Abstract
Advances in genetic engineering have led to the synthesis of protein-based block copolymers with control of chemistry and molecular weight, resulting in unique physical and biological properties. The benefits from incorporating peptide blocks into copolymer designs arise from the fundamental properties of proteins to adopt ordered conformations and to undergo self-assembly, providing control over structure formation at various length scales when compared to conventional block copolymers. This review covers the synthesis, structure, assembly, properties, and applications of protein-based block copolymers.
Keywords: Block copolymers, proteins, genetic engineering, self-assembly
1. Introduction
Protein-based block copolymers have drawn attention for their ability to undergo microphase separation resulting in complex morphologies and sequence-directed control over material structure and properties 1. The motivation behind research in the field of protein-based block copolymers extends from fundamental issues in polymer science and engineering to applications ranges from novel polymer designs of new biomaterials for the pharmaceutical industry to utility in regenerative medicine. Fundamental insight into mechanisms of nanostructure self-assembly of biological polymers is of interest as a complement to the extensive literature on synthetic block copolymer systems. Engineering protein-based block copolymers provides an accessible route to study hierarchical, structural and morphological order and desired functions that can be programmed into peptide block primary sequence/chemistry 2. Through the appropriate selection and positioning of amino acid residues, polymers can be designed with control of hydrophobicity patterns and secondary structures, leading to bioengineered tailor-made biomaterials 3. Several reviews have focused on self-assembled block copolymers containing peptide segments in the solution state 4-6. The structure-function relationships in these types of polymers remain poorly understood and this fundamental gap can result in limitations on the use of these novel materials.
In this article, we aim to provide physical principles of block copolymer separation applicable to protein-based block copolymers, compare design strategies (i.e. genetic engineering vs. chemical synthesis), and highlight key research publications in the field since 2000. In addition, the article provides a survey of peptide-based block copolymers, with the main focus on the interplay between structure, architecture, and function. Current research strategies employed to engineer peptide-based block copolymers are reviewed, along with the properties and possible applications. Moreover, guidelines and approaches to current designs in the field of protein-based block copolymers are provided to assist the materials science community in engineering customized fine-tuned biomaterials. We divide protein-based block copolymers into two groups: (i) synthetic polymer-peptide (hybrid) block copolymers, and (ii) protein/peptide block copolymers, to demonstrate the main differences in design strategies, synthesis techniques, and applications. However, the principles that guide phase separation in both synthetic and natural block copolymers will first be reviewed, as they provide the overall guide to chemistry designs.
2. Physical Properties of Block Copolymers
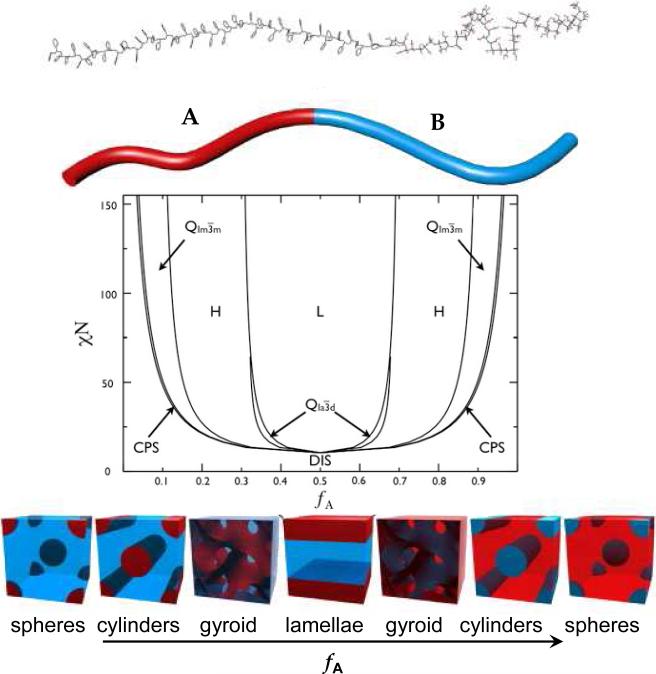
Block copolymers are a special type of polymer in which two or more chemically distinct sequences (blocks) are covalently linked. In block copolymers, both synthetic and protein-based, microstructures (morphologies) are the result of microphase separation, since blocks are covalently attached and therefore cannot undergo two-phase separation (macroseparation). According to Fredrickson and Bates (1997) 7, 8, three main parameters are responsible for the microphase separation: (1) the overall degree of polymerization (the number, N, of monomeric units in a block copolymer chain composed of polymers A and B (i.e. N = NA + NB), (2) the volume fraction, fA, of a hydrophobic block (the volume occupied by polymer A, VA, in the context of the total volume, V, i.e., fA = VA/V), and (3) the Flory-Huggins segment-segment interaction parameter, χAB in a case of a diblock copolymer. The first parameter, the degree of polymerization, influences block copolymer behavior through entropic contributions to the system and can be controlled and assigned by the researcher 8-10. It is known that at a large N value, conformational and translational entropy lead to local ordering by minimizing the A-B monomer contacts. The second parameter, the volume fraction of hydrophobic block A, is responsible for the ordered-state symmetry and can also be controlled by the researcher. This parameter can be also derived from a degree of polymerization parameter (i.e. fA = NA/(NA+NB) in the case of a diblock copolymer). Different classes of structures emerge from the microphase separation dependent on the ratio between the degrees of polymerization of the A and B blocks (Figure 1). For NA << NB spherical inclusions of A in a B-matrix are formed and the spheres form a body-centered cubic lattice. When NA < NB, the A-blocks assume a cylindrical shape and are arranged in a hexagonal lattice. Under symmetric conditions NA ≈ NB layered lamellar-type lattices are observed. Finally, when NA > NB the phases are inverted and the A-blocks constitute the matrix. The third parameter, the Flory-Huggins interaction parameter, χAB, is the decisive parameter to consider, when designing block copolymers. It is dimensionless and describes in an empirical manner the free energy cost per monomer in a situation when two unlike units want to segregate 11. When a χAB value has a positive value, this means that the dissimilar monomers A and B have repulsive interactions and microphase separation occurs. A negative χAB value indicates that a phase transition does not take place and the system continues to stay in a homogeneous phase 12. Additionally, the Flory-Huggins parameter has a temperature dependence, χ ~ 1/T. As temperature increases, the block copolymers become miscible, whereas phase separation occurs at a low temperature 11. The microphase separation leads to different classes of structures dependent on the volume fraction of block A and product NχAB. The length scale of the microstructures is comparable to the size of the block copolymer molecules (5-100 nm). Therefore, the microstructures possess all chemical and physical characteristics of “the building block molecules”. By controlling these parameters via precise block designs, it is possible to control the microstructure of engineered materials. Figure 1 illustrates a mean-field phase diagram and six morphologies that have been observed in synthetic diblock copolymer of poly(styrene-block-methylmethacrylate) (PS-b-PMMA) 13.
Figure 1.
Diblock copolymer phase diagram and morphologies observed for synthetic diblock copolymers. Phases are labeled: L (lamellar), H (hexagonal cylinders), QIa3d (gyroid), QIm3m (body centered cubic spheres), CPS (close-packed spheres), and DIS (disordered). The A–B diblock copolymer is represented by the PS-b-PMMA molecule at the top and depicted as a two-color chain for simplicity. Reproduced with permission from reference 11, 13. Copyright 1996 American Chemical Society. Copyright 2007 ELSEVIER.
Spheres, hexagonally packed cylinders, and lamellae are three classic microphases; but recently, hexagonally modulated lamellae, hexagonally perforated layers, and bicontinuous gyroids have been reported 13, 14. The findings give the impression that almost all synthetic block copolymers composed of two segments exhibit qualitatively similar phase behavior. Analogous with synthetic diblock copolymer systems, triblock copolymers exhibit similar microphase separated states. Castelletto and Hamley (2004) 14 published a review on the various morphologies observed in synthetic ABC triblock copolymers (each letter represents an individual block) and rod-coil block copolymer melts. For more detailed information on ordering in thin films of synthetic block copolymers the reader is directed to a recent review by Hamley (2009) 15.
In contrast to synthetic block copolymers where microstructure formation is dictated by the volume fraction of a hydrophobic block and product NχAB, microstructure formation in peptide-based block copolymers is not solely dependent on phase separation parameters. Instead, other factors such as intra- and intermolecular bonding and chain conformation exert an influence. Hence, one can expect significant divergence from traditional phase behavior observed in the synthetic block copolymer systems due to the rich hydrogen bonding, hydrophobic and electrostatic interactions, peptide backbone rotational restrictions, and complex chemistries in proteins that are for the most part absent from the synthetic polymer block design systems. The main structural elements responsible for the final supramolecular organization in proteins are α-helices and β- sheets. The a-helix is formed by winding the polypeptide backbone into a right-handed helix with a periodicity of 3.6 amino acids per turn and a pitch of 0.56 nm 16. In the helix, the backbone is arranged in such way that the carbonyl oxygen atom of the nth residue points along the helix axis toward the amide hydrogen atom of the (n+4)th residue. The core of the helix is tightly packed and stabilized by hydrophobic and van der Waals’ forces. An example of helix-helix packing is a-helical coiled-coils that are widely used in peptide block copolymer designs. In contrast to a-helices, β-sheets are formed by extensive intermolecular backbone hydrogen bonding between the amide hydrogen atoms of one strand and carbonyl oxygen atoms of the adjacent strands. β-sheets are responsible for mechanical functions of many proteins including silks 17. Moreover, peptides and proteins are amphiphilic. When intramolecularly folded, these macromolecules can display specific faces or solvent-exposed regions that are either hydrophobic or hydrophilic 1. These properties of peptides and proteins add another factor to consider when predicting morphological states in protein-based block copolymers. Several reviews have been published recently focusing on the preparation routes 18, solid-state structure and organization 19, and applications of peptide-based block copolymers 4, 20. However, conditions under which microphase separation occurs and subsequent phase diagram relationships in protein-polymer block copolymers have not been determined. Thus, it is essential to continue to bridge the gap in structure–architecture–function relationships in protein-based block copolymers and learn how to design for function, using peptides and proteins.
3. Design Strategies: Genetic Engineering vs. Conventional Polymer Synthesis
While polymer chemistry is considered a mature discipline, comparisons to the performance of natural systems suggest a wide gap remains with respect to structure-function 21. Significant advances in polymer chemistry include the development of living/controlled polymerization techniques for the synthesis of polymers with defined molecular weights, low polydispersities (Mw/Mn less than 1.1), and reasonable control of architecture. However, the synthetic polymers still lack control over polymer chain length and distribution, as well as access to defined block architectures. Additionally, synthetic polymers do not exhibit the hierarchical structural organization that permeates biological polymers. Moreover, synthetic polymers often require the use of organic solvents for processing into materials in order to perform their functions, whereas biopolymers are designed to be processed in an aqueous environment. In protein-based biopolymers, the primary sequence is responsible for the formation of well-defined secondary structures (e.g., α-helices, β-sheets, β-turns) and folding domains (e.g., calcium-binding domain, E-selectin domain), which then assemble into supramolecular structures 22. As a result of these properties, precise control over the structure at different length scales is achieved. Since structure relates to function, by programming the primary sequence of the polymer predictable three-dimensional structures and desired properties can be engineered 23.
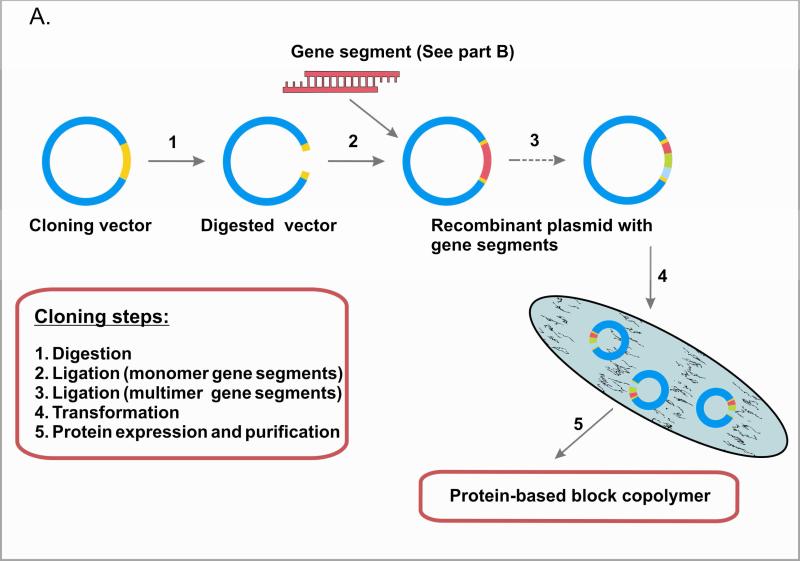
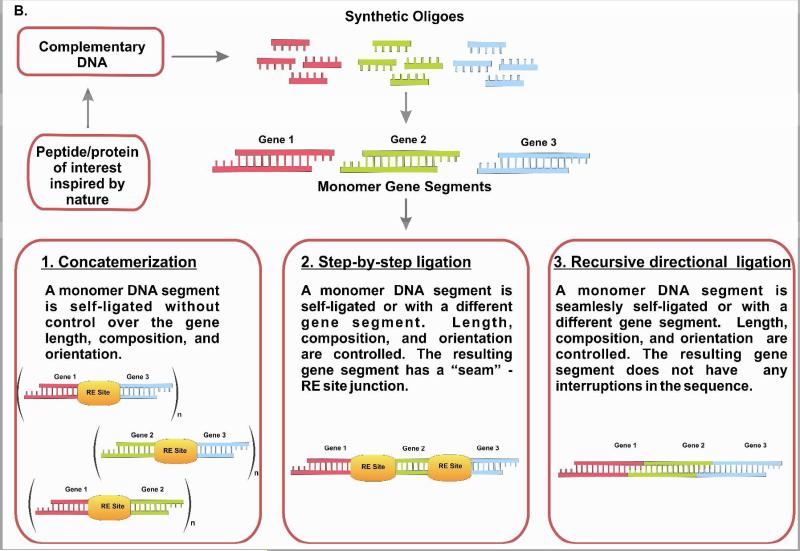
Based on the advantages of biopolymers, protein-based block copolymers have received increasing attention as a strategy to engineer for function 24-26. A number of studies have utilized recombinant DNA technology for the synthesis of protein-based block copolymers, including elastin-like and silk-like block copolymers 27; spider silk block copolymers 28, 29; elastin-like cartilage oligomeric matrix protein block copolymers 24; and block copolymers with coiled-coil domains 30. The recombinant DNA approach enables the formation of block copolymers with programmed sequences, secondary structures, architectures, and precise molecular weight 31, 32. In contrast, chemical synthesis does not achieve this level of control. For example, ring opening polymerization of protected α-amino acid-N-carboxyanhydrides (α-NCA), is limited to a low degree of polymerization (about 100 residues), high polydispersities, and lack of control over the protein sequence and chain architecture 33. Moreover, large repetitive sequences can be constructed by using concatemerization, step-by-step directional approach, and recursive ligation 29, 34-36). Concatemerization is a useful method when a library of genes of different sizes is desired, but has limitations in the preparation of genes with specific sizes 37. To overcome limitations of concatemerization, recursive directional ligation or a step-by-step ligation are employed 35, 38. Recursive directional ligation allows for facile modularity, where control over the size of the genetic cassettes is achieved. Moreover, recursive directional ligation eliminates the restriction sites at the junctions between monomeric genetic cassettes without interrupting key gene sequences with additional base pairs that makes it different from the step-by-step ligation approach 35. Figure 2 summarizes these recombinant DNA approaches.
Figure 2.
Genetic engineering of polypeptides (A). Gene multimerization approaches (B).
The advantages of using recombinant DNA approaches when working with protein based block copolymers include: (i) the formation of biopolymers with defined primary sequence, stereochemistry, and precise molecular weight by employing genetic templates; (ii) reasonable quantities of polymers generated by utilizing bacterial expression systems; (iii) targeted secondary and tertiary structures by using in vivo folding machinery; and (iv) lack of harsh organic solvents used in synthesis and purification. Moreover, (v) purification of recombinant proteins is accomplished with the use of water as solvent under ambient temperature and pressure that makes proteins readily available for biomedical applications. When moving recombinant DNA technology concept from research into commercial applications, optimization steps (e.g. media composition, induction time, concentration of oxygen, others) are required to produce the highest amount of desired polymers per unit media volume per unit time. Several research groups successfully demonstrated gram-level expression of recombinant biomaterials 38, 39. It is also important to recognize that the quality control of recombinant materials varies substantially between research and medical grades, thus biocompatibility becomes an important issue when genetically engineered material is used as a tissue scaffold. Recombinant DNA technology provides the means to generate these materials through the careful design of constructs and selection of appropriate expression systems. Examples of successful recombinant proteins currently approved by the Food and Drug Administration (FDA) include vaccines, such as recombinant human papillomavirus and hepatitis B vaccines, and protein drugs, such as recombinant human erythropoietin and recombinant antithrombin.
The approach has disadvantages, such as the time involved in gene assembly and the limitations to generating biopolymers with unnatural amino acids, such as those that have either the D-form, those not coded by conventional codons, or those with chemical modifications 40. Collagen hydroxy-proline, ornithine, and D-serine-46 in omega-agatoxin of the funnel web spider, Agelenopsis aperta, are examples of unnatural amino acids. Currently, there are up to 300 unnatural amino acids available for research, including β-amino acids, homo-amino acids, cyclic amino acids, aromatic amino acids, and alanine and glycine derivatives.
