposon from the trypanosomatid Crithidia fasciculata. Proc Natl Acad Sci U S A. 1991 Nov 1;88(21):9794–9798. doi: 10.1073/pnas.88.21.9794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel A., Yen T. J., Schwartz D. C., Smith C. L., Boeke J. D., Sollner-Webb B., Cleveland D. W. A rapidly rearranging retrotransposon within the miniexon gene locus of Crithidia fasciculata. Mol Cell Biol. 1990 Feb;10(2):615–624. doi: 10.1128/mcb.10.2.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould S. J., Subramani S., Scheffler I. E. Use of the DNA polymerase chain reaction for homology probing: isolation of partial cDNA or genomic clones encoding the iron-sulfur protein of succinate dehydrogenase from several species. Proc Natl Acad Sci U S A. 1989 Mar;86(6):1934–1938. doi: 10.1073/pnas.86.6.1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson L. E., Copeland T. D., Sowder R. C., Smythers G. W., Oroszlan S. Primary structure of the low molecular weight nucleic acid-binding proteins of murine leukemia viruses. J Biol Chem. 1981 Aug 25;256(16):8400–8406. [PubMed] [Google Scholar]
- Iovannisci D. M., Beverley S. M. Structural alterations of chromosome 2 in Leishmania major as evidence for diploidy, including spontaneous amplification of the mini-exon array. Mol Biochem Parasitol. 1989 May 1;34(2):177–188. doi: 10.1016/0166-6851(89)90009-1. [DOI] [PubMed] [Google Scholar]
- Ivanov V. A., Melnikov A. A., Siunov A. V., Fodor I. I., Ilyin Y. V. Authentic reverse transcriptase is coded by jockey, a mobile Drosophila element related to mammalian LINEs. EMBO J. 1991 Sep;10(9):2489–2495. doi: 10.1002/j.1460-2075.1991.tb07788.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakubczak J. L., Burke W. D., Eickbush T. H. Retrotransposable elements R1 and R2 interrupt the rRNA genes of most insects. Proc Natl Acad Sci U S A. 1991 Apr 15;88(8):3295–3299. doi: 10.1073/pnas.88.8.3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakubczak J. L., Xiong Y., Eickbush T. H. Type I (R1) and type II (R2) ribosomal DNA insertions of Drosophila melanogaster are retrotransposable elements closely related to those of Bombyx mori. J Mol Biol. 1990 Mar 5;212(1):37–52. doi: 10.1016/0022-2836(90)90303-4. [DOI] [PubMed] [Google Scholar]
- Jensen S., Heidmann T. An indicator gene for detection of germline retrotransposition in transgenic Drosophila demonstrates RNA-mediated transposition of the LINE I element. EMBO J. 1991 Jul;10(7):1927–1937. doi: 10.1002/j.1460-2075.1991.tb07719.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchner J., Connolly C. M., Sandmeyer S. B. Requirement of RNA polymerase III transcription factors for in vitro position-specific integration of a retroviruslike element. Science. 1995 Mar 10;267(5203):1488–1491. doi: 10.1126/science.7878467. [DOI] [PubMed] [Google Scholar]
- Lake J. A., de la Cruz V. F., Ferreira P. C., Morel C., Simpson L. Evolution of parasitism: kinetoplastid protozoan history reconstructed from mitochondrial rRNA gene sequences. Proc Natl Acad Sci U S A. 1988 Jul;85(13):4779–4783. doi: 10.1073/pnas.85.13.4779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin H. L., Weaver D. C., Boeke J. D. Two related families of retrotransposons from Schizosaccharomyces pombe. Mol Cell Biol. 1990 Dec;10(12):6791–6798. doi: 10.1128/mcb.10.12.6791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan D. D., Korman M. H., Jakubczak J. L., Eickbush T. H. Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: a mechanism for non-LTR retrotransposition. Cell. 1993 Feb 26;72(4):595–605. doi: 10.1016/0092-8674(93)90078-5. [DOI] [PubMed] [Google Scholar]
- Mathias S. L., Scott A. F., Kazazian H. H., Jr, Boeke J. D., Gabriel A. Reverse transcriptase encoded by a human transposable element. Science. 1991 Dec 20;254(5039):1808–1810. doi: 10.1126/science.1722352. [DOI] [PubMed] [Google Scholar]
- Muhich M. L., Hughes D. E., Simpson A. M., Simpson L. The monogenetic kinetoplastid protozoan, Crithidia fasciculata, contains a transcriptionally active, multicopy mini-exon sequence. Nucleic Acids Res. 1987 Apr 10;15(7):3141–3153. doi: 10.1093/nar/15.7.3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pélisson A., Finnegan D. J., Bucheton A. Evidence for retrotransposition of the I factor, a LINE element of Drosophila melanogaster. Proc Natl Acad Sci U S A. 1991 Jun 1;88(11):4907–4910. doi: 10.1073/pnas.88.11.4907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villanueva M. S., Williams S. P., Beard C. B., Richards F. F., Aksoy S. A new member of a family of site-specific retrotransposons is present in the spliced leader RNA genes of Trypanosoma cruzi. Mol Cell Biol. 1991 Dec;11(12):6139–6148. doi: 10.1128/mcb.11.12.6139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver D. C., Shpakovski G. V., Caputo E., Levin H. L., Boeke J. D. Sequence analysis of closely related retrotransposon families from fission yeast. Gene. 1993 Sep 6;131(1):135–139. doi: 10.1016/0378-1119(93)90682-s. [DOI] [PubMed] [Google Scholar]
- Wellauer P. K., Dawid I. B., Tartof K. D. X and Y chromosomal ribosomal DNA of Drosophila: comparison of spacers and insertions. Cell. 1978 Jun;14(2):269–278. doi: 10.1016/0092-8674(78)90113-7. [DOI] [PubMed] [Google Scholar]
- Williams S. M., Robbins L. G. Molecular genetic analysis of Drosophila rDNA arrays. Trends Genet. 1992 Oct;8(10):335–340. doi: 10.1016/0168-9525(92)90277-b. [DOI] [PubMed] [Google Scholar]
- Xiong Y., Eickbush T. H. Origin and evolution of retroelements based upon their reverse transcriptase sequences. EMBO J. 1990 Oct;9(10):3353–3362. doi: 10.1002/j.1460-2075.1990.tb07536.x. [DOI] [PMC free article] [PubMed] [Google Scholar]