To incorporate unnatural amino acids, native chemical ligation is a useful tool. This approach allows one to link synthetic amino acids to yield proteins that would be difficult or impossible to prepare otherwise 41. This ligation technique allows the coupling of unprotected synthetic peptides in aqueous solution at neutral pH, preserving the secondary or tertiary structure. Native amide bond formation at the site of ligation results from the spontaneous rearrangement of a thiol exchange product, which is chemo-selectively formed between ligated peptides during a reaction of a C-terminal thioester with a thiol group of an N-terminal cysteine residue.
Recently, a novel approach was developed by Liu et al. (2007) that allowed unnatural amino acids to be genetically encoded in mammalian cells. An orthogonal suppressor tRNA and a mutant Escherichia coli aminoacyl-tRNA synthetase were used to introduce the unnatural amino acid in response to a unique nonsense or frameshift codon in the gene of interest 42. Six unnatural amino acids (e.g. p-methoxyphenylalanine (pMpa), p-acetylphenylalanine (pApa), p-benzoylphenylalanine (pBpa), p-iodophenylalanine (pIpa), p-azidophenylalanine (pAzpa), and p-propargyloxyphenylalanine (pPpa) were successfully incorporated into green fluorescent protein (GFP) expressed in Chinese Hamster Ovary (CHO) cells with efficiencies up to 1 Qg protein per 2×107 cells 42. This approach provides new options for genetic engineering to overcome obstacles in the incorporation of unnatural amino acids.
4. Synthetic polymer peptide block copolymers
Synthetic polymer-peptide conjugates, also known as hybrid block copolymers, have being studied since they were first synthesized by Gallot group in 1976 43. Ring-opening polymerization (ROP) of protected α–amino acid-N-carboxyanhydrides (α-NCAs) initiated by a primary amino end-functionalized polymer was used to synthesize a series of polybutadiene-b-poly(γ-benzyl-L-glutamate) and polybutadiene-b-poly(N-hydroxypropyl-L-glutamine) diblock copolymers 43, 44. The solid state structures of the block copolymers were investigated using circular dichroism and infrared spectroscopy together with X-ray scattering and electron microscopy, and well-ordered lamellar structures were found for those samples with the volume fractions close to 50-50 ratio.
Recently, conjugation of peptides or proteins and synthetic polymers has led to new materials with properties that overcome some of the disadvantages of the individual components 6, 45-47. For example, the peptide/protein segment can allow enhanced control over nanoscale structure formation of the synthetic component. The synthetic segment can also reduce toxicity and immunogenicity, and prevent enzymatic degradation or loss of function due to unfolding of the peptide/protein component 48, 49.
There are diverse routes to prepare hybrid block copolymers 46-48, 50, 51. Controlled radical polymerization, ring opening polymerization, polymerization of macromonomers, and convergent synthesis of peptide-polymer hybrids are some examples. The most frequently used synthetic methodology to prepare homopolypeptide blocks is ROP of protected α-NCAs 18. The technique allows multi-gram scale synthesis, but has disadvantages, such as: (i) chain breaking and termination reactions; (ii) precipitation of the growing polypeptide chain at a certain molecular weight; and (iii) formation of unwanted secondary structures making it difficult to prepare homopolypepties with defined molecular weights and low polydispersity indices 52. For detailed information on the assessment of synthesis methods to prepare peptide-based block copolymers readers are directed to reviews by Marsden 18 and Nikos 53.
Below, we provide a survey of varied hybrid block copolymers that were synthesized using the synthetic approach with an emphasis on preparation routes, structures, morphologies, and potential applications.
4.1 Hybrid block copolymers with homopolypeptide block
Hybrid block copolymers with homopolypeptide blocks are of interest as building blocks for the development of novel self-assembled materials 54-57. The construction of a number of stimuli-sensitive self-assembled materials with various architectures has been reported 54-57. For example, poly(butadiene)m/poly(L-lysine)n block copolymers formed spherical and rod-like micelles in aqueous media with diameters of 82 and 250-350 nm, respectively. Ionization of the lysine side chains at a low pH led to a helix-coil transition and caused swelling of the assemblies. Additionally, at low pH, poly(butadiene)m and poly(L-lysine)n block copolymer underwent transitions from rod-like micelles to spherical micelles 56.
Self-assembly of poly(butadiene)-b-poly(-L-glutamic acid) (PB-b-PGA) diblock copolymers in aqueous solution was studied and formed well-defined vesicles in water with the size of the polymersomes dependent on pH and ionic strength 54. Depending on the pH of the aqueous solution, the hydrodynamic radii of the vesicles varied from 100 nm to 150 nm 54. Rod – coil types of diblock copolymers comprising an α–helical oligopeptide block (γ-benzyl-L-glutamate or ε-benzylocarbonyl-L-lysine) and a short oligostyrene (n=10) block have been prepared 55. The attractive features of this type of block copolymer were that self-assembly was driven not only by microphase separation, but also by the aggregation of the rod segments. Moreover, the α–helical segment was sensitive to temperature and block length ratio suggesting that this system would be useful for stimuli-sensitive self-assembled materials.
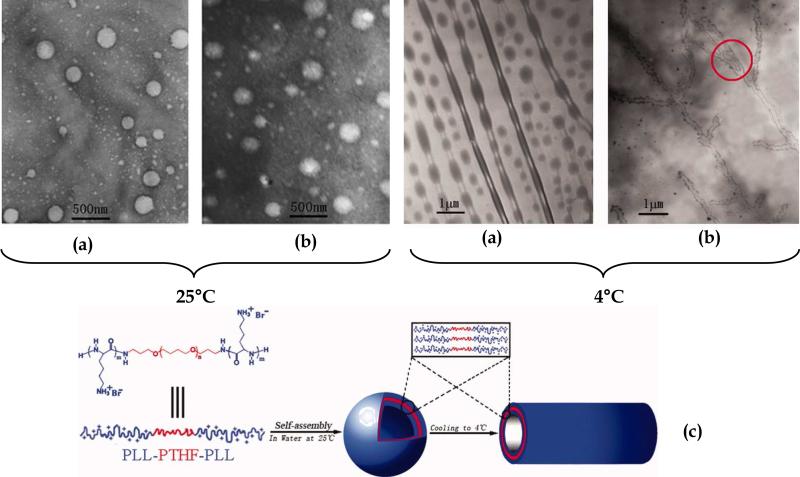
The preparation of amphiphilic triblock copolymers poly(L-lysine)-b-poly(tetrahydrofuran)-b-poly(L-lysine) (PLL-b-PTHF-b-PLL) that self-assembled into vesicles in pure water at room temperature has been explored 57. By reducing the temperature to 4°C, the block copolymers adopted tubular architectures 230 nm in diameter and several μm in length. Figure 3 contains TEM images of vesicles and tubular structures along with the proposed model of the self-assembly of (PLL-b-PTHF- b-PLL) block copolymer. Based on the TEM images, the authors concluded that vesicles were precursors of the tubules based on the existence of intermediate structures, which were nanotubes consisting of necklace-like structures. These block copolymers were proposed as promising candidates as vehicles for gene transfer therapy and drug delivery.
Figure 3.
TEM images of (a) PLL18-b-PTHF14-b-PLL18 and (b) PLL30-b-PTHF14-b-PLL30 aggregates obtained in water at 25°C (left hand images) and then cooled to 4°C for 24 h (right hand images) at a concentration of 0.1 wt %, (c) schematic description of the self-assembly of triblock copolymer PLL-b-PTHF-b-PLL into vesicular and tubular structures. Red circle represents branches and networks of tubular structures. Reproduced with permission from reference 57. Copyright 2007 John Wiley and Sons.
Biofunctional scaffolds that provide both support for cells and biochemical clues for tissue formation are of interest for medical needs. To address this issue, a novel biodegradable triblock copolymer, poly(ethylene glycol)-b-poly(L-lactide)-b-poly(L-lysine) (PEG-PLA-PLL), was synthesized 58. The lysine residues of this block copolymer were further modified with RGD peptide that is a well-known modulator of cell functions 59. The engineered triblock copolymer formed spherical micelles with a diameter of about 120 – 150 nm based on field emission scanning electron microscopy (ESEM) data. Micelle formation was induced by solvent exchange (DMF/water). Moreover, cell experiments with thin films of PEG-PLA-PLL/RGD block copolymer suggested non-toxicity, biocompatibility, and biodegrability of the hybrid polymer. As a result, the authors proposed that this block copolymer system was suitable for biomedical applications for sutures, artificial tissues, implants and drug delivery 58. Table 1 summarizes structures, morphologies, and potential applications of the representative examples of hybrid block copolymers with a homopolypeptide block.
Table 1.
Summary of block chemistries, structures, morphologies, and potential applications of hybrid block copolymers with homopolypeptide blocks.
| A Block | B Block | Mw (number of units or kDa) | Morphologies | Secondary Structure | Solvent | Techniques | Size | Applications | References | |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | poly(butadien)m | poly(L-lysine)n | m=107; n=200 | spherical micelles | α-helix | aqueous solution | CD, DLS, SLS, TEM | 82 ± 2 nm (low pH); 62± 2 nm (high pH) | Drug delivery | Gebhardt et al., 2007 |
| m=107; n=100 | elongated micelles | coil | aqueous solution | 226 ± 2 nm (low pH); 140 ± 2 nm (high pH) | Drug delivery | Gebhardt et al., 2007 | ||||
| m=60; n=50 | elongated micelles | coil | aqueous solution | 155 ± 2 nm (low pH); 112 ± 2 nm (high pH) | Drug delivery | Gebhardt et al., 2007 | ||||
| 2 | poly(butadien)m (PB) | poly(L-lysine)n (P(Lys)) | m=107; n=27 | vesicles | α-helix/β-sheet | aqueous solution | CD, DLS, SLS, TEM | 72 - 92 nm | Drug delivery | Gebhardt et al., 2008 |
| 3 | poly(butadiene)m (PB) | b-poly(-L-glutamic acid)n (PGA) | m=40; n=100 | polymersomes or peptosomes | α-helix | aqueous solution | SLS, DLS, fluorescence spectroscopy, TEM | 100 - 150 nm | Drug delivery | Checot et al., 2003 |
| 4 | poly(styrene)10 (PS) | b-(γ-benzyl-L-glutamate)n (Pblg) | n = 10 - 80 | columnar hexagonal arrangement/lamellae (rod-coil architecture) | α-helix/β-sheet | THF | DSC, FTIR, SAXS, NMR | 16 - 43 A | stimuli-sensitiv materials | Lecommandoux et al., 2001 |
| 5 | poly(styrene)10 (PS) | b-(ε-benzyloxycarbonyl-L-lysine)r | n = 10 - 80 | lamellae | α-helix/β-sheet | THF | DSC, FTIR, SAXS, NMR | 20 A | stimuli-sensitiv materials | Lecommandoux et al., 2001 |
| 6 | poly(tetrahydrofuran)m | b-poly(L-lysine)n | m=14; n=30; n = 18 | vesicles and nanotubes | α-helix | aqueous solution | CD, DLA, TEM | 80 - 117 nm | gene transfer therapy | Tian et al., 2008 |
| 7 | PEG 750 | b-poly(lysine)n | < 3.5 kDa | micelles | α-helix | DMF/water | NMR, XPS, FTIR, DLS, ESEM | 120 - 150 nm | sutures, artificial tissues, implants and drug delivery | Deng et al., 2008 |
4.2. Hybrid block copolymers with complex peptide blocks
Hybrid block copolymers containing biologically-inspired peptide blocks offer significant potential for bioactive molecular recognition and structural hierarchy when compared with simple homopolypeptide block copolymers discussed above 60. Peptide/protein blocks offer a protein with intrinsic ability to fold into predictable three-dimensional tertiary structures that are uniform in size and shape based on the block primary sequence. Biological motifs, such as cell-binding sites, adhesion molecules, growth factors, and enzyme recognition sequences, can be easily incorporated into a complex design providing the materials with advantages over those made with unmodified synthetic polymers 61-63. Additionally, by controlling intra- and intermolecular interactions of peptide blocks such as hydrogen bonding, it is possible to program the primary sequence to form materials with enhanced control over structure and architecture 64.
4.2.1. Block copolymers containing β sheet forming sequences
In Nature, β-sheet forming peptides have a tendency to form fibrils and fibers as well as protein aggregates. The ability of β-sheet forming peptides to form fibers comes from the intrinsic properties of β-strands to establish an extensive hydrogen bond network with neighboring strands in which the amide groups in the backbone of one strand form hydrogen bonds with the carbonyl groups in the backbone of the adjacent strands. The resulting hydrogen-bond arrangement produces the strongest inter-strand stability because it allows the inter-strand hydrogen bonds between carbonyls and amines to be planar 16.
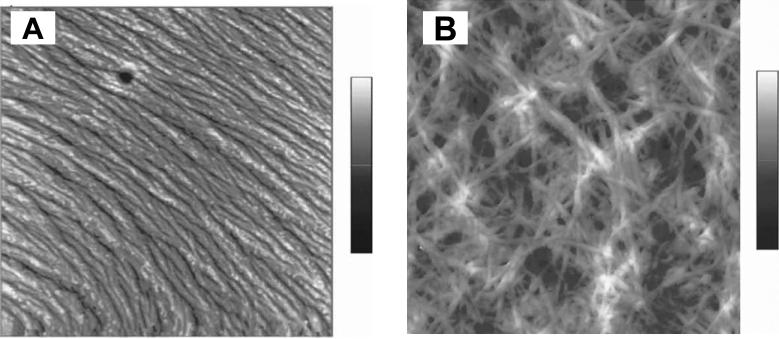
Classic examples of β-sheet forming proteins are silks (spider and silkworm) and amyloid proteins 17, 48. Naturally occurring silks are known for their excellent mechanical properties 31, whereas amyloid peptides are of interest due to their role in prion-related diseases, such as Alzheimer and Creutzfeldt- Jakob 65. Thus, in search for clues from Nature, β-sheet forming peptides have been used to enhance control over molecular architecture in hybrid block copolymers to produce materials with novel properties and functions. Three peptide-poly(ethylene glycol) (PEG) conjugates containing fragments of amyloid β-peptide Aβ, KLVFF, and modified variants of this peptide, AAKLVFF, and FFKLVFF were prepared and characterized 48. PEG was chosen because of its water solubility and biocompatibility. Moreover, it has been widely used as a polymeric conjugation partner for proteins in the pharmaceutical industry. It was observed that only the KLVFF-PEG3300 conjugate formed fibers (Figure 4). PEG attachment to other peptides prevented fiber formation, possibly due to the steric constraints 48.
Figure 4.
AFM height images of (a) FFKLVFF-PEG3300 in methanol- the height scale is 0-14 nm); and (b) FFKLVFF in methanol-the height scale is 0-200 nm. The scan size of both images is 5×5 μm2. Reproduced with permission from reference 48. Marta J. Krysmann, Sérgio S. Funari, Elisabetta Canetta, Ian W. Hamley: The Effect of PEG Crystallization on the Morphology of PEG/Peptide Block Copolymers Containing Amyloid β - Peptide Fragments. Macromolecular Chemistry and Physics. 2009. 209. 883-889. Copyright Wiley-VCH Verlag GmbH & Co. KGaA.
A series of PEG hybrid block copolymers were prepared that contained de novo designed amphiphilic β-strand peptide sequences 2. The conjugation of PEG stabilized the β-strand secondary structure in the PEG hybrid block copolymers and reduced sensitivity of the peptide secondary structure to variations in pH. The formation of rod-like aggregates was observed by SAXS, irrespective of peptide secondary structure. In the search for a cure of Alzheimer's disease, hybrid block copolymers with two blocks were combined, where the polymer block was represented by PEG and the peptide block came from a fragment of the β-amyloid precursor protein βA(10-35). The block copolymer formed fibers, where the formation could be reversed, and was then used to characterize the steps involved in fibrillogenesis 66. The addition of PEG resulted in the inhibition of irreversible fibril-fibril associations of the β-amyloid blocks due to a shielding effect of PEG on the hydrophobic domains in a βA(10-35). These hybrid materials may have potential use in the development of specific inhibitors of fibrillogenesis 66.
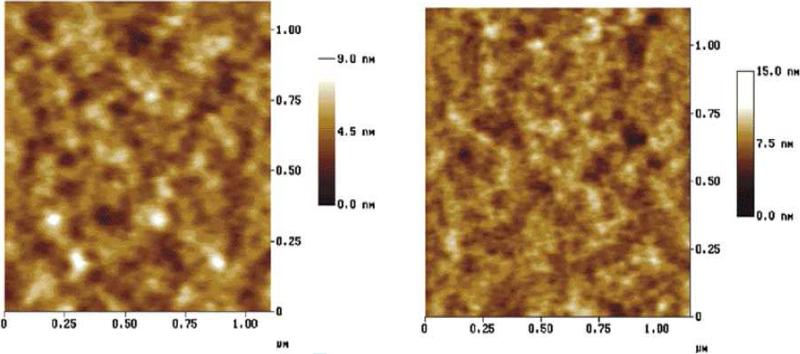
Inspired by the structure of spider silk of Nephila clavipes, multiblock copolymers were prepared by replacement of the amorphous peptide domains of a spider silk with PEG 51. The PEG-poly(alanine) block copolymers retained the antiparallel β-sheet structure that was confirmed by FTIR and X-ray diffraction studies 51. AFM studies revealed phase-separated architectures with the poly(alanine) domains in the range of 100-200 nm (Figure 5).
Figure 5.
Tapping mode AFM topological plots of silk-inspired block copolymers (a) block copolymer with four alanine residues and (b) block copolymer with six alanine residues in each block. Number of PEG units equals to 16 residues per block. Reproduced with permission from reference 51. Copyright 2001 American Chemical Society.
The authors concluded that the hard blocks consisted of the poly(alanine) sequences and the soft blocks were represented by the PEG chemistry. GAGA sequences of Bombyx mori were used to prepare similar multiblock copolymers. The β-sheet structures were retained and phase-separated morphologies with islands of a polypeptide-rich phase (20−50 nm) dispersed in a continuous polyether-rich matrix were formed. Moreover, the larger superstructures, on the order of 100−150 nm, consisted of agglomerations (clusters in which particles stick to each other) of smaller peptide particles with the polyether dispersed between them 50. It was concluded that the PEG segment does not disturb the β-sheet structure and thus can be used to make hybrid silk-like materials.
A similar approach was used to study the effect of polyisoprene in spider silk-like block copolymers. The synthesis and characterization of segmented multiblock copolymers was accomplished in which the poly(alanine) blocks were preserved and the glycine-rich blocks were replaced with polyisoprene blocks to improve solubility 67. Fourier transform infrared spectroscopy, wide angle X-ray diffraction (WAXD), and nuclear magnetic resonance (NMR) spectroscopy supported the presence of β-sheets in the poly(alanine) block. After casting the polymer in solvents such as chloroform and 2-chloroethanol, micellar-like aggregation was observed by TEM 67. The authors observed that only the block copolymers with shorter isoprene blocks formed micellar-like aggregates and concluded that by varying the length of a soft block it is possible to direct the assembly in silk-like block copolymers.
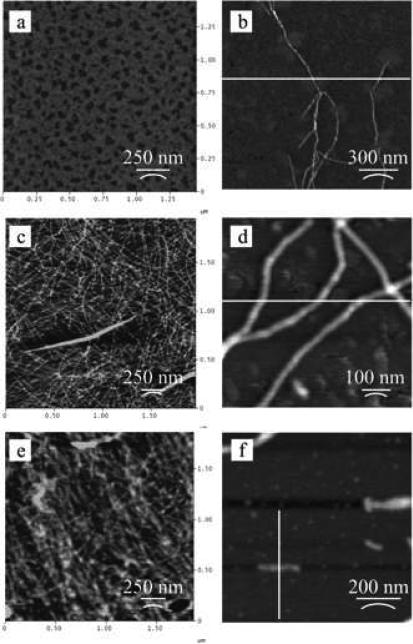
To investigate the effect of PEG chain length on the assembly behavior of block copolymers containing a β-sheet forming block, PEG based triblock copolymers consisting of [(AG)3EG]10 repeats were prepared 68. The β-sheet forming block was prepared by genetic engineering and expressed in E. coli. PEG blocks were coupled with a polypeptide block with N- and C-terminal cysteine residues via conjugation of maleimide-functionalized PEGs with molecular weights of 750, 2,000 and 5,000 g/mol. Infrared spectroscopy showed no major effect of PEG chain length on polypeptide folding as indicated by the strong bands at 1623 and 1522 cm-1 and the weak band at 1697 cm-1 that are indicative for the antiparallel β-sheet conformation. A fibrillar microstructure was observed by AFM for all conjugates, with fibrillar heights of 2 nm (Figure 6). PEG (5,000 g/mol) influenced the assembly of the shorter fibers, when compared to the block copolymers containing the lower molecular weight PEG chains. The poly(alanine) blocks were capable of fibril formation through β-sheet assembly, and the PEG chains prevented further side-to-side aggregation without strong interference of the β-sheet interactions.
Figure 6.
AFM of dried films of [(AG)3EG]10 conjugated with (a) α-maleimidocaproic acid, (b) PEG-750, (c, d) PEG-2,000, and (e, f) PEG-5,000. Reproduced with permission from reference 68. Copyright 2006 American Chemical Society
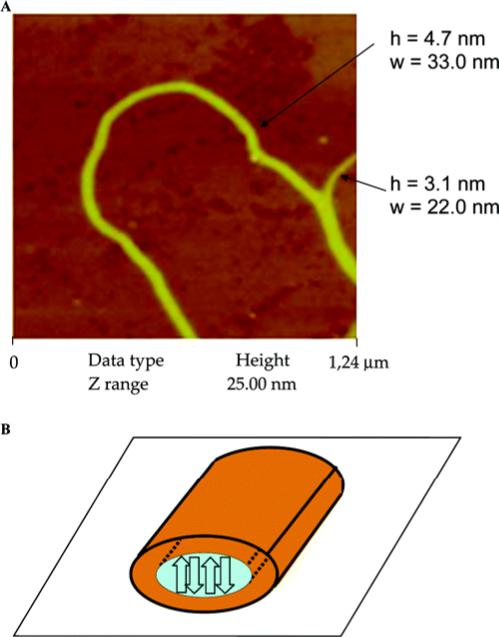
Aside from silk proteins, others also assume a β-sheet conformation. Histidine-containing β-sheet forming peptides, HPKFKIIEFEPPH, formed nanofibers when triggered by metal ions 69. The peptide assumed a random coil conformation in a neutral buffer and underwent a structural transition to a β-sheet conformation in the presence of Cu2+, Zn2+ and Ni2+ 69. Although the peptide-copper complex is not a “true” block copolymer, such designs provide inspiration for block copolymer systems. Polyferrocenylsilane (PFS) graft and block copolymers containing a β-sheet forming GAGA sequence were recently synthesized 70. The GAGA sequence retained its ability to form the antiparallel β-sheet structure in PFS-AGAG conjugated block copolymers. In toluene, PFS-GAGA conjugated block copolymers formed a fibrous network (Figure 7). AFM experiments revealed the dimensions of the fibers with heights of 3-5 nm and widths of 22-50 nm.
Figure 7.
(A) AFM image of PFS-b-AGAG in toluene (0.2 mg/mL). (B) Model presentation of fiber. The orange corona represents the organometallic PFS. The blue core contains the peptide, assembled into antiparallel β-sheets oriented perpendicular to the surface. Reprinted with permission from reference 70. Copyright 2006 American Chemical Society.
The driving force for fiber formation was the self-assembly of the antiparallel-sheet peptides, whereas PFS prevented uncontrolled lateral aggregation of fibers 70. Introduction of inorganic metals to the peptide-based block copolymers offers access to novel biomaterials with new structures and functions. Moreover, conjugation of β-sheet forming peptides to synthetic polymers provides a useful strategy to enhance nanostructure formation and blend the properties of the biological polymers with those of the synthetic polymers. More examples of the block copolymers containing β-sheet forming sequences are listed in Table 2. For more information on β-sheet-forming peptides and their analogues possessing the self-organizing features, readers are referred to a prior review 3.
Table 2.
Summary of block chemistries, synthesis and properties of conjugated block copolymers with β-sheet forming sequences.
| A Block | B Block | Synthesis | Mw, kDa | Morphologies | Secondary Structure | Solvent | Techniques | Size | |
|---|---|---|---|---|---|---|---|---|---|
| 1 | (PEG)3300 | β-amiloid peptides: a) KLVFF | Fmoc solid phase synthesis | 3.9 | non-organized | random coil | methanol (0.25 %) | WAXS, SAXS, FTIR, AFM | N/A |
| b) AAKLVFF | 4.1 | non-organized | random coil | methanol (0.25 %) | N/A | ||||
| c) FFKLVFF | 4.2 | fibrils | β -sheet | methanol (0.25 %) | H = 39 ± 17 nm; W = 56 ± 11 nm | ||||
| 2 | (PEG)3000 | de novo designed amphiphilic β-strand peptides (GKLKKLKQQELELELELG) | Fmoc solid phase synthesis | 5 | fibrillar rods | α-helix and β-strand | aqueous solution | CD, SAXS, TEM | nm scale |
| 3 | Cu2+, Zn2+, Ni2+ | HPKFKIIEFEPPH | Fmoc solid phase synthesis | 1.6 | tape-like fibers | β -sheet | 15 % ethanol/water (20mM Tris-HCl, pH7.4) | CD, TEM | nm scale |
| 4 | PEG | (A)4 and (A)6 | Ring opening polymerization | 15-25 | nanostructures with hard and soft blocks | antiparallel β-sheets | 40 % (w/v) HFIP | NMR, FTIR, AFM, X-ray diffraction | 100-200 nm |
| 5 | PEO | GAGA | Step-growth polymerization | N/A | islands of a polypeptide-rich phase in a continuous polyether-rich | N/A | trifluoroethanol (TFE) | FTIR, X-Ray, 13C NMR, DSC, TEM, AFM | 20-50 nm and 100-150 nm |
| 6 | polyferrocenylsilane (PFS) | GAGA | Fmoc solid phase synthesis | 0.2 | fibrous network | antiparallel β-sheets | toluene | NMR, FTIR, DCS, TEM | H = 3 - 5 nm; W = 22 - 50 nm |
| 7 | PEG | [(AG)3EG]10 | Genetic Engineering | 12 | fibers | β-sheets and γ-turns | methanol | FTIR, AFM, TEM | H = 2 nm; W = 7.5-12 nm |
| 8 | PEG | β-amiloid peptide (YEVHHQKLVFFAEDVGSNKGAIIGL) | Fmoc solid phase synthesis | 6.2 | solid fibers | random coil to β-sheet transition | aqueous solution | NMR, CD, TEM | nm scale |
| 9 | poly(n-butyl acrylate) (pnBA) | (TV)5nFG | Fmoc solid phase synthesis | 1.2 + pnBA | left-handed helical superstructures | antiparallel β-sheets | diethyl ether/methanol (85 %v/v) | CD, FTIR, AFM | H = 3 nm; W = 10 nm; P = 37nm |
4.2.2. Block copolymers containing coiled-coil forming sequences
In Nature, the coiled-coil motif is found in up to 10% of eukaryotic proteins, including transcription factors, chaperon proteins, and keratins. This structure represents an example of helix–helix packing. A coiled-coil is a bundle of two or more right-handed amphiphilic helices wrapping around each other into a slightly left-handed super-helix 71. This motif is characterized by a seven-residue pseudo-repeat, abcdefg, where residues in positions a and d tend to be nonpolar and are positioned in the hydrophobic core of coiled coil bundles. The hydrophobic interactions are the main forces involved in stabilization of coiled coil structure. Amino acids in the e and g positions are involved in electrostatic interactions between strands. These residues are important in specifying the orientation of the α-helix (parallel vs. antiparallel). Residues in the b, c, and f positions are usually exposed to the solvent and involved in hydrogen bonding 72. Coiled-coil motifs have been employed to prepare self-assembled hydrogels 30, 33, long fibers 73, and hybrid biomaterials 45. In these examples, self-assembly was driven by coiled-coil peptide interactions. Coiled-coil interactions were not disrupted by PEG, allowing several research groups to generate hybrid block copolymers based on PEG and coiled-coil blocks 46, 71, 72, 74.
High molecular weight multiblock copolymers comprising PEG and coiled-coil peptide segments were prepared via NHS-activated amide bond formation 49. Two coiled-coil forming peptides (glutamic acid-rich and arginine-rich peptides) and homobifunctional polyethylene glycol N-hydroxysuccinimide esters were utilized in the synthesis of (AB)m-type peptide-PEG polymers with m>15, through the f-position of the peptides and termini of the PEG. These block copolymers formed heterooligomeric assemblies in the form of hydrogels, as well as homooligomeric micellar structures, suggesting that the peptides retained their coiled-coil capacity 49. The architectures (micelles and hydrogels) can be utilized as drug delivery reservoirs and controlled release systems, as well as chemically and thermally responsive hydrogels.
In addition to coiled-coil forming peptides, coiled-coil structures can be generated by leucine zippers. In natural peptides, the leucine zipper consists of a periodic repetition of leucine at every seventh position over eight helical turns. The segments containing these periodic arrays of leucine residues are in the α-helical conformation. The leucine side chains extend from one α-helix and interact with those from a similar α-helix of a second polypeptide, facilitating the formation of a coiled-coil structure that is held together by hydrophobic interactions between the leucine residues. Genetically engineered triblock copolymers AC10A, consisting of two leucine zipper end blocks (A) and a random coil multiblock (C10) composed of nonpeptide sequence (-AG)3-PEG- have been prepared 75. The block copolymer formed soft hydrogels that exhibited a high-frequency plateau in storage modulus, and thus is a promising candidate for biomedical applications.
Besides building blocks with known assembly behavior, de novo designed peptide blocks have become increasingly important for the construction of new self-assembling systems.
The synthesis and self-assembly of hybrid block copolymers based on the peptide sequence, GEAK(LAEIAK)2LAEIYA, derived from the α-helical coiled-coil motif, has been described 46. This amino acid sequence was derived from a de novo designed coiled-coil 76. The conjugation of a peptide block with a synthetic block (PEG) did not disrupt the self-organization of the peptide sequence and led to the formation of discrete nano-objects that were formed into thin films. These features coarsened and eventually disappeared upon decreasing the PEG molecular weight and removing the PEG chains.
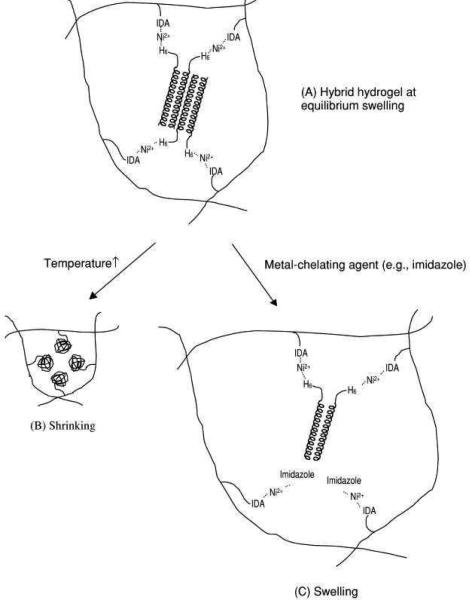
As an alternative to the synthesis of hybrid block copolymers, the de novo design of a peptide sequence, (VSSLESK)6, was used to genetically engineer α-helical coiled coil structures 33, 71. The hybrid hydrogels formed by cross-linking with water-soluble N-(2-hydroxypropyl) methacrylamide and N-(N',N'-dicarboxymethylaminopropyl)-methacrylamide (poly(HPMA-co-DAMA)) containing a metal chelating group iminodiacetic acid (IDA) in the presence of Ni2+, through IDA-Ni2+-His complexation 33, 71. Figure 8 demonstrates this hybrid hydrogel assembly from synthetic polymer and coiled coil protein blocks. The hydrogels were biocompatible and responsive to strong metal-chelating ligands such as imidazole 33. However, in both examples the hybrid block copolymers did not demonstrate higher-order assembly.
Figure 8.
Schematic of a hybrid hydrogel assembled from synthetic polymer and coiled coil protein domains (A), and the volume transition in response to temperature (B) and chemicals (C) (IDA is iminodiacetic acid). Reprinted with permission from reference 71. Copyright 2001 Elsevier.
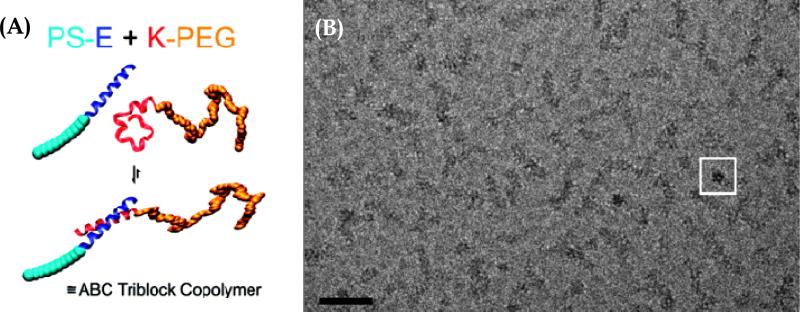
Larger assemblies of peptide-polymer hybrids occurred due to both coiled-coil formation and hydrophobic-block-induced aggregation 72. The authors employed an α-helical coiled-coil pair of peptides, G(EIAALEK)3 (peptide E) and (KIAALKE)3G (peptide K), in combination with polystyrene (PS) and PEG blocks, respectively, as the synthetic polymers 72. These block copolymer underwent two levels of self-assembly upon dispersion in solution: the specific association of the peptide pair led to the formation of the new amphiphilic hybrid ABC triblock copolymer PS-E/K-PEG, which subsequently self-assembled into rod-like micelles with dimensions of 42 ± 10 nm in length and 8 ± 1 nm in width 72. Figure 9 shows the hierarchical self-assembly of the PS−E and K−PEG hybrid block copolymers (A) and block copolymer cryo-TEM images. Above 50°C the reversible dissociation of the coiled coil segments was induced, resulting in the transition of the rod-like micelles into spherical micelles 72. Such reversible transitions together with the potential to further modify peptide chemistry make the E and K peptide motifs promising building blocks for bottom-up approaches for materials formation.
Figure 9.
A) Schematic representation of the hierarchical self-assembly of the hybrids PS−E and K−PEG containing complementary peptide blocks. (B) Cryo-TEM image of PS−E/K−PEG, with 50 nm scale bar. The rods of PS−E/K−PEG are formed by small dots organized along the rod. The white box depicts a rod that is perpendicular to the page, viewed down the cylinder axis. Reproduced with permission from reference 72. Copyright 2008 American Chemical Society.
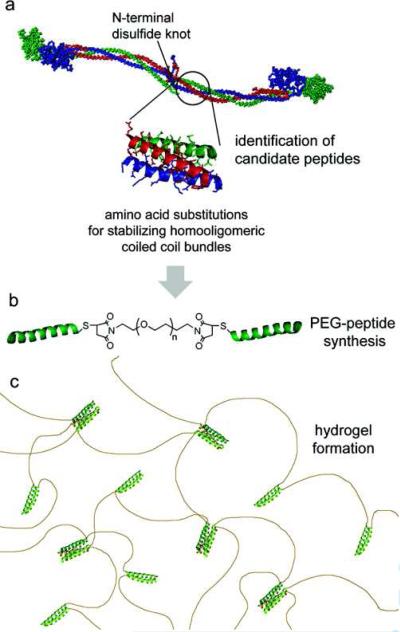
Peptide-polymer conjugates inspired by coiled-coil domains of human fibrin have been prepared using the sequence: IDFISTYITKIDKKIQSIEDIIHQIENKISEIKQLIK) 74. A triblock copolymer composed of a central PEG block flanked by two coiled-coil forming sequences (γKI-PEG-γKI) self-assembled into viscoelastic hydrogels. Analytical ultracentrifugation revealed the presence of dimeric and tetrameric bundles (Figure 10).
Figure 10.
Schematic for fibrin-inspired coiled coil biomaterials. (A) Short peptides from the coiled coil domain of chicken fibrin (α chain (red), β chain (blue), γ chain (green). (B) Substitutions are made to stabilize homooligomeric coiled coil formation, and designed peptides are conjugated to short PEG chains to form triblock peptide-PEG-peptides. (C) Triblock copolymers self-assemble in appropriate buffers to produce hydrogels. Reprinted with permission from reference 74. Copyright 2008 American Chemical Society.
Fibrin-inspired coiled coil biomaterials demonstrated no cytotoxicity to primary human endothelial cells 74. In terms of mechanical properties, the triblocks exhibited moduli similar to fibrin gels, although fibrin gels can attain the same stiffness values at significantly lower concentrations. The attractive mechanical properties, absence of cytotoxicity, and ease of synthesis make γKI-PEG-γKI triblock copolymers promising candidates for biomedical technologies, such as for scaffolds for regenerative medicine 74. Table 3 summarizes the hybrid-block copolymer systems with coiled-coil forming sequences.
Table 3.
Summary of block chemistries, synthesis and properties of conjugated block copolymers with coiled-coil forming sequences.
| A Block | B Block | Synthesis | Mw, kDa | Morphologies | Secondary Structure | Solvent | Techniques | Size | Applications | References | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | PEG77 | (KIAALKE)3G | Fmoc solid phase synthesis | 6 | soluble protein N/A | random coil | DMF/PBS | CD, DLS, cryo-TEM | N/A | nanomaterials | Marsden et al., 2008 |
| 2 | PS11 | G(EIAALEK)3 | Fmoc solid phase synthesis | 3.3 | spherical | α-helix | DMF/PBS | CD, DLS, cryo-TEM | 15 ± 2 nm | nanomaterials | Marsden et al., 2008 |
| 3 | PS - PEG | G(EIAALEK)3/(KIAALKE)3G | Fmoc solid phase synthesis | 9.1 | rod-like | coiled-coil | DMF/PBS | CD, DLS, cryo-TEM | L = 42 ± 10 nm; W = 8 ± 1 nm | nanomaterials | Marsden et al., 2008 |
| 4 | PEG | IDFISTYITKIDKKIQSIEDIIHQIENKISEIKQLIK | Fmoc solid phase synthesis followed by cystein-maleimide chemistry | 8.1 and 1.2 | hydrogel | α-helix to coiled-coil | PBS | CD, AUC | N/A | engineering, controlled therapeutic release, and in vitro cell expansion | Jing et al., 2008 |
| 5 | PEG | GEAK(LAEIAK)2LAEIYA | Fmoc solid phase peptide synthesis followed by Pegylation | 3.3 and 4.2 | thin layers of aggregates with nanosized features | α-helical coiled-coil motif | PBS | CD, AUC, EPR, AFM | H = 1 nm; D = 10 nm | nanomaterials | Vandermeulen et al., 2004; Vandermeulen et al., 2003 |
| 6 | PEG3400 | IEEEVEKEVQRLEFEVQALEKEVAEYQEGI and TEEEVRKRVQRLRFRVQALRKRVAEYQEGI | Fmoc solid phase peptide synthesis followed by PEGylation | m = 15; Mw >100kDa | micelles, hydrogels | α-helix, coiled-coil | PBS | CD, AUC, DLS, SLS | Rh = 55 nm; Rg = 62 nm | hydrogels, drug delivery systems | Sahin and Kiick, 2009 |
| 7 | (HPMA) and (DAMA) | 6H-(random coil block)-(VSSLESK)6 | Genetic Engineering | 1 block 1 peptide | hydrogel | α-helix to coiled coil | PBS | CD | N/A | drug delivery systems | Tang et al., 2001 |
As demonstrated above, the presence of coiled-coil forming sequences in block copolymer designs offers more specific and complementary interactions resulting in the formation of more stable and specific macromolecular architectures. These features suggest that these hybrid systems are promising building blocks for the development of supramolecular materials for a range of potential applications.
5. Protein/Peptide Block Copolymers
Protein-based block copolymers are emerging as a new class of biomaterials due to their unique physical, chemical, and biological properties. Protein-based polymers have many advantages over conventional synthetic polymers because they are able to assemble hierarchically into stable, ordered conformations 16, 77. This ability depends on the structures of protein chains as well as the microenvironment. Protein-based block copolymers have the ability to form varied nanostructures in aqueous solution that provides potential benefits for biomedical applications, such as therapeutic delivery, tissue engineering, and medical imaging 78. Some prototypical examples of engineered protein-based block copolymers are silk-like 29, 79, resilin-like 36 and elastin-like polymers 80-82, coiled-coil and leucine zipper domains 73, 83, and various peptide amphiphiles 84.
5.1. Elastin like Block Copolymers
Protein-block copolymers with elastin-like sequences, known as ELP, have been investigated as biomaterials for tissue engineering and delivery of therapeutics 85-90. This interest is fueled by the properties of elastin, such as high elasticity, high fatigue lifetime, and self-assembly properties. Elastin is an extracellular matrix protein that provides elasticity to many tissues and organs. The sequence of native elastin consists mostly of alternating hydrophobic and cross-linking domains, with distinct exons coding for each of these domains 91. Elastin hydrophobic domains are rich in glycine (G), valine (V), proline (P), leucine (L) and other nonpolar amino acids. 35 The hydrophobic core of elastin comprises tandem repeats with the following sequences: PGGV, PGVGV, PGVGVA, and GGLGV 27, 92. In contrast, the crosslink domains are rich in lysine (K) and alanine (A) residues that are capable of forming covalent crosslinks stabilizing fibers formed of elastin.
The biocompatibility of elastin-based biopolymers, such as poly(GVGVP) and its cross-linked matrix, has been supported by a series of tests, such as Ames mutagenicity, cytotoxicity-agarose overlay, acute systemic toxicity, intracutaneous toxicity, muscle implantation, acute intraperitoneal toxicity, and systemic antigenicity 93. Elastin-based polymers are soft, compliant, water-containing, biocompatible matrices that have potential in a number of biomedical applications 94. In addition to this, ELPs are easy to purify. They do not require using column chromatography. Instead, purification is accomplished by inverse temperature transition (ITC) cycling as follows: (1) the cell lysate is centrifuged at 4°C to precipitate the insoluble fraction of cell lysate, (2) the supernatant, containing soluble ELP, is heated to 37°C, (3) once the solution becomes turbid, it is centrifuged at 37°C to precipitate aggregated, insoluble ELP, and then, (4) the pellet is resuspended in cold, low ionic strength buffer (usually PBS). Typically, 3-5 rounds of ITC are enough to attain >95% purity93.
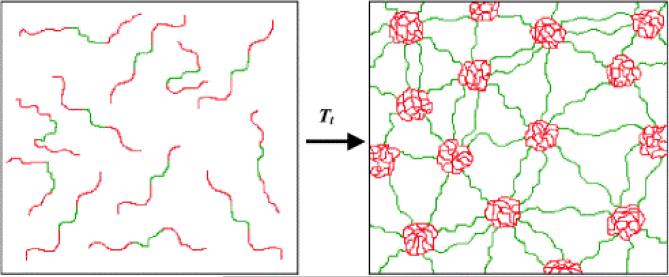
Temperature responsive elastin-like block copolymers were prepared where the hydrophilic blocks (A) were derived either from the tetrapeptide sequence (APGG) or pentapeptide sequences (APGVG and APAVG) that demonstrated elevated values of lower critical solution temperature (i.e. the temperature below which a mixture is miscible in all proportions) 38, 93, 95, 96. The hydrophobic block (B) was derived from the pentapeptide sequences [(V/I)PXYG], where X is a G or A residue, and Y is a G or V residue. The sequence for the hydrophobic block was chosen so the lower critical solution temperature was below 37°C 95. An inverse temperature transition was used to control assembly. The process converts the polypeptide from a soluble, extended state to a collapsed, aggregated state above a lower critical solution temperature, Tt. As a result of this design, elastin-like block copolymers undergo reversible temperature-dependent hydrophobic assembly in aqueous solutions, resulting in the formation of protein nanoparticles with varied morphologies.
A diblock copolymer with the A block sequence [VPGEG(IPGAG)4]14 and B block sequence [VPGFG(IPGVG)4]14 (polymer 1) formed spherical nanoparticles with an average diameter of 50–90 nm above the critical solution temperature 80. The potential for encapsulation of small molecules with the hydrophobic core of a particle was investigated using a fluorescence probe, 1-anilinophthalene-8-sulfonic acid, in a temperature dependent manner 80. The group found that the hydrophobic block was exclusively responsible for binding the florescent probe. Triblock BAB elastin-like copolymers, in which hydrophobic blocks comprised the end blocks, were able to form thermoplastic elastomeric hydrogels above a lower critical solution temperature (Figure 11) 38, 95. Thus, elastin-like block copolymers were able to undergo reversible self-assembly in aqueous solution under physiologically- relevant conditions demonstrating potential for use in nanomedicine 81.
Figure 11.
Schematic representation of the formation of a water-swollen network through micellization of hydrophobic end-block domains of a BAB triblock copolymer. Elastin-mimetic triblock copolymers occur as unassociated monomers in aqueous solution below the phase transition of the end-block domains (left). However above the phase transition (right), the hydrophobic elastin domains (red) undergo desolvation and associate into micellar aggregates that act as virtual cross-links between the central elastin domains (green). Reprinted with permission from reference 38. Copyright 2002 Elsevier.
The stimuli-responsible self-assembly capability of elastin-like block copolymers, together with sequence-derived control over structure and properties via genetic engineering, led to studies that added multifunctionality to elastin-like block copolymers, while preserving the phase-transition properties 32, 97, 98. One of the examples is a bacterial metalloregulatory protein (MerR), known for its high affinity and selectivity to mercury, that was fused to the elastin-like block 98. As a result, selective binding of mercury was demonstrated at a molar ratio of 0.5 mercury/biopolymer and the sequestered mercury was recovered. Moreover, minimal binding of competing heavy metals, such as cadmium, nickel, and zinc, was observed; an example of “green” technology based on proteins 98. In a different study, a hexahistidine metal-binding motif was fused with an elastin-like block to generate selective gel Elastin sequences have also been used to prepare biomaterials for tissue engineering and implantation 99. Three different block architectures were synthesized; A, ABA, and BABA; where the A block was represented by cross-linkable, hydrophobic ELP blocks with periodic lysine residues, and the B block was represented by aliphatic, hydrophilic ELP blocks with no cross-linking sites. All ELP block copolymers were rapidly cross-linked with hydroxymethylphosphines in aqueous solution. The length ratio of uncross-linked block vs. cross-linked block, and the block copolymer architecture, had a significant effect on swelling ratio of the cross-linked hydrogels, their microstructure, and mechanical properties 99. Mouse fibroblasts embedded in the ELP hydrogels survived the cross-linking process and remained viable for at least 3 days in vitro 99. Moreover, enhanced cell viability within triblock ELP hydrogels was observed at day 3, based on total DNA. This study further confirmed that the mechanical properties of ELP hydrogels and the microenvironment they present to cells can be tuned by the design of the block copolymer sequence.
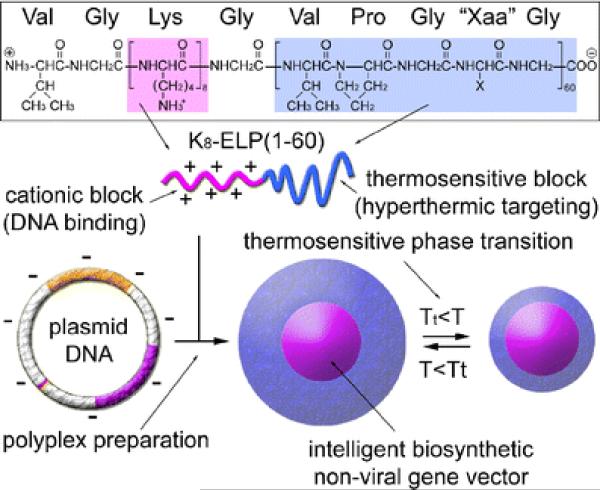
The elastin-like block copolymers can be useful for gene therapy 81, 100. Nanomaterials composed of electrostatically condensing oligolysine block and thermosensitive, particle stabilizing ELP blocks have been reported for this application 47, 100. Cationic elastin diblock copolymers, K8-ELP(1–60), have been prepared by recursive directional ligation. In this system, K8 was denoted as oligolysine, (VGK8G) and ELP was the elastin-like polypeptide with 60 repetitive pentapeptide units, [(VPGXG)60, where X is valine (V), alanine (A), and glycine (G) in a 5:2:3 ratio]. Figure 12 depicts the chemical structure of the biosynthesized K8-ELP(1–60) diblock copolymers for thermosensitive nonviral gene vectors. These elastin-like block copolymers were condensed and released pDNA based on agarose gel electrophoresis, and no cytotoxic effects were found 100. The K8-ELP(1–60)/pDNA polyplex formed particles between 32 and 115 nm in diameter based on dynamic light scattering (DLS) and had dependency on the a cation to anion (N/P) ratio of the polyplexes. The cationic elastic block copolymers offer potential as gene carriers. Additionally, Bae et al. (2007) conjugated geldanamycin, a heat shock protein 90 inhibitor, with thermosensitive poly(K)8-poly(VPGXG)60 block copolymers [K8-ELP(1-60)]. 47 The conjugates formed nanoparticles with a size ranging from 50 to 200 nm, were soluble in PBS, and were effective in hyperthermic combination chemotherapy with facile heat modulation.
Figure 12.
The chemical structure of biosynthesized K8-ELP(1–60) diblock copolymers for thermosensitive nonviral gene vectors. Reprinted with kind permission from reference 100. Copyright 2008 Springer Science+Business Media.
In the search for smart biomaterials, three block copolymers comprised of elastin (E) and cartilage oligomeric matrix protein (COMP) coiled-coil domains have been prepared 24. The blocks were fused in two orientations (EC and CE) and an additional E block was appended to the final construct. Although nearly identical in composition, the EC and CE diblock copolymers exhibit differences in secondary structure and behaved differently when subjected to changes in temperature. The EC, a random coil-like structure was observed at low temperatures, which then transformed to a predominantly helical and β-conformation at higher temperatures. The CE diblock revealed an overall random and β-structure at low temperatures and exhibited a transition to a predominantly β-conformation as a function temperature. The ECE triblock showed similar behavior as the EC diblock copolymer at low and high temperatures 24. This study further demonstrates that block orientation plays a significant role in protein-based block copolymers, in contrast to synthetic block copolymers. Table 5 presents examples of elastin-based block copolymers and their use in biomedical applications. The biomedical applications of elastin-based polymers are described in detail elsewhere 4.
Table 5.
Examples of elastin-based block copolymers.
| Origin | A Block | B Block | Synthesis | Mw, kDa | Morphologies | Secondary Structure | Solvent | Techniques | Size | Applications | References | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Elastin-based | ||||||||||||
| 1 | [VPGEG(IPGAG)4]14 | [VPGFG(IPGVG)4]14 | Genetic Engineering | 58 | core/shell nanoparticles | random coil | aqueous solution | DLS, cryo-HRSEM, TEM | 50 - 90 nm | encapsulation of small drugs | Wright and Conticello, 2002 | |
| 2 | [VPGVG(IPGVGVPGVG)2]19 | [VPAVG(IPAVG)4]16 | Genetic Engineering | 64 | hydrogels | random coil | aqueous solution | DLS, DSC, NMR | N/A | controlled release materials | Wright and Conticello, 2002 | |
| 3 | MGP(GVGVP)153 | MerR - metalloregulatory protein | Genetic Engineering | 80 | hydrogels | N/A | Tris-Cl, potassium phosphate and sodium | Turbidity measurements, binding experiments | N/A | mercury recyclable systems | Kostal et al., 2003 | |
| 4 | G3SGGTGH6/H6G3SGGTG | [(VPGVG)2VPGEG(VPGVG)2]20 | Genetic Engineering | 100 | hydrogels | N/A | aqueous solution | LCSM, SEM | N/A | heavy metal removal | Lao et al., 2007 | |
| 5 | (VPGXG)72 [X is K:F:V in 1:7:1 ratio] | (VPGXG)32 [X is V:G:A in 1:7:8 ratio] | Genetic Engineering | 61 - 85 | hydrogels | N/A | aqueous solution | microrheology, shear modules studies, SEM, cell viability assay | chamber diameters: monoblock (20 30 μm), triblock (100 - 200 μm), tetrablock (35 - 90 μm) | tissue engineering | Lim et al., 2008 | |
| 6 | (VPGXG)60 [X is V:A:G in 5:2:3 ratio] | VGK8G | Genetic Engineering | 25 | microparticles | N/A | PBS | DLS, | 30 - 115 nm | gene therapy | Chen et al., 2008 |
5.2. Silk like Block Copolymers
Silk-based block copolymers have attracted interest due to their mechanical properties and self-assembling features related to the biomaterials field. A unique combination of high elasticity, toughness, and mechanical strength, along with biological compatibility and biodegrability make silks useful candidates for biomaterials 17. Silks are natural block copolymers with a characteristic crystalline β-sheet secondary structure encoded by hydrophobic peptide blocks. β-sheets form through natural hydrogen bonding of amino acid sequences, which, in spider and silkworm silks, consist of multiple repeats of mainly alanine, glycine-alanine, or glycine-alanine-serine. The hydrophilic, non-crystalline regions of silk are commonly composed of: (i) β-spirals similar to a β-turn of GPGXX repeats (where X is mostly glutamine) and (ii) helical structures of GGX 101. The fundamental process of silk protein self-assembly into functional materials, known as the natural spinning process, takes place in the spinning duct, where β-sheet formation is achieved by the progressive loss of water in the gland and alignment of the hydrophobic regions during flow 17. In contrast, artificial spinning is based on the chemical/mechanical transformation of recombinant proteins or reprocessed/resolubilized native proteins. The first step is to dissolve silk proteins in harsh solvents (e.g. highly concentrated LiBr or 100% hexafluoroisopropanol) and then extrude the protein through a thin needle/spinneret into an organic-based coagulation bath (e.g. 90% isopropanol or methanol) to form solid fibers 102.
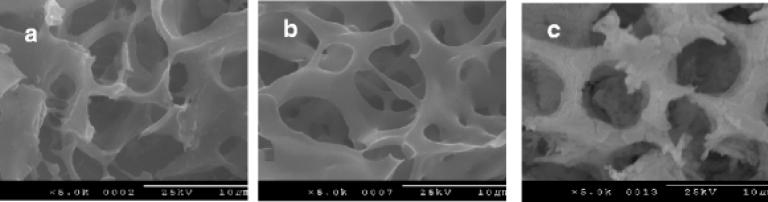
One of the first examples of silk-based block copolymers consisted of the crystalline segment of B. mori fibroin (GAGAGS) and the cell attachment domain of human fibronectin 103. The block copolymer was a product of genetic engineering and had the following composition: HEAD-(Silk-like)9-Fibronectin)12-(Silk-like)2-TAIL. The material was designed to be used to coat artificial surfaces like polystyrene culture dishes to promote both the adhesion and spreading of cells. Wide angle X-ray scattering (WAXS) and selected-area electron diffraction (SAED) data verified that the polymer had semi-crystalline structure. Transmission electron microscopy (TEM) demonstrated that silk-fibronectin-like block copolymers cast from 88% formic acid had a microstructure formed of woven bundles. The bundles were composed of well-defined whisker crystallites with the width of the whiskers of 11.8 - 2.2 nm. This width correlated to the length of the silk-like segment in the block copolymer. It was suggested that the silk-like block copolymers had the silk I structure that was not disrupted by the fibronectin sequence.
Recently, a pH-responsive silk-like block copolymer with a sequence of [(GA)3GE(GA)3GL)]28 was produced in the yeast Pichia pastoris at the g/L level 104. A model peptide, the silk-like peptide ([(GA)3GE(GA)3GE]24), was used and is water soluble above the isoelectric point and forms insoluble stacks of β–sheets in a dried state 105. Every second octapeptide had a hydrophobic leucine residue in place of glutamate. This change made the peptide more amphiphilic based on tensiometry and resulted in added amphiphilicity to the silk-like polymer. The peptide rendered a hydrophobic substrate more hydrophilic and, conversely, a hydrophilic substrate more hydrophobic 104. The block copolymer was capable of fiber formation upon crystallization in 70% (v/v) formic acid under vapor diffusion of methanol, as shown by AFM, with the average fibril height of 2.7 nm and width of 49 nm. The CD spectra of the negatively charged polymer in aqueous solution at high pH indicated a dominance of random and extended helical (silk III-like) structures, whereas the spectra of a coating prepared from such a solution showed a conformation rich in β-turns. Given the biocompatibility of silk-like materials, the polymer may be useful for biomedical applications, such as the coating of surgical implants or pH- responsive controlled drug release.
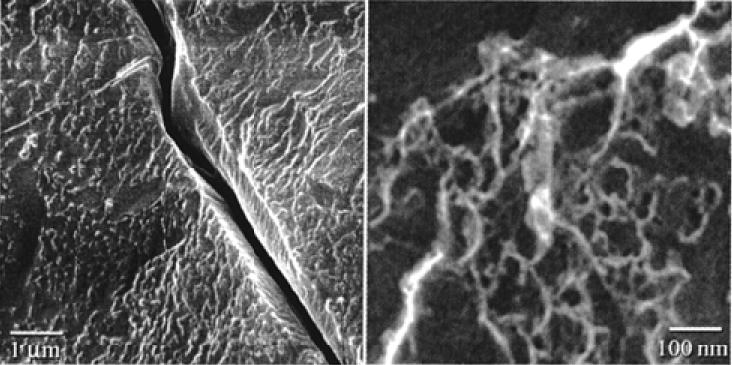
The construction of a biosynthetic multiblock protein polymer based on the sequences of dragline silk from Araneus diadematus [(AEAEAKAK)2AG(GPGQQ)6GS]9 was reported 106. The block copolymer spontaneously formed self-supporting macroscopic films via rearrangement of segments within the polypeptide from α-helices to β-strands. The primary structure of the polymer consisted of the amphiphilic peptide, (AEAEAKAK)2, that was capable of adopting a β-strand conformation over a wide range of pHs and temperatures. The glycine-rich segment, GPGQQ, was derived from Araneus diadematus dragline silk fibroin. The central proline-glycine (PG) unit in the glycine-rich block had a high propensity for the formation of type II β-turns. High resolution field emission scanning electron microscopy (HRSEM) of cryo-immobilized, water-swollen membranes revealed a network of fibrils, approximately 10-20 nm in diameter, interspersed within a less structured matrix as shown in Figure 13. Formation of β-sheet structures induced irreversible aggregation of the polypeptide into a hydrogel network through inter-strand hydrogen bonding interactions between chain segments 106.
Figure 13.
Cryo-HRSEM images of a water-swollen membrane derived from self-assembly of silk multiblock copolymer. Reprinted with permission from reference 106. Copyright 2000 American Chemical Society.
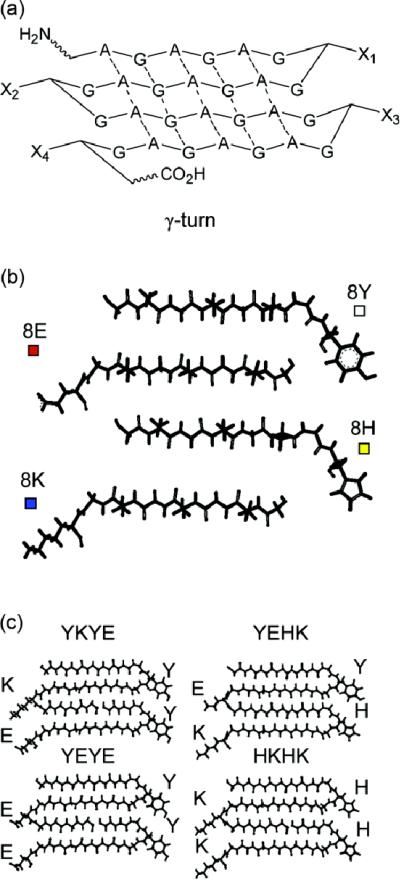
Libraries of genes coding β-sheet forming repetitive sequences, and block-copolymers bearing various C- and N-terminal sequences, were constructed 37. The authors plan to employ these polypeptide libraries for the systematic investigation of protein folding and misfolding, tertiary peptide interactions, and amyloidogenesis. The design was based on the assembly of DNA cassettes coding for the (GA)3GX amino acid sequence. The edges of this β-sheet were functionalized by the turn-inducing amino acids (GX). Figure 14 depicts the design of repetitive unit architectures.
Figure 14.
Design of repetitive unit architecture. (a) General design of polypeptide repetitive unit. (b) Peptide fragments which are coded by the smallest DNA building blocks (strand + turn). (c) Representative repetitive units YKYE, YEHK, YEYE, HKHK. Reprinted with permission from reference 37. Copyright 2007 American Chemical Society.
The polypeptides were expressed in E. coli using conventional vectors and purified by Ni-nitriloacetic acid chromatography. The influence of the polypeptide structure on the physical properties of silk-like block copolymers using gel electrophoresis was investigated under denaturing conditions. The number of repetitive units, as well as side chain modifications (e.g., glycosylation, phosphorylation), significantly affected gel mobility of the peptides. For example, negatively charged and carbamylated peptides showed reduced gel mobility then compared with unmodified peptides, positively charged, and neutral polypeptides 37.
Inspired by the material properties of spider silks, spider silk block copolymers were prepared based on the assembly of individual spider silk modules, in particular poly-alanine (A) and glycine-rich (B) blocks that display different phase behavior in aqueous solution 28, 107. In this study, the interplay between the silk block copolymer sequence, composition, length and self-assembly behavior was assessed to generate materials with precise structural and morphological features 28. The A block consisted of one polyalanine/polyglycine repeat (GAGAAAAAGGAG) responsible for β-sheet formation. The B block was composed of four GGX repeats, separated by the GSQGSGR sequence. The GGX repeats adopted a helical conformation and served as a hydrophilic link between crystalline β-sheet regions as well as neighboring GGX helices in adjacent protein molecules that helped reinforce fiber alignment. Based on FTIR and SEM analysis, trends in block copolymer assembly behavior into specific morphologies, as a function of the number of hydrophobic blocks (A) and solvent effects, were defined. In terms of structure, as the size of the hydrophobic block increased, the content of β-sheets increased in the silk block copolymers. In terms of morphological features, the increase in hydrophobicity (increase in the number of A blocks) was connected with a transformation from thin films to micelles (with diameters around 1-3 μm) and finally to larger compound micelles (diameters ~ 50 μm) in water. In terms of solvent selection, when 2-propanol was chosen as a solvent, a transformation from thin films to nanofibers (50-200 nm) and large compound micelles was observed (Figure 15).
Figure 15.
Molecular assemblies of spider silk-like block copolymers in water and 2-propanol. Reprinted with permission from reference 28. Copyright 2009 American Chemical Society.
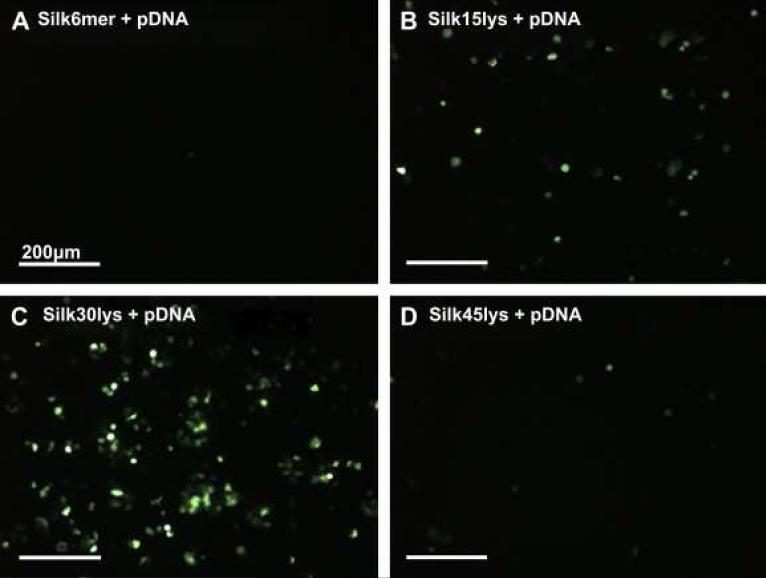
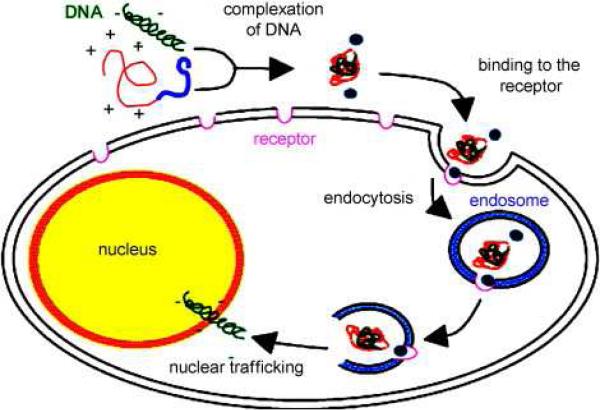
As a continuation of this study, five types of silk-based block copolymers carrying poly(L-lysine) sequence and the cell binding motif (RGD) were genetically engineered and tested for gene delivery to human embryonic kidney cells (HEK) by our group 108. The complexes vary in the position and the number of RGD binding sequences (e.g. one, two, or eleven RGD sequences). Silk block copolymers self-assembled in solution and complexed plasmid DNA encoding green fluorescent protein through ionic interactions. DNA-block copolymer complex formation was characterized by AFM, DLS, and agarose gel electrophoresis (Table 6). To evaluate the feasibility of using the pDNA-block copolymer complexes for gene delivery, in vitro transfection experiments were carried out with HEK cells. The experiments revealed that the pDNA complex of Silk6mer-30lys and 11 RGD sequences (i.e. six silk monomeric gene sequences with 30 lysine residues) with a diameter 380 nm was the most efficient complex as indicated by fluorescent microscopy (Figure 16) 108. Moreover, the MTT assay demonstrated that silk block copolymers had no toxicity to HEK cells at the concentrations used in the transfection experiments (0.76 mg/ml). Thus recombinant silk containing polylysine sequences have demonstrated feasibility for applications in gene delivery.
Table 6.
Summary of block chemistries, synthesis and properties of protein-based block copolymers.
| Origin | A Block | B Block | Synthesis | Mw, kDa | Morphologies | Secondary Structure | Solvent | Techniques | Size | Applications | References | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Silk-based | ||||||||||||
| 1 | silkworm silk block: [(GAGAGS)9]12 | Cell attachment domain of human fibronectin | Genetic Engineering | 95 | woven sheaves microstructures | crystalline β-sheets | formic acid (88 %) | WAXS, TEM, SAED | W= 12 ± 2 nm | tissue engineering and regenerative medicine | Anderson et al., 1994 | |
| [GAAVTGRGDSPASAAGY]12 | ||||||||||||
| 2 | silk-like block I: [(GA)3GE]28 | silk-like block II: [(GA)3GL]28 | Genetic Engineering | 32 | fibrils | stacks of β-sheets, β-turns | formic acid (70 %) | FTIR, CD, tensiometry, AFM | H = 1.5 - 7.5 nm; W = 20 - 30 nm | biomedical applications | Werten et al., 2008 | |
| silk-like block I: [(GA)3GE]24 | silk-like block II: [(GA)3GE]24 | Genetic Engineering | 28 | fibrils | β-sheets and β-turns | formic acid (70 %) | FTIR, CD, tensiometry, AFM | H = 1.5 - 7.5 nm; W = 20 - 30 nm | biomedical applications | Werten et al., 2008 | ||
| 3 | spider silk block from A. diadematus: (AEAEAKAK)2 | spider silk block from A. diadematus: (GPGQQ)6 | Genetic Engineering | 44 | fibril network | antiparallel β-sheets and β-turns | aqueous solution | HRSEM, FTIR, CD, NMR | D = 10 - 20 nm | tissue engineering | Qu et al., 2000 | |
| spider silk block from N. clavipes: (SGRGGLGGQGAGAAAAAGGAGQGGYGGLGSQGT)6 | (K)15,30,45 | Genetic Engineering | 23, 25, and 27 | films | β-sheets | HFIP/water | DLS, AFM, cell viability assay | gene delivery | Numata et al., 2009 | |||
| 4 | hydrophobic block from N. clavipes dragline silk: (GAGAAAAAGGAG)1-6 | hydrophilic block from N. clavipes dragline silk: QGGYGGLGSQGSGRGGLGGQ | Genetic Engineering | 8 - 13 | nanofibers, bowl-shapwed micelles, polymerosomes | antiparallel β-sheets | aqueous solution | FTIR, AFM, SEM | D1 = 1-3 μm; D2 = 70μm; W= 400 nm | controlled drug delivery, tissue engineering, and biosurface engineering | Rabotyagova et al., 2009 | |
| Silk-Elastin | ||||||||||||
| 1 | [GAGAGS]11 | [GXGVP)9]11 | Genetic Engineering | 47 | hydrogels | N/A | PBS | microrheology | N/A | drug delivery | Nagarsekar et al., 2002 | |
| 2 | [(GAGAGS)4]12(GAGAGS) | [(GXGVP)8]13 | Genetic Engineering | 70 | hydrogels | N/A | aqueous solution | turbidity assay, DNA release study | N/A | controlled gene delivery system | Megeed et al., 2002 | |
| 3 | GAGAGS | GVGVP | Genetic Engineering | 55 - 87 | hydrogels | N/A | PBS | microrheology, DSC | N/A | controlled gene delivery system | Haider et al., 2005 | |
| Others | ||||||||||||
| 1 | spider silk block: GGAGQGGYGGLGGQGAGRGGLGGQGAGAAAA | Collagen block: (GXY)r | Genetic Engineering | 57 - 60 | fibers* (ongoing study) | N/A | N/A | N/A | N/A | biomedical applications | Teule et al., 2003 | |
| 2 | (KHKHKHKHKK)6 | FGF2 represents human fibroblast growth factor 2 | Genetic Engineering | 27 | microparticles | N/A | PBS | cell proliferation assay, cell toxicity assays, photon correlation spectroscopy (PCS) | D = 230; 500; 800 nm* | non-viral gene delivery | Hatefi et al., 2006 | |
| 3 | coiled-coil block: (ISSLESK)-(IYYLEYK)2-(ISSLESK) | random coil: [(AG)3PEG]10 | Genetic Engineering | 14 - 20 | hydrogels | α-helical coiled-coil | PBS | CD, AUC, SEM, microrheology | N/A | drug delivery systems | Xu and Kopecek, 2008 | |
| COMP block: DLAPQMLRELQETNAALQDVRELLRQQVKEITFLKNTVMESDASG | elastin block: [(VPGVG)2VPGFG(VPGVG)2]5 | Genetic Engineering | 22, 23, and 35 | aggregates | random coils or β-spirals | PBS | far-UV CD, DLS, SALS | Rh = 60-80 nm | “smart” biomaterials | Haghapanah et al., 2009 | ||
| 4 | leucine zipper block: LGHELAEHKKKLAQLKSELAALKKELAEWE | random coil block: (GAGAGAGPE)10 | Genetic Engineering | 18 | hydrogels | disorderd central domain, helical conformation of the end blocks | PBS | CD, confocal microscopy, surface absorption and cell response assays | N/A | cell-cpecific surface coatings | Fischer et al., 2007 | |
| 5 | (APQMLRELQETNAALQDVRELLRQQVKEITFLKNTVMESDAS) and coiled-coil leucine zipper block (SGDLENEVAQLEREVRSLEDEAAELEQKVSRLKNEIEDLKAE) | (AGAGAGPEG)10 | Genetic Engineering | 20 - 22 | hydrogels | α-helical coiled-coil | aqueous solution | DLS, microrheology | N/A | tissue engineering materials | Shen et al., 2006 | |
Figure 16.
Transfection results in loading pDNA complexes with different polylysine sequences in HEK cells. Fluorescence microscopy images of cells incubated on the silk films containing pDNA complexes of Silk6mer (A), Silk6mer-15lys (B), Silk6mer-30lys (C), and Silk6mer-45lys (D). The green in the images represents successfully transfected cells. Reprinted with permission from reference 108. Copyright 2009 ELSEVIER.
To enhance cell-binding and transfection efficiency a cell binding sequence, RGD, was added to the silk-based block copoymers. Ionic complexes of these silk-polylysine-RGD block copolymers with pDNA were prepared, characterized and utilized for gene delivery to HeLa cells and HEK cells. It was demonstrated that pDNA was transferred to the nucleus with the recombinant silk proteins via integrin-mediated endocytosis 109. Table 6 summarizes chemistries, synthesis, and properties of silk-based block copolymers.
5.3. Silk Elastin Block Copolymers
Silk-elastin block copolymers are another type of protein-based block copolymer that are widely studied as drug and gene delivery carriers 110. Silk-elastin polymers consist of tandem repeats of silkworm silk- like sequence (GAGAGS) 111 and human elastin-like sequence (GVGVP) 88. Silk blocks are insoluble in water and spontaneously precipitate from aqueous solution through the formation of hydrogen bonded β-sheets 31. Addition of elastin blocks to silk-like block copolymers disrupts the crystalline structure in silks that makes silk-elastin block copolymer water soluble 112. Water solubility is an important property, as it is essential for manufacturing, purifying, and processing silk-like materials. Increasing the number of silk blocks within a domain in a silk-elastin like block copolymer increases the rate of gelation, and decreases the rate of resorption of the polymer 86. Capello et al. named silk-elastin block copolymers in their studies “ProLastins” 113. Molecular weights of ProLastins ranged from 60kDa to 85kDa and may be defined using the general formula [(GAGAGS)n(GVGVP)m]o. The composition of ProLastins used in the study is described in Table 6. It was confirmed by thermal analysis and rheological investigation that ProLastins undergo self-assembly resulting in gelation due to crystallization event. Moreover, this process can be precisely controlled by adjusting copolymer composition and crystallization conditions that makes this system suitable for drug delivery 113.
Silk-elastin block copolymers are currently under investigation for controlled delivery of bioactive agents 81, 114-116. Stimuli-sensitive silk-elastin block copolymers were prepared for loading and release of bioactive agents in response to changes in the environmental conditions such as pH, temperature, and ionic strength 86, 117, 118. A series of silk-elastin block copolymers was biosynthesized with one silk unit (GAGAGS) and varied numbers of elastin units. The elastin units varied in length (11 and 16 repeats) and sequence (presence or absence of glutamic amino acid) 86. Evaluation of these polymers at various ionic strengths showed a decrease in Tt (the temperature at which half-maximal turbidity is observed) with increasing ionic strength. Silk-elastin block copolymers were sensitive to pH as detected from the changes in their Tt. For example, Tt of silk-elastin block copolymers with glutamic acid residues increased with an increase in the pH range 3.0 – 7.0, unlike block copolymers containing nonpolar valine residues. These studies suggested that silk-elastin block copolymers with glutamic acid or valine substitutions were differentially sensitive to pH, temperature, ionic strength, and concentration, features that can be exploited for controlled drug loading and release. Moreover, by substituting charged amino acids for neutral amino acids at precise locations along the polymer backbone, and by control over polymer length, it is possible to control the sensitivity of silk-elastin block copolymers to various environmental stimuli 86, 117.
Silk-elastin block copolymers with several silk units are capable of forming hydrogels as a function of temperature, due to the formation of β–sheets as well as self-assembly of the elastin units. Release of plasmid DNA was demonstrated using silk-elastin block copolymers with lysine residues, known as SELP-47 26. The copolymer was able to form hydrogels as demonstrated by X-ray diffraction, DSC, and rotational viscometry studies. The system was capable of releasing plasmid DNA with molecular weight 2.59 MDa containing the Renilla luciferase gene under the control of a cytomegalovirus promoter. Recently, SELP-47K was fabricated into microdiameter fibers using a wet-spinning technique 119. Upon chemical cross-linking, hydrated fibers had a tensile mechanical strength up to 20 MPa, and at the same time high deformability was not compromised 119.
The degree of cross-linking plays an important role in the release of nucleic acids from hydrogels 81. In turn, the degree of cross-linking of silk-elastin hydrogels is a function of several parameters, including the polymer concentration, length, sequence, and composition. By varying these parameters it is possible to control gelation, release, and biodegradation of silk-elastin matrices 81. Megged et al. provided a further review of the literature on genetically engineered silk-elastin-like hydrogels as matrices for cancer gene therapy and readers are referred to this text for more information 120.
5.4. Other Protein based Block Copolymers
Other block copolymers such, as poly(glutamic acids) 121, silk-collagen polymers 122, lysine–histidine copolymers 123, and coiled-coil block copolymers 30 have been prepared and explored. Inspired by collagen-silk copolymers from natural fiber proteins in the byssus thread of marine mussels, combinations of collagen and silk building blocks were prepared to develop biomaterials 122. Three collagen-spidroin block copolymers were engineered with variable amounts of alanine motifs to see the effect on block copolymer behavior and fiber formation. The synthetic genes were introduced into yeast (Pichia pastoris) for protein production and block copolymer characterization and fiber production 122.
To address the needs of intratumor gene delivery, a protein-based block copolymer composed of six lysine and histidine repeats fused with fibroblast growth factor 2 (FGF-2) was engineered and characterized 116, 123. In the design, lysine residues formed complexes with plasmid DNA 124, histidine residues promoted endosomal escape of DNA 125, 126, and FGF-2 was capable to target cells over-expressing fibroblast growth factor receptor, such as cancer cells 127. The block copolymer, named as (KH)6-FGF2, was genetically engineered, expressed, and purified. The purity and expression was determined by SDS-PAGE and western blot analysis. Primary sequence of (KH)6-FGF2 and representation of block copolymer mediated gene transfection are shown in Figure 17. (KH)6-FGF2 block copolymers interacted with plasmid DNA in a dose-dependent manner, had no deleterious effects on cell proliferation (i.e. NIH 3T3 cell line) regardless of the dose, and mediated gene transfer in varied cell lines (i.e. NIH 3T3, COS-1, and T-4-D) via active targeting of the fibroblast growth factor receptor and thus are potentially useful gene carriers 116, 123.
Figure 17.
Schematic of (KH)6-FGF2 fusion-protein mediated gene transfection. (i) Nucleic acid condensed by the positively charged fusion protein, (ii) vector recognition at the FGFR, (iii) internalization by receptor-mediated endocytosis , (iv) endosomal escape by proton-sponge mechanism, and (v) nuclear trafficking ensuing in transgene expression. Reprinted with permission from reference 114. Copyright 200- Elsevier.
The preparation of peptide-based diblock (AB) and triblock (ABA) copolymers were reported based on coiled-coil sequences capable of forming hydrogels by physical crosslinking after interactions of the coiled-coil segments 30. The hydrogel microstructure was investigated by SEM and interconnected network structures were observed (Figure 18).
Figure 18.
SEM images of hydrogels self-assembled from triblock polypeptides: (a) 16 wt.% A4BA4; (b) 20 wt.% A5BA5; (c) 30 wt.% A6BA6. Scale bar is 10 μm. Reprinted with kind permission from reference 30. Copyright 2007 Springer Science+Business Media.
The A block was represented by the coiled coil forming sequence, (ISSLESK)-(IYYLEYK)2-(ISSLESK) and the B block was represented by a random coil sequence, [(AG)3PEG]10, (Table 6). The block copolymers were expressed in E. coli and purified by immobilized metal affinity chromatography. All diblock and triblock copolymers showed predominant α-helical secondary structures based on CD. The thermal stability of coiled-coil block copolymers was also evaluated by CD. The change of ellipticity at 222 nm (α-helix negative peak) with temperature showed that the peptides with five or six repeat units in the coiled-coil domains had a high degree of thermal stability and proposed that the peptides with longer coiled coil block had stronger hydrophobic interactions than peptides with shorter sequences 30. The study demonstrated that hydrogel formation and physical properties can be manipulated by choosing the structure and the length of the coiled coil blocks.
The erosion rate of artificial protein hydrogels from a triblock copolymer bearing dissimilar and similar helical coiled-coil end domains was reported 75. The triblock copolymers consisted of a non-helical C10 block (AGAGAGPEG)10, the coiled-coil P block derived from the (N)-terminal fragment of rat cartilage oligomeric matrix protein and the coiled-coil leucine zipper A block (Table 6). The erosion rate can be tuned by harnessing selective molecular recognition, discrete aggregation number, and orientational discrimination of coiled-coil protein domains 75. Hydrogels formed from a triblock protein bearing dissimilar helical coiled-coil end domains (P and A) eroded more than one hundred-fold slower than hydrogels formed from those bearing the same end domains (either P or A). The reduced erosion rate was a consequence of the looped chains that are suppressed because P and A tend not to associate with each other 75. Thus, by manipulation of the block composition in protein based block copolymers the erosion rate can be tuned over several orders of magnitude in artificial protein hydrogels.
Protein block copolymers are capable of functioning as cell-specific surface coatings that are critical to investigating the role of cell–substrate interactions in regulating cell adhesion, viability, migration, proliferation and differentiation 64. A novel triblock copolymer called CRC comprising an unstructured polyelectrolyte domain flanked by two amphiphilic leucine zipper domains was synthesized and tested. The CRC block design was based on prior designs 128 and 129 for the leucine zipper and disordered polyelectrolyte domains, respectively (Table 6). The block copolymer formed hydrogel coatings through interactions of the amphiphilic end domains of CRC with several surfaces (e.g., glass, polystyrene, or polyester) and was non-adhesive to cells. Additionally, integrin-binding sequence (RGDS) was inserted into the middle block of the CRC polymer. The incorporation of RGD-binding sequence promoted the attachment and spreading of human foreskin fibroblasts and did not affect secondary structure, hydrogel formation behavior, or surface adsorption characteristics of the triblock copolymers. Based on the experimental results, authors believe that the coating is sufficiently robust to support serum-free cell culture and, in particular, could offer a series of new substrates for the ex vivo expansion of rat neural stem cells 64.
The above studies provide proof-of-concept for subsequent designs of “smart” protein- based block copolymers with repetitive sequences to study structure-architecture-property relationships. By altering the sequence, composition and number of the repeating motifs, and systematic physicochemical and biological conditions, it is possible to bioengineer materials tailored for specific applications.
Conclusions
Peptide-based block copolymers have emerged as a new family of block copolymers that possess unique properties when compared to synthetic block copolymers. The understanding of the interplay between primary structure of building blocks, composition, assembly behavior, and properties in protein-based block copolymers is crucial for bringing these materials from research to applications. We have summarized trends in protein building block designs, approaches to the preparations, characterization methodologies, and potential applications. It is apparent that recombinant technology has become a useful approach to engineer protein-based block copolymers, as it provides advantages to the field of biomaterials in terms of sequence control, composition, and molecular orientation. Conjugation of a peptide block with a synthetic block results in the formation of novel biomaterials with advanced functions. Moreover, by using Nature as a source of inspiration, the design of novel biomaterials has become possible by applying the concepts of polymer engineering. Biological peptide motifs possess excellent building blocks, from which materials with desired structure, architecture and functions can be constructed. In peptide-based block copolymers, the conjugation of peptide blocks with synthetic or biological block results in the formation of biomaterials with programmed structures and functions. Despite many successful examples discussed in this review, the understanding of how to design for function has only begun receiving attention and fundamental roles remain to be elucidated. As a result, there remain many challenges and opportunities to implement concepts from both Nature and polymer science to adapt natural materials for a range of applications.
Table 4.
Sequences of diblock and triblock elastin-like protein copolymers. Reprinted with permission from reference38. Copyright 2002 Elsevier.
| Polymer | B block | A blocka | B block |
|---|---|---|---|
| 1 | – | [{VPGEG(IPGAG)4}]14 | [{VPGFG(IPGVG)4}]14 |
| 2 | – | [(APGGVPGGAPGG)2]x | [{VPAVG(IPAVG)4}]16 |
| 3 | [{VPAVG(IPAVG)4}]16 | [{VPGVG(IPGVGVPGVG)2}]19 | [{VPAVG(IPAVG)4}]16 |
| 4 | [{VPAVG(IPAVG)4}]16 | [{VPGEG(VPGVG)4}]30 | [{VPAVG(IPAVG)4}]16 |
| 5 | [{VPAVG(IPAVG)4}]16 | [{VPGEG(VPGVG)4}]48 | [{VPAVG(IPAVG)4}]16 |
| 6 | [{VPAVG(IPAVG)4}]16 | [(APGGVPGGAPGG)2]22 | [{VPAVG(IPAVG)4}]16 |
| 7 | [{VPAVG(IPAVG)4}]16 | [{VPGMG)5}]x | [{VPAVG(IPAVG)4}]16 |
The letter ‘x’ indicates that the degree of concatemerization of the indicated block has not been determined unequivocally.
ACKNOWLEDGMENT
We thank the AFOSR, the NIH and the NSF for support of these studies.
References
- 1.Branco MC, Schneider JP. Self-assembling materials for therapeutic delivery. Acta Biomaterialia. 2009;5(3):817–831. doi: 10.1016/j.actbio.2008.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hamley IW, Ansari IA, Castelletto V, Nuhn H, Rosler A, Klok H-A. Solution self-assembly of hybrid block copolymers containing poly(ethylene glycol) and amphiphilic β-strand peptide sequences. Biomacromolecules. 2005;6(3):1310–1315. doi: 10.1021/bm049286g. [DOI] [PubMed] [Google Scholar]
- 3.König HM, Kilbinger A. Learning from Nature: β-Sheet-Mimicking Copolymers Get Organized. Angewandte Chemie International Edition. 2007;46(44):8334–8340. doi: 10.1002/anie.200701167. [DOI] [PubMed] [Google Scholar]
- 4.Chow D, Nunalee ML, Lim DW, Simnick AJ, Chilkoti A. Peptide-based biopolymers in biomedicine and biotechnology. Materials Science and Engineering: R: Reports. 2008;62(4):125–155. doi: 10.1016/j.mser.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harada A, Kataoka K. Supramolecular assemblies of block copolymers in aqueous media as nanocontainers relevant to biological applications. Progress in Polymer Science. 2006;31(11):949–982. [Google Scholar]
- 6.Vandermeulen GW, Klok H-A. Peptide/protein hybrid materials: enhanced control of structure and improved performance through conjugation of biological and synthetic polymers. Macromolecular Bioscience. 2004;4(4):383–398. doi: 10.1002/mabi.200300079. [DOI] [PubMed] [Google Scholar]
- 7.Bates FS, Wayne W. Maurer WW, Lipic PM, Hillmyer MA, Almdal K, Mortensen K, Fredrickson GH, Lode TP. Polymeric bicontinuous microemulsions. Phys. Rev. Lett. 1997;79:849–852. [Google Scholar]
- 8.Fredrickson GH, Bates FS. Design of bicontinuous polymeric microemulsions. Journal of Polymer Science, Part B: Polymer Physics. 1997;35(17):2775–2786. [Google Scholar]
- 9.Khandpur AK, Foerster S, Bates FS, Hamley IW, Ryan AJ, Bras W, Almdal K, Mortensen K. Polyisoprene-polystyrene diblock copolymer phase diagram near the order-transition. Macromolecules. 1995;28(26):8796–8806. [Google Scholar]
- 10.Fredrickson GH, Bates FS. Dynamics of block copolymers: Theory and experiment. Annual Review of Materials Science. 1996;26(1):501–550. [Google Scholar]
- 11.Matsen MW, Bates FS. Unifying weak- and strong-segregation block copolymer theories. Macromolecules. 1996;29(4):1091–1098. [Google Scholar]
- 12.Strobl GR. The Physics of Polymers Concepts for Understanding Their Structures and Behavior. 3rd ed. Springer; Berlin: 2007. p. 518. [Google Scholar]
- 13.Darling SB. Directing the self-assembly of block copolymers. Progress in Polymer Science. 2007;32(10):1152–1204. [Google Scholar]
- 14.Castelletto V, Hamley IW. Morphologies of block copolymer melts. Current Opinion in Solid State and Materials Science. 2004;8(6):426–438. [Google Scholar]
- 15.Hamley IW. Ordering in thin films of block copolymers: Fundamentals to potential applications. Progress in Polymer Science. 2009;34(11):1161–1210. [Google Scholar]
- 16.Voet D, Voet JG. Biochemistry. 3rd ed. Wiley and Sons, Inc.; 2005. p. 1616. [Google Scholar]
- 17.Kluge JA, Rabotyagova O, Leisk GG, Kaplan DL. Spider silks and their applications. Trends in Biotechnology. 2008;26(5):244–251. doi: 10.1016/j.tibtech.2008.02.006. [DOI] [PubMed] [Google Scholar]
- 18.Marsden HR, Kros A. Macromolecular Bioscience. Polymer-peptide block copolymers - an overview and assessment of synthesis methods. 2009;9(10):939–951. doi: 10.1002/mabi.200900057. [DOI] [PubMed] [Google Scholar]
- 19.Klok HA, Lecommandoux S. Solid-state structure, organization and properties of peptide-synthetic hybrid block copolymers. Peptide Hybrid Polymers In Advances in Polymer Science. 2006;202:75–111. [Google Scholar]
- 20.Upadhyay KK, Agrawal HG, Upadhyay C, Schatz C, Le Meins JF, Misra A, Lecommandoux S. Role of block copolymer nanoconstructs in cancer therapy. Crit Rev Ther Drug Carrier Syst. 2009;26(2):157–205. doi: 10.1615/critrevtherdrugcarriersyst.v26.i2.20. [DOI] [PubMed] [Google Scholar]
- 21.Hecht S. Construction with macromolecules. Materials Today. 2005;8(3):48–55. [Google Scholar]
- 22.Koehl P, Delarue M. Polar and nonpolar atomic environments in the protein core: Implications for folding and binding. Proteins: Structure, Function, and Genetics. 1994;20(3):264–278. doi: 10.1002/prot.340200307. [DOI] [PubMed] [Google Scholar]
- 23.Elemans J, Rowan A, Nolte R. Mastering molecular matter. Supramolecular architectures by hierarchical self-assembly. Journal of Materials Chemistry. 2003;13(11):2661–2670. [Google Scholar]
- 24.Haghpanah JS, Yuvienco C, Civay DE, Barra H, Baker PJ, Khapli S, Voloshchuk N, Gunasekar SK, Muthukumar M, Montclare JK. Artificial protein block copolymers blocks comprising two distinct self-assembling domains. ChemBioChem. 2009;10(17):2733–2735. doi: 10.1002/cbic.200900539. [DOI] [PubMed] [Google Scholar]
- 25.Osborne JL, Farmer R, Woodhouse KA. Self-assembled elastin-like polypeptide particles. Acta biomaterialia. 2008;4(1):49–57. doi: 10.1016/j.actbio.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 26.Megeed Z, Cappello J, Ghandehari H. Genetically engineered silk-elastin-like protein polymers for controlled drug delivery. Advanced Drug Delivery Reviews. 2002;54(8):1075–1091. doi: 10.1016/s0169-409x(02)00063-7. [DOI] [PubMed] [Google Scholar]
- 27.Cappello J, Crissman J, Dorman M, Mikolajczak M, Textor G, Marquet M, Ferrari F. Genetic engineering of structural protein polymers. Biotechnology Progress. 1990;6(3):198–202. doi: 10.1021/bp00003a006. [DOI] [PubMed] [Google Scholar]
- 28.Rabotyagova OS, Cebe P, Kaplan DL. Self-assembly of genetically engineered spider silk block copolymers. Biomacromolecules. 2009;10(2):229–236. doi: 10.1021/bm800930x. [DOI] [PubMed] [Google Scholar]
- 29.Prince JT, McGrath KP, DiGirolamo CM, Kaplan DL. Construction, cloning, and expression of synthetic genes encoding spider dragline silk. Biochemistry. 1995;34(34):10879–10885. doi: 10.1021/bi00034a022. [DOI] [PubMed] [Google Scholar]
- 30.Xu C, Kopecek J. Genetically engineered block copolymers: Influence of the length and structure of the coiled-coil blocks on hydrogel self-assembly. Pharmaceutical Research. 2008;25(3):674–682. doi: 10.1007/s11095-007-9343-z. [DOI] [PubMed] [Google Scholar]
- 31.Kaplan DL, Mello CM, Arcidiacono S, Fossey S, Senecal K, Muller W. Silk. In: Protein Based Materials. McGrath K, Kaplan DL, editors. Birkhauser; Boston: 1997. p. 429. [Google Scholar]
- 32.Lao UL, Sun M, Matsumoto M, Mulchandani A, Chen W. Genetic engineering of self-assembled protein hydrogel based on elastin-like sequences with metal binding functionality. Biomacromolecules. 2007;8(12):3736–3739. doi: 10.1021/bm700662n. [DOI] [PubMed] [Google Scholar]
- 33.Wang C, Stewart RJ, Kopecek J. Hybrid hydrogels assembled from synthetic polymers and coiled-coil protein domains. Letters to Nature. 1999;397(6718):417–420. doi: 10.1038/17092. [DOI] [PubMed] [Google Scholar]
- 34.Chilkoti A, Dreher MR, Meyer DE. Design of thermally responsive, recombinant polypeptide carriers for targeted drug delivery. Advanced Drug Delivery Reviews Engineered Protein Polymers for Drug Delivery and Biomedical Applications. 2002;54(8):1093–1111. doi: 10.1016/s0169-409x(02)00060-1. [DOI] [PubMed] [Google Scholar]
- 35.Meyer DE, Chilkoti A. Genetically encoded synthesis of protein-based polymers with precisely specified molecular weight and sequence by recursive directional ligation: Examples from the elastin-like polypeptide system. Biomacromolecules. 2002;3(2):357–367. doi: 10.1021/bm015630n. [DOI] [PubMed] [Google Scholar]
- 36.Lyons RE, Lesieur E, Kim M, Wong DCC, Huson MG, Nairn KM, Brownlee AG, Pearson RD, Elvin CM. Design and facile production of recombinant resilin-like polypeptides: gene construction and a rapid protein purification method. Protein Engineering, Design and Selection. 2007;20(1):25–32. doi: 10.1093/protein/gzl050. [DOI] [PubMed] [Google Scholar]
- 37.Higashiya S, Topilina NI, Ngo SC, Zagorevskii D, Welch JT. Design and preparation of β-sheet forming repetitive and block-copolymerized polypeptides. Biomacromolecules. 2007;8(5):1487–1497. doi: 10.1021/bm061098y. [DOI] [PubMed] [Google Scholar]
- 38.Wright ER, Conticello VP. Self-assembly of block copolymers derived from elastin-mimetic polypeptide sequences. Advanced Drug Delivery Reviews Engineered Protein Polymers for Drug Delivery and Biomedical Applications. 2002;54(8):1057–1073. doi: 10.1016/s0169-409x(02)00059-5. [DOI] [PubMed] [Google Scholar]
- 39.Kim M, Elvin C, Brownlee A, Lyons R. High yield expression of recombinant pro-resilin: Lactose-induced fermentation in E. coli and facile purification. Protein Expression and Purification. 2007;52(1):230–236. doi: 10.1016/j.pep.2006.11.003. [DOI] [PubMed] [Google Scholar]
- 40.Paramonov SE, Gauba V, Hartgerink JD. Synthesis of collagen-like peptide polymers by native chemical ligation. Macromolecules. 2005;38(18):7555–7561. [Google Scholar]
- 41.Dawson P, Muir T, Clark-Lewis I, Kent S. Synthesis of proteins by native chemical ligation. Science. 1994;266(5186):776–779. doi: 10.1126/science.7973629. [DOI] [PubMed] [Google Scholar]
- 42.Liu W, Brock A, Chen S, Chen S, Schultz PG. Genetic incorporation of unnatural amino acids into proteins in mammalian cells. Nature Methods. 2007;4(3):239–244. doi: 10.1038/nmeth1016. [DOI] [PubMed] [Google Scholar]
- 43.Billot J-P, Douy A, Gallot B. Synthesis and structural study of block copolymers with a hydrophobic polyvinyl block and a hydrophilic polypeptide block: Copolymers polystyrene/poly-L-lysine) and polybutadiene/poly(L-lysine). Die Makromolekulare Chemie. 1976177(6):1889–1893. [Google Scholar]
- 44.Billot J-P, Douy A, Gallot B. Preparation, fractionation, and structure of block copolymers polystyrene-poly(carbobenzoxy-L-lysine) and polybutadiene-poly(carbobenzoxy-L-lysine). Die Makromolekulare Chemie. 1977;178(6):1641–1650. [Google Scholar]
- 45.Vandermeulen GWM, Hinderberger D, Xu H, Sheiko SS, Jeschke G, Klok H-A. Structure and dynamics of self-assembled poly(ethylene glycol) based coiled-coil nano-objects. ChemPhysChem. 2004;5(4):488–494. doi: 10.1002/cphc.200301079. [DOI] [PubMed] [Google Scholar]
- 46.Vandermeulen GWM, Tziatzios C, Klok H-A. Reversible self-organization of poly(ethylene glycol)-based hybrid block copolymers mediated by a de novo Four-Stranded α-helical hoiled coil motif. Macromolecules. 2003;36(11):4107–4114. [Google Scholar]
- 47.Bae Y, Buresh RA, Williamson TP, Chen T-HH, Furgeson DY. Intelligent biosynthetic nanobiomaterials for hyperthermic combination chemotherapy and thermal drug targeting of HSP90 inhibitor geldanamycin. Journal of Controlled Release. 2007;122(1):16–23. doi: 10.1016/j.jconrel.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 48.Krysmann MJ, Funari S, Canetta E, Hamley IW. The effect of peg crystallization on the morphology of peg/peptide block copolymers containing amyloid β-peptide fragments. Macromolecular Chemistry and Physics. 2008;209(9):883–889. [Google Scholar]
- 49.Sahin E, Kiick KL. Macromolecule-induced assembly of coiled-coils in alternating multiblock polymers. Biomacromolecules. 2009;10(10):2740–2749. doi: 10.1021/bm900474k. [DOI] [PubMed] [Google Scholar]
- 50.Rathore O, Sogah DY. Nanostructure formation through β-sheet self-assembly in silk-based materials. Macromolecules. 2001;34(5):1477–1486. [Google Scholar]
- 51.Rathore O, Sogah DY. Self-assembly of β-sheets into nanostructures by poly(alanine) segments incorporated in multiblock copolymers inspired by spider silk. Journal of the American Chemical Society. 2001;123(22):5231–5239. doi: 10.1021/ja004030d. [DOI] [PubMed] [Google Scholar]
- 52.Smeenk JM, Schon P, Otten MBJ, Speller S, Stunnenberg HG, van Hest JCM. Fibril Formation by Triblock Copolymers of Silklike β-Sheet Polypeptides and Poly(ethylene glycol). Macromolecules. 2006;39(8):2989–2997. [Google Scholar]
- 53.Hadjichristidis NP, Stergios, Floudas, George . Block Copolymers. Wiley-Interscience; Hoboken, NJ: 2003. p. 419. [Google Scholar]
- 54.Checot F, Lecommandoux S, Klok H-A, Gnanou Y. From supramolecular polymersomes to stimuli-responsive nano-capsules based on poly(diene-β-peptide) diblock copolymers. The European Physical Journal E - Soft Matter. 2003;10(1):25–35. doi: 10.1140/epje/e2003-00006-1. [DOI] [PubMed] [Google Scholar]
- 55.Lecommandoux S, Achard M-F, Langenwalter JF, Klok H-A. Self-assembly of rod-coil diblock oligomers based on α-helical peptides. Macromolecules. 2001;34(26):9100–9111. [Google Scholar]
- 56.Gebhardt KE, Ahn S, Venkatachalam G, Savin DA. Rod-sphere transition in polybutadiene-poly(l-lysine) block copolymer assemblies. Langmuir. 2007;23(5):2851–2856. doi: 10.1021/la062939p. [DOI] [PubMed] [Google Scholar]
- 57.Tian Z, Li H, Wang M, Zhang A, Feng Z. Vesicular and tubular structures prepared from self-assembly of novel amphiphilic ABA triblock copolymers in aqueous solutions. Journal of Polymer Science Part A: Polymer Chemistry. 2008;46(3):1042–1050. [Google Scholar]
- 58.Deng C, Chen X, Yu H, Sun J, Lu T, Jing X. A biodegradable triblock copolymer poly(ethylene glycol)-β-poly(l-lactide)-β-poly(l-lysine): Synthesis, self-assembly, and RGD peptide modification. Polymer. 2007;48(1):139–149. [Google Scholar]
- 59.Foo CWP, Patwardhan SV, Belton DJ, Kitchel B, Anastasiades D, Huang J, Naik RR, Perry CC, Kaplan DL. Novel nanocomposites from spider silk-silica fusion (chimeric) proteins. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(25):9428–9433. doi: 10.1073/pnas.0601096103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hamley IW. Block Copolymers in Solution: Fundamentals and Applications. Wiley and Sons, Ltd; Chichester: 2005. p. 288. [Google Scholar]
- 61.Yamaguchi N, Zhang L, Chae B-S, Palla CS, Furst EM, Kiick KL. Growth factor mediated assembly of cell receptor-responsive hydrogels. Journal of the American Chemical Society. 2007;129(11):3040–3041. doi: 10.1021/ja0680358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bini E, Foo CWP, Huang J, Karageorgiou V, Kitchel B, Kaplan DL. RGD-functionalized bioengineered spider dragline silk biomaterial. Biomacromolecules. 2006;7(11):3139–3145. doi: 10.1021/bm0607877. [DOI] [PubMed] [Google Scholar]
- 63.George PA, Doran MR, Croll TI, Munro TP, Cooper-White JJ. Nanoscale presentation of cell adhesive molecules via block copolymer self-assembly. Biomaterials. 2009;30(27):4732–4737. doi: 10.1016/j.biomaterials.2009.05.039. [DOI] [PubMed] [Google Scholar]
- 64.Fischer SE, Liu X, Mao H-Q, Harden JL. Controlling cell adhesion to surfaces via associating bioactive triblock proteins. Biomaterials. 2007;28(22):3325–3337. doi: 10.1016/j.biomaterials.2007.03.026. [DOI] [PubMed] [Google Scholar]
- 65.Makin OS, Serpell LC. Structures for amyloid fibrils. FEBS Journal. 2005;272(23):5950–5961. doi: 10.1111/j.1742-4658.2005.05025.x. [DOI] [PubMed] [Google Scholar]
- 66.Burkoth TS, Benzinger TLS, Jones DNM, Hallenga K, Meredith SC, Lynn DG. C-Terminal PEG Blocks the Irreversible Step in β-Amyloid(10-35) Fibrillogenesis. J. Am. Chem. Soc. 1998;120(30):7655–7656. [Google Scholar]
- 67.Zhou C, Leng B, Yao J, Qian J, Chen X, Zhou P, Knight DP, Shao Z. synthesis and characterization of multiblock copolymers based on spider dragline silk proteins. Biomacromolecules. 2006;7(8):2415–2419. doi: 10.1021/bm060199t. [DOI] [PubMed] [Google Scholar]
- 68.Smeenk JM, Schon P, Otten MBJ, Speller S, Stunnenberg HG, van Hest JCM. Fibril formation by triblock copolymers of silk-like β-sheet polypeptides and poly(ethylene glycol). Macromolecules. 2006;39(8):2989–2997. [Google Scholar]
- 69.Matsumura S, Uemura S, Mihara H. Metal-triggered nanofiber formation of his-containing β-sheet peptide. Supramolecular Chemistry. 2006;18(5):397–403. [Google Scholar]
- 70.Vandermeulen GWM, Kim KT, Wang Z, Manners I. Metallopolymer-peptide conjugates: synthesis and self-assembly of polyferrocenylsilane graft and block copolymers containing a β-sheet forming Gly-Ala-Gly-Ala tetrapeptide segment. Biomacromolecules. 2006;7(4):1005–1010. doi: 10.1021/bm050732p. [DOI] [PubMed] [Google Scholar]
- 71.Tang A, Wang C, Stewart RJ, Kopecek J. The coiled coils in the design of protein-based constructs: hybrid hydrogels and epitope displays. Journal of Controlled Release. 2001;72(173):57–70. doi: 10.1016/s0168-3659(01)00262-0. [DOI] [PubMed] [Google Scholar]
- 72.Marsden HR, Korobko AV, van Leeuwen ENM, Pouget EM, Veen SJ, Sommerdijk N, Kros A. Noncovalent triblock copolymers based on a coiled-coil peptide motif. J. Am. Chem. Soc. 2008;130(29):9386–9393. doi: 10.1021/ja800254w. [DOI] [PubMed] [Google Scholar]
- 73.Lomander A, Hwang W, Zhang S. Hierarchical self-assembly of a coiled-coil peptide into fractal structure. Nano Letters. 2005;5(7):1255–1260. doi: 10.1021/nl050203r. [DOI] [PubMed] [Google Scholar]
- 74.Jing P, Rudra JS, Herr AB, Collier JH. Self-assembling peptide-polymer hydrogels designed from the coiled coil region of fibrin. Biomacromolecules. 2008;9(9):2438–2446. doi: 10.1021/bm800459v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shen W, Zhang K, Kornfield JA, Tirrell DA. Tuning the erosion rate of artificial protein hydrogels through control of network topology. Nature Materials. 2006;5(2):153–158. doi: 10.1038/nmat1573. [DOI] [PubMed] [Google Scholar]
- 76.Su JY, Hodges RS, Kay CM. Effect of chain length on the formation and stability of synthetic α-helical coiled coils. Biochemistry. 1994;33(51):15501–15510. doi: 10.1021/bi00255a032. [DOI] [PubMed] [Google Scholar]
- 77.Duncan R. The dawning era of polymer therapeutics. Nature Reviews. Drug Discovery. 2003;2(5):347–360. doi: 10.1038/nrd1088. [DOI] [PubMed] [Google Scholar]
- 78.Smart T, Lomas H, Massignani M, Flores-Merino MV, Perez LR, Battaglia G. Block copolymer nanostructures. Nano Today. 2008;3(374):38–46. [Google Scholar]
- 79.Huemmerich D, Scheibel T, Vollrath F, Cohen S, Gat U, Ittah S. Novel Assembly Properties of Recombinant Spider Dragline Silk Proteins. Current Biology. 2004;14(22):2070–2074. doi: 10.1016/j.cub.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 80.Lee T, Cooper A, Apkarian RP, Conticello VP. Thermo-reversible self-assembly of nanoparticles derived from elastin-mimetic polypeptides. Advanced Materials. 2000;12(15):1105–1110. [Google Scholar]
- 81.Haider M, Leung V, Ferrari F, Crissman J, Powell J, Cappello J, Ghandehari H. Molecular engineering of silk-elastinlike polymers for matrix-mediated gene delivery: Biosynthesis and characterization. Mol. Pharm. 2005;2(2):139–150. doi: 10.1021/mp049906s. [DOI] [PubMed] [Google Scholar]
- 82.Pechar M, Brus J, Kostka L, Konak C, Urbanova M, Slouf M. Thermoresponsive self- assembly of short elastin-like polypentapeptides and their poly(ethylene glycol) derivatives. Macromolecular Bioscience. 2007;7(1):56–69. doi: 10.1002/mabi.200600196. [DOI] [PubMed] [Google Scholar]
- 83.Kopecek J. Smart and genetically engineered biomaterials and drug delivery systems. European Journal of Pharmaceutical Sciences. 2003;20(1):1716. doi: 10.1016/s0928-0987(03)00164-7. [DOI] [PubMed] [Google Scholar]
- 84.Gustafson J, Greish K, Frandsen J, Cappello J, Ghandehari H. Silk-elastinlike recombinant polymers for gene therapy of head and neck cancer: From molecular definition to controlled gene expression. Journal of Controlled Release. 2009;140(3):256–61. doi: 10.1016/j.jconrel.2009.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cabello CR, Reguera J, Girotti A, Alonso M, Testera AM. Developing functionality in elastin-like polymers by increasing their molecular complexity: the power of the genetic engineering approach. Progress in Polymer Science. 2005;30(11):1119–1145. [Google Scholar]
- 86.Nagarsekar A, Crissman J, Crissman M, Ferrari F, Cappello J, Ghandehari H. Genetic synthesis and characterization of pH- and temperature-sensitive silk-elastin-like protein block copolymers. Journal of Biomedical Materials Research. 2002;62(2):195–203. doi: 10.1002/jbm.10272. [DOI] [PubMed] [Google Scholar]
- 87.Ayres L, Vos MRJ, Adams P, Shklyarevskiy IO, van Hest JCM. Elastin-based side-chain polymers synthesized by ATRP. Macromolecules. 2003;36(16):5967–5973. [Google Scholar]
- 88.Daamen WF, Veerkamp JH, van Hest JCM, van Kuppevelt TH. Elastin as a biomaterial for tissue engineering. Biomaterials. 2007;28(30):4378–4398. doi: 10.1016/j.biomaterials.2007.06.025. [DOI] [PubMed] [Google Scholar]
- 89.Bellingham CM, Lillie M, John M. Gosline JM, Glenda M. Wright GM, Starcher BC, Bailey AJ, Woodhouse KA, Keeley FW. Recombinant human elastin polypeptides self-assemble into biomaterials with elastin-like properties. Biopolymers. 2003;70(4):445–455. doi: 10.1002/bip.10512. [DOI] [PubMed] [Google Scholar]
- 90.Heslot H. Artificial fibrous proteins: A review. Biochimie. 1998;80(1):19–31. doi: 10.1016/s0300-9084(98)80053-9. [DOI] [PubMed] [Google Scholar]
- 91.Bellingham CM, Woodhouse KA, Robson P, Rothstein SJ, Keeley FW. Self-aggregation characteristics of recombinantly expressed human elastin polypeptides. Biochimica et Biophysica Acta (BBA) - Protein Structure and Molecular Enzymology. 2001;1550(1):6–19. doi: 10.1016/s0167-4838(01)00262-x. [DOI] [PubMed] [Google Scholar]
- 92.Martino M, Tamburro AM. Chemical synthesis of cross-linked poly(KGGVG), an elastin-like biopolymer. Biopolymers. 2001;59(1):29–37. doi: 10.1002/1097-0282(200107)59:1<29::AID-BIP1003>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 93.MacEwan SR, Chilkoti A. Elastin-like polypeptides: Biomedical applications of tunable biopolymers. Peptide Science. 2010;94(1):60–77. doi: 10.1002/bip.21327. [DOI] [PubMed] [Google Scholar]
- 94.Urry DW, Parker TM, Reid MC, Gowda DC. biocompatibility of the bioelastic materials, poly(gvgvp) and its γ-irradiation cross-linked matrix: summary of generic biological test results. Journal of Bioactive and Compatible Polymers. 1991;6(3):263–282. [Google Scholar]
- 95.Wright ER, McMillan RA, Cooper A, Apkarian RP, Conticello VP. Thermoplastic elastomer hydrogels via self-assembly of an elastin-mimetic triblock polypeptide. Advanced Functional Materials. 2002;12(2):149–154. [Google Scholar]
- 96.Urry DW. Physical chemistry of biological free energy transduction as demonstrated by elastic protein-based polymers. The Journal of Physical Chemistry B. 1997;101(51):11007–11028. [Google Scholar]
- 97.Lao UL, Mulchandani A, Chen W. Simple conjugation and purification of quantum dot-antibody complexes using a thermally responsive elastin-protein scaffold as immunofluorescent agents. J. Am. Chem. Soc. 2006;128(46):14756–14757. doi: 10.1021/ja065343x. [DOI] [PubMed] [Google Scholar]
- 98.Kostal J, Mulchandani A, Gropp KE, Chen W. A temperature responsive biopolymer for mercury remediation. Environ. Sci. Technol. 2003;37(19):4457–4462. doi: 10.1021/es034210y. [DOI] [PubMed] [Google Scholar]
- 99.Lim DW, Nettles DL, Setton LA, Chilkoti A. In situ cross-linking of elastin-like polypeptide block copolymers for tissue repair. Biomacromolecules. 2008;9(1):222–230. doi: 10.1021/bm7007982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Chen T-H, Bae Y, Furgeson D. intelligent biosynthetic nanobiomaterials (ibns) for hyperthermic gene delivery. Pharmaceutical Research. 2008;25(3):683–691. doi: 10.1007/s11095-007-9382-5. [DOI] [PubMed] [Google Scholar]
- 101.Lewis RV. spider silk: ancient ideas for new biomaterials. Chem. Rev. 2006;106(9):3762–3774. doi: 10.1021/cr010194g. [DOI] [PubMed] [Google Scholar]
- 102.Heim M, Keerl D, Scheibel T. Spider silk: from soluble protein to extraordinary fiber. Angewandte Chemie International Edition. 2009;48(20):3584–3596. doi: 10.1002/anie.200803341. [DOI] [PubMed] [Google Scholar]
- 103.Anderson JP, Cappello J, Martin DC. Morphology and primary crystal structure of a silk-like protein polymer synthesized by genetically engineered Escherichia coli bacteria. Biopolymers. 1994;34(8):1049–1058. doi: 10.1002/bip.360340808. [DOI] [PubMed] [Google Scholar]
- 104.Werten MWT, Moers APHA, Vong T, Zuilhof H, van Hest JCM, de Wolf FA. Biosynthesis of an amphiphilic silk-like polymer. Biomacromolecules. 2008;9(7):1705–1711. doi: 10.1021/bm701111z. [DOI] [PubMed] [Google Scholar]
- 105.Krejchi MT, Atkins EDT, Waddon AJ, Fournier MJ, Mason TL, Tirrel DA. Chemical sequence control of β-sheet assembly in macromolecular crystals of periodic polypeptides. Science. 1994;265(5177):1427–1432. doi: 10.1126/science.8073284. [DOI] [PubMed] [Google Scholar]
- 106.Qu Y, Payne SC, Apkarian RP, Conticello VP. self-assembly of a polypeptide multi-block copolymer modeled on dragline silk proteins. J. Am. Chem. Soc. 2000;122(20):5014–5015. [Google Scholar]
- 107.Rabotyagova OS, Cebe P, Kaplan DL. role of polyalanine domains in β-sheet formation in spider silk block copolymers. Macromolecular Bioscience. 2010;10(1):49–59. doi: 10.1002/mabi.200900203. [DOI] [PubMed] [Google Scholar]
- 108.Numata K, Subramanian B, Currie HA, Kaplan DL. Bioengineered silk protein-based gene delivery systems. Biomaterials. 2009;30(29):5775–5784. doi: 10.1016/j.biomaterials.2009.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Numata K, Hamasaki J, Subramanian B, Kaplan DL. Gene delivery mediated by recombinant silk proteins containing cationic and cell binding motifs. Journal of Controlled Release. In Press, Corrected Proof. [DOI] [PMC free article] [PubMed]
- 110.Bhowmick AK. Current topics in elastomeric research. CRC Press; Boca Raton, FL: 2008. [Google Scholar]
- 111.Hofmann S, Wong C, Rossetti F, Textor M, Vunjak-Novakovic G, Kaplan DL, Merkle HP, Meinel L. Silk fibroin as an organic polymer for controlled drug delivery. Journal of Controlled Release. 2006;111(172):219–227. doi: 10.1016/j.jconrel.2005.12.009. [DOI] [PubMed] [Google Scholar]
- 112.Leach JB, Wolinsky JB, Stone PJ, Wong JY. Crosslinked α-elastin biomaterials: towards a processable elastin mimetic scaffold. Acta Biomaterialia. 2005;1(2):155–164. doi: 10.1016/j.actbio.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 113.Cappello J, Crissman JW, Crissman M, Ferrari FA, Textor G, Wallis O, Whitledge JR, Zhou X, Burman D, Aukerman L, Stedronsky ER. In-situ self-assembling protein polymer gel systems for administration, delivery, and release of drugs. Journal of Controlled Release. 1998;53(173):105–117. doi: 10.1016/s0168-3659(97)00243-5. [DOI] [PubMed] [Google Scholar]
- 114.Dandu R, Ghandehari H. Delivery of bioactive agents from recombinant polymers. Progress in Polymer Science (Oxford) 2007;32(879):1008–1030. [Google Scholar]
- 115.Herrero-Vanrell R, Rincon AC, Alonso M, Reboto V, Molina-Martinez IT, Rodriguez-Cabello JC. Self-assembled particles of an elastin-like polymer as vehicles for controlled drug release. Journal of Controlled Release. 2005;102(1):113–122. doi: 10.1016/j.jconrel.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 116.Hatefi A, Cappello J, Ghandehari H. Adenoviral gene delivery to solid tumors by recombinant silk-elastinlike protein polymers. Pharmaceutical Research. 2007;24(4):773–779. doi: 10.1007/s11095-006-9200-5. [DOI] [PubMed] [Google Scholar]
- 117.Nagarsekar A, Crissman J, Crissman M, Ferrari F, Cappello J, Ghandehari H. Genetic engineering of stimuli-sensitive silkelastin-like protein block copolymers. Biomacromolecules. 2003;4(3):602–607. doi: 10.1021/bm0201082. [DOI] [PubMed] [Google Scholar]
- 118.Chow DC, Dreher MR, Trabbic-Carlson K, Chilkoti A. Ultra-high expression of a thermally responsive recombinant fusion protein in E. coli. Biotechnology Progress. 2006;22(3):638–646. doi: 10.1021/bp0503742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Qiu W, Teng W, Cappello J, Wu X. Wet-spinning of recombinant silk-elastin-like protein polymer fibers with high tensile strength and high deformability. Biomacromolecules. 2009;10(3):602–608. doi: 10.1021/bm801296r. [DOI] [PubMed] [Google Scholar]
- 120.Megeed Z, Haider M, Li D, O'Malley J, Bert W, Cappello J, Ghandehari H. In vitro and in vivo evaluation of recombinant silk-elastinlike hydrogels for cancer gene therapy. Journal of Controlled Release. 2004;94(273):433–445. doi: 10.1016/j.jconrel.2003.10.027. [DOI] [PubMed] [Google Scholar]
- 121.Zhang G, Fournier MJ, Mason TL, Tirrell DA. Biological synthesis of monodisperse derivatives of poly(α,L-glutamic acid): model rod-like polymers. Macromolecules. 1992;25(13):3601–3603. [Google Scholar]
- 122.Teule F, Aube C, Ellison M, Abbott A. Biomimetic manufacturing of customized novel fibre proteins for specialised applications. Autex Research Journal. 2003;3(4):160–165. [Google Scholar]
- 123.Hatefi A, Megeed Z, Ghandehari H. Recombinant polymer-protein fusion: A promising approach towards efficient and targeted gene delivery. Journal of Gene Medicine. 2006;8(4):468–476. doi: 10.1002/jgm.872. [DOI] [PubMed] [Google Scholar]
- 124.Chen DJ, Majors BS, Zelikin A, Putnam D. Structure-function relationships of gene delivery vectors in a limited polycation library. Journal of Controlled Release. 2005;103(1):273–283. doi: 10.1016/j.jconrel.2004.11.028. [DOI] [PubMed] [Google Scholar]
- 125.Chen Q-R, Zhang L, Stass SA, Mixson AJ. Branched co-polymers of histidine and lysine are efficient carriers of plasmids. Nucleic Acids Research. 2001;29(6):1334–1340. doi: 10.1093/nar/29.6.1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Midoux P, Monsigny M. Efficient gene transfer by histidylated polylysine/pDNA complexes. Bioconjugate Chemistry. 1999;10(3):406–411. doi: 10.1021/bc9801070. [DOI] [PubMed] [Google Scholar]
- 127.Blanckaert VD, Hebbar M, Louchez M-M, Vilain M-O, Schelling ME, Peyrat J-P. Basic fibroblast growth factor receptors and their prognostic value in human breast cancer. Clinical Cancer Research. 1998;4(12):2939–2947. [PubMed] [Google Scholar]
- 128.Lombardi A, Bryson JW, DeGrado WF. De novo design of heterotrimeric coiled coils. Peptide Science. 1996;40(5):495–504. doi: 10.1002/(SICI)1097-0282(1996)40:5%3C495::AID-BIP7%3E3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 129.Petka WA, Harden JL, McGrath KP, Wirtz D, Tirrell DA. Reversible hydrogels from self-assembling artificial proteins. Science. 1998;281(5375):389–392. doi: 10.1126/science.281.5375.389. [DOI] [PubMed] [Google Scholar